Abstract
We constructed a new large-scale database of tRNA genes by analyzing 534 complete genomes of prokaryotes and 394 draft genomes in WGS (Whole Genome Shotgun) division in DDBJ/EMBL/GenBank and approximately 6.2 million DNA fragment sequences obtained from metagenomic analyses. This exhaustive search for tRNA genes was performed by running three computer programs to enhance completeness and accuracy of the prediction. Discordances of assignment among three programs were found for ∼4% of the total of tRNA gene candidates obtained from these prokaryote genomes analyzed. The discordant cases were manually checked by experts in the tRNA experimental field. In total, 144 061 tRNA genes were registered in the database ‘tRNADB-CE’, and the number of the genes was more than four times of that of the genes previously reported by the database from analyses of complete genomes with tRNAscan-SE program. The tRNADB-CE allows for browsing sequence information, cloverleaf structures and results of similarity searches among all tRNA genes. For each of the complete genomes, the number of tRNA genes for individual anticodons and the codon usage frequency in all protein genes and the positioning of individual tRNA genes in each genome can be browsed. tRNADB-CE can be accessed freely at http://trna.nagahama-i-bio.ac.jp.
INTRODUCTION
Metagenome analyses, which consist of shotgun sequencing of mixed genomes from environmental uncultured microorganisms, have been developed to study novel genes from poorly characterized microorganisms, such as unculturable species. A massive number of fragment sequences obtained from metagenomic analyses should contain a large number of tRNA genes derived from a wide variety of novel microorganisms, but practically no information on tRNA genes has been annotated in the current database. The tRNA genes found from the fragment sequences obtained from metagenomic analyses should provide new insights into tRNA sequences and their gene organization.
After completion of a conventional type of genome sequencing, tRNA genes from each genome have been predicted primarily using one computer program and inscribed on the flat file of the genome sequence data for registration to the International DNA Data Banks (DDBJ/EMBL/GenBank). However, in ∼5% of the completely sequenced genomes, the annotation data of tRNA genes have not been inscribed. There also exist many cases where an important information of anticodon (and thus of amino acid) has not be added while the tRNA gene annotation is present.
Using tRNAscan-SE (1), a tRNA gene database has been constructed for the complete genomes (http://lowelab.ucsc.edu/GtRNAdb/). In addition, the database for tRNA sequences including modified nucleosides (2) and that for RNA modification pathways (3) were reported.
We have constructed a new large-scale database for tRNA genes not only from the complete genomes of prokaryotes but also from draft sequences of prokaryote genomes in WGS (Whole Genome Shotgun) division in DDBJ/EMBL/GenBank and fragment sequences obtained from metagenomic analyses of environmental microorganisms mixtures, which contain a wide range of novel unculturable microorganisms. In the present database, therefore, more than four times of tRNA genes than those compiled by the previous database, which was constructed for the complete genomes using tRNAscan-SE, could be stored.
MATERIALS
The following three sources of DNA sequences were used: the complete genomes of 543 prokaryotes published by Genome Information Broker (http://gib.genes.nig.ac.jp/) of DDBJ up to July 2007; the draft genome sequences of 394 prokaryotes published by WGS division of DDBJ/EMBL/GenBank up to July 2007; the 6 225 284 sequence fragments obtained from metagenomic analyses of environmental microbe mixtures and released by DDBJ/EMBL/GenBank up to April 2008.
RESULTS
Search for tRNA genes
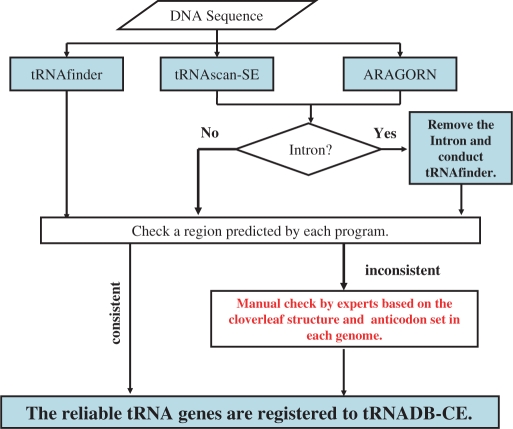
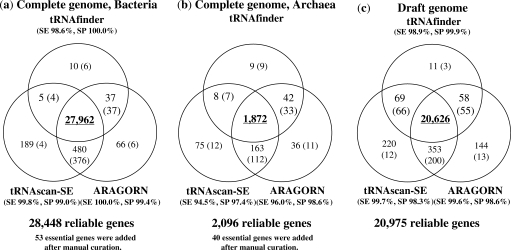
Figure 1 shows the workflow of searching for tRNA genes. In order to enhance the completeness and accuracy of prediction, three computer programs, tRNAscan-SE (1), ARAGORN (4) and tRNAfinder (5), were used in combination since their algorithms are partially different and render somewhat different results. First, we checked to what degree the predicted regions and the anticodons of individual tRNA genes from the complete genomes of bacteria were consistent with each other (Figure 2a). The total of tRNA gene candidates predicted by the three programs was 28 749, and the three programs consistently rendered results of 27 962 genes (97.3%). These tRNA genes concordantly found by all three programs were stored in tRNADB-CE without further checks. Then, the residual 787 discordant cases (∼3% of the total of bacterial gene candidates) were checked manually by three experts (Y.Y., A.M. and H.I.) in the tRNA experimental field independently and were classified into three categories after their discussion: (i) reliable tRNA gene (O), (ii) not tRNA gene (X) and (iii) ambiguous case (Δ). This manual check was conducted with reference to the consensus sequences of various cloverleaf structures and the various characteristics of functional tRNAs, such as characteristics specific to individual amino acid groups, which are difficult to include in computer algorithms. Knowledge from literatures was also utilized. Out of the discordant 787 candidates, 433 genes (55.0%) were predicted as reliable tRNA genes (O) and stored in the database with short comments from the experts. Users can download either the reliable tRNA genes or all candidate genes by choosing ‘The reliable tRNA genes’ or ‘All candidate genes’ in ‘Target’ section in ‘tRNA gene data download’ page. Most pseudogenes may be found from the ‘All candidate genes’ category.
Figure 1.
Workflow of searching for tRNA genes.
Figure 2.
Result of tRNA gene search. Numbers in Venn diagrams show numbers of tRNA genes found by individual programs and those in parenthesis show numbers after manual checks by experts. Abbreviations and definitions are the followings: SE, sensitivity level = 100 × (the number of reliable tRNAs predicted by one program)/(the number of all reliable tRNAs); SP, specificity level = 100 × (the number of reliable tRNAs predicted by one program)/(the number of candidate tRNAs predicted by the respective program). Because tRNAfinder can not predict tRNAs with intron, SE and SP are not presented for archaea.
Next, by investigating the minimum anticodon set most likely essential for translation system of each complete genome (6), the candidate genes for the lacking anticodons were searched in the following ways. First, tRNA gene candidates previously classified into categories B and C were reexamined to consider the possibility that some of them might be functional, although they differed in structure from the standard cloverleaf model. Second, a search was conducted for tRNA genes on the plasmids present along with the genome of the respective species. Nine cases in which the essential tRNA genes were on plasmids were detected. Finally, we examined the possibility that even bacterial tRNAs might have introns (7,8), in the following ways. For example, tRNALeu (TAA-anticodon) genes were missing in 14 species of genus Burkholderia, and we found tRNALeu (TAA) genes with introns in all 14 species. All together, the number of reliable tRNA genes rose to 28 448, as shown in Figure 2a.
In the case of CAT-anticodon tRNAs, three types (initiator tRNA-Met, elongator tRNA-Met and tRNA-Ile whose anticodon is enzymatically converted to read ATA codon) are known, and TFAM program (9,10) can discriminate the three types. After TFAM execution followed by manual checks by experts, reliable cases are noted in ‘Comments’ part in ‘The detailed information of tRNA gene sequence’ page.
Figure 2b shows the tRNA genes found for the complete genomes of archaea. Because a significant portion of archaea tRNA genes have introns, the level of discordance in the predictions among the computer programs increased, as compared with searches of the bacterial genomes. Since tRNAfinder cannot predict tRNA genes with intron, intron regions were predicted with tRNAscan-SE and ARAGORN, and after removal of intron regions, tRNAfinder was used to examine correctness as mature tRNA sequences. The manual check by experts was conducted in combination with the prediction program SPLITS (11,12) for the tRNA genes with introns. Furthermore, based on knowledge of experts derived from literatures, the splitted-tRNAs (tRNAHis (GTG), tRNAGlu (CTC), tRNAGlu (TTC), tRNAiMet (CAT) and tRNATrp (CCA)) in Nanoarchaeum equitans reported by Randeau et al. (13) were registered, and positions and orientations of two segments for each splitted-tRNA were listed in the ‘Comments’ part of these genes in ‘The detailed information of tRNA gene sequence’. By checking a minimum set of the essential anticodons for archaea genomes, 40 tRNA genes were additionally included, and the number of reliable tRNA genes was 2096.
The searches for tRNA genes in the 394 draft genomes in WGS division of DDJB/EMBL/GenBank were conducted in the same manner, except for a check of the requirement of the minimum anticodon set. Figure 2c shows the result of the draft genomes. The number of reliable tRNA genes was 20 975.
In the case of fragment sequences obtained from metagenome analyses of environmental samples, only tRNA genes concordantly found by the three programs were registered at the present stage. Approximately twice as many tRNA genes were detected as detected in both the complete and draft genomes, and the number is divided and separately listed according to categories of environments (Supplementary Table 1). Because a significant portion of environmental DNA sequences are thought to be from unculturable microbes, tRNA genes of highly novel microbes should exist. This may enable us to acquire new knowledge likely very different from the current knowledge of tRNA sequences and their gene structures. It is known that a minor portion of metagenome sequences were derived from eukaryotic microorganisms (14,15).
Functions of tRNADB-CE and data access
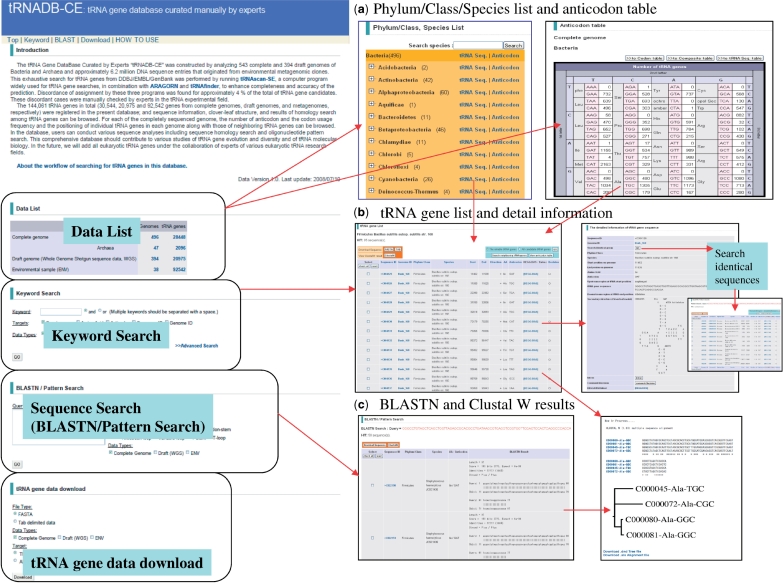
The tRNADB-CE was implemented under the Apache/Perl/PostgreSQL environment on the Linux platform. The basic functionalities aim to browse the stored data and to search the database with a user-specified input. A browse page is presented in Figure 3.
Figure 3.
tRNADB-CE interface snapshots.
First, a list of tRNA genes and anticodons can be browsed depending on the numbering of genomes (i.e. genome ID) or DNA fragments of environmental samples stored in the database. The statistical information for copy numbers of tRNA genes stored in each phylotype/species and the anticodon type in each amino acid group can also be browsed (Figure 3a). Furthermore, the tRNA gene data can be linked to KEGG-DAS (http://das.hgc.jp/) and NCBI Entrez (16) for obtaining data on other genes, including protein genes around the selected tRNA (Figure 3b). A list of codon usage in protein genes of each complete or draft genome can be browsed.
When clicking ‘Search neighboring tRNA genes’, tRNA genes found within 100 bp can be listed to detect candidates for tRNA gene clusters. Likewise, by clicking sequence ID of each tRNA gene, detailed information on the selected tRNA genes can be browsed, e.g. tRNA gene sequences, their upstream and downstream sequences (10 nt), information on the secondary structures for the tRNAs, and curation comments on individual tRNAs.
If ‘Search identical group’ in ‘The detailed information of tRNA gene sequence’ (Figure 3b) is clicked, tRNA genes that have sequences identical with the selected tRNA can be searched for among all the tRNA genes stored in this database. A ‘Keyword Search’ can also be conducted using retrieved items such as Species name, Amino Acid, Anticodon, Sequence ID and Genome ID. This can be done by using multiple keywords in combination.
Two types of sequence search—a sequence similarity search ‘BLASTN’ (17) and a Pattern Search—can be conducted. In the Pattern Search (i.e. oligonucleotide sequence search), we can focus the search area on the stems/loops of cloverleaf structures and combine the areas in various patterns. After selecting tRNA genes of interest using the sequence search procedures, multiple alignments with ClustalW (18) and downloading of the aligned sequences and of the obtained dendrograms are available (Figure 3c).
Rfam (19) compiles a large collection of multiple sequence alignments of noncoding RNA families derived mainly from complete genome sequences for aiming to facilitate the identification of new members of the known families. While tRNA is not a main target of Rfam, a systematic comparison of their tRNA data with those of the present database for complete genome sequences may provide an additional strategy of tRNA finding, especially from archaea genomes. Multiple alignments in our website can be created for the data set selected on the user-specified criteria for support of their knowledge discovery.
DISCUSSIONS
We found novel bacterial tRNA genes with an intron in 14 species of genus Burkholderia. We also found two ochre and five amber suppressor-type anticodons in the environmental DNA sequences. Because only one tRNA sequence was found for each of the seven suppressor-type anticodons at the present stage, it is not clear whether the anticodon is attributed to nonuniversal codon or sequencing error. There are also novel bacterial tRNA genes with the following A-start anticodons: leucine AAG, threonine AGT, serine AGA and isoleucine AAT (Figure 3a). The AGA or AAT anticodon was found in only one case and therefore, might be attributed to sequencing errors. However, the tRNAs with either the AAG or AGT anticodon were found for different species, indicating that these anticodons must be real.
By analyzing both environmental fragment sequences and genome sequences of most (if not all) prokaryotes currently available, four times the numbers of genes as the number registered by the previous database (1; http://lowelab.ucsc.edu/GtRNAdb/) have been found. This is promising for new discoveries, such as tRNA genes related with non-universal codon table (5) or tRNAs with novel functions as yet undiscovered.
When we focused on tRNA groups with identical sequences for the complete and draft genomes, we often found groups of phylotype-specific sequences: i.e. particular types of tRNA sequences found only in particular lineages. Moreover, identical sequences belonging to these groups could be found in environmental DNA fragments, showing that the identical tRNA groups, especially phylotype-specific groups, may provide good phylogenetic, diagnostic markers. In the cases of novel (or poorly characterized) protein genes even with high potential usefulness obtained from metagenomic analyses, their phylogenetic origins can not be predicted properly because of the lack of the orthologous sequence set essential for making a reliable evolutionary tree, except for the case of coexistence with good phylogenetic markers such as rDNAs (rRNA genes) in the same DNA fragment. Because the size of rDNAs (or well characterized protein genes usable for phylogenetic markers) is large, the probability of coexistence of such genes with other protein genes is low because of shortness of genomic fragments sequenced by metagenomic approaches (primarily <1 kb). In the case of tRNA, however, the coexistence with protein genes is highly probable. We are now analyzing the ‘phylotype-specific identical tRNA groups’, which have the potential for use as phylogenetic markers.
In the future, we will expand the database by analyzing most (if not all) available genomic sequences. An additional revision will be made by updating the sequence information, including eukaryotic, viral and plasmid sequences, in collaboration with experts in the various experimental fields of tRNA research.
ACCESS TO THE DATABASE
tRNADB-CE can be accessed freely from http://trna.nagahama-i-bio.ac.jp.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Integrated Database Project; Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Funding for open access charge: Integrated Database Project from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Ms Yukari Yonemori for article preparation.
REFERENCES
- 1.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprinzl M, Vassilenko SK. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunin-Horkawicz S, Czerwoniec A, Gajda JM, Feder M, Grosjean H, Bujnicki MJ. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinouchi M, Kurokawa K. tRNAfinder: a software system to find all tRNA genes in the DNA sequence based on the cloverleaf secondary structure. J. Comp. Aided Chem. 2006;7:116–126. [Google Scholar]
- 6.Osawa S. Evolution of the Genetic Code. NY: Oxford University Press; 1995. [Google Scholar]
- 7.Biniszkiewicz D, Cesnavicienel E, Shub DA. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paquin B, Kathe SD, Nierzwicki-Bauer SA, Shub DA. Origin and evolution of group I introns in cyanobacterial tRNA genes. J. Bacteriol. 1997;179:6798–6806. doi: 10.1128/jb.179.21.6798-6806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ardell DH, Andersson SGE. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006;34:893–904. doi: 10.1093/nar/gkj449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva FJ, Belda E, Talens SE. Differential annotation of tRNA genes with anticodon CAT in bacterial genomes. Nucleic Acids Res. 2006;34:6015–6022. doi: 10.1093/nar/gkl739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugahara J, Yachie N, Sekine Y, Soma A, Matsui M, Tomita M, Kanai A. SPLITS: a new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 2006;6:411–418. [PubMed] [Google Scholar]
- 12.Sugahara J, Yachie N, Arakawa K, Tomita M. In silico screening of archaeal tRNA-encoding genes having multiple introns with bulge-helix-bulge splicing motifs. RNA. 2007;13:671–681. doi: 10.1261/rna.309507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Randau L, Munch R, Hohn MJ, Jahn D, Soll D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 14.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 15.Abe T, Sugawara H, Kinouchi M, Kanaya S, Ikemura T. Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res. 2005;12:281–290. doi: 10.1093/dnares/dsi015. [DOI] [PubMed] [Google Scholar]
- 16.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]