Abstract
NCBI's Conserved Domain Database (CDD) is a collection of multiple sequence alignments and derived database search models, which represent protein domains conserved in molecular evolution. The collection can be accessed at http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml, and is also part of NCBI's Entrez query and retrieval system, cross-linked to numerous other resources. CDD provides annotation of domain footprints and conserved functional sites on protein sequences. Precalculated domain annotation can be retrieved for protein sequences tracked in NCBI's Entrez system, and CDD's collection of models can be queried with novel protein sequences via the CD-Search service at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. Starting with the latest version of CDD, v2.14, information from redundant and homologous domain models is summarized at a superfamily level, and domain annotation on proteins is flagged as either ‘specific’ (identifying molecular function with high confidence) or as ‘non-specific’ (identifying superfamily membership only).
INTRODUCTION
Visualization of protein domain architecture and highlighting the presence of protein domains conserved in molecular evolution have become de facto standards in protein sequence analysis and are routinely provided by protein information resources. A conserved domain footprint may reveal aspects of a protein's molecular or cellular function, and its domain architecture may pinpoint that function more precisely, grouping the protein with only a few others that are known to share the particular architecture. When a protein sequence of interest is found to contain an evolutionarily conserved domain, functional annotation may be transferred from that domain model to the protein sequence. This annotation may be very generic, such as ‘binds nucleic acids’, or very specific, such as ‘decarboxylates orotidine 5′-monophosphate to form uridine 5′-phosphate’. If the domain assignment is based on a residue-by-residue alignment of the protein sequence to a domain model, the annotation defines a footprint region, and it is possible to transfer information about the presence and location of functional sites from model to sequence as well.
There are numerous information resources that provide computational annotation for protein sequences and protein domains. Many of them are well maintained, and CDD imports various collections, Pfam (1), SMART (2), COGs (3), Protein Clusters (4), to provide comprehensive coverage of protein databases and genomic sequence collections. Actively maintained collections such as Pfam (1) continue to grow, as more sequence data become available, and it can be expected that over time the fraction of newly discovered protein domains that are truly ancient and therefore present in a diverse set of taxonomic lineages will diminish, and that most newly discovered protein domains will be specific to relatively narrow taxonomic lineages.
NCBI's Conserved Domain Database (CDD) has been established to annotate protein sequences with footprints of ancient conserved domains (5). To this end, CDD does not, for the most part, attempt to discover new domain families. CDD relies on Pfam and other sources to provide comprehensive coverage. It attempts, however, to reconcile the conservation of protein sequence, as reflected in alignment models imported from various collections, with what is now known about domain 3D structure. CDD also attempts to reflect the diversity of protein domain families and represents many families as structured hierarchies of related models instead of single all-encompassing models or collections of related models without explicit relationships. CDD also attempts to annotate the presence of functional sites, such as catalytic or binding sites, in protein domain families, so that these annotations can be transferred computationally onto protein sequences. CDD provides web interfaces and software tools that assist in the interpretation of protein domain annotation and the classification of user protein query sequences within existing NCBI-curated domain hierarchies, via live or precalculated database search results.
Domain models imported from external sources are processed so that they fit into the CDD framework, and the derived database search models (Position-specific scoring matrices or PSSMs) do not, in general, behave exactly like corresponding database search models in the data providers’ resources (such as Hidden Markov Models or HMMs). While the content of the models is determined by the providers, the sequence alignments are processed in an automated way upon import, in order to identify the sequence fragments in NCBI's Entrez database and provide links to 3D structure. Occasionally, sequence fragments that cannot be matched or identified are omitted or substituted for closely related matches. The content imported from SMART and COGs is static and has not been updated in several years except for a small set of revised descriptions and links to literature in the set derived from COGs. The import process has been discussed in greater detail in a previous manuscript (5).
Recently, CDD has introduced three major changes to how conserved domain data and annotations are presented:
Both NCBI-curated domain hierarchies and models imported from outside sources are clustered into groups of domain models that are presumed to describe homologous sequence fragments. These groups are labeled ‘superfamilies’, and are now indexed in the Entrez/CDD query and retrieval system.
Domain annotation on protein sequences now refers to those superfamilies instead of referring to the best scoring models from within the superfamilies. However, if domain annotation is obtained via an NCBI-curated model, and if the match exceeds a stringent threshold, the annotation is derived from that particular model only and labeled ‘specific’.
Information about conserved sites, such as active sites and binding interfaces, is now displayed by CD-Search (6), the web service that visualizes domain annotation in CDD. Site annotation is also transferred to protein sequences in the Entrez/Protein database and may be displayed together with protein records.
CONSERVED DOMAIN SUPERFAMILIES
Starting with the development of the CDART resource (7), domain models in CDD have been clustered in order to deal with redundancy. CDD has been a redundant collection ever since its conception, as it contains sets of models imported from several sources with overlapping scope, has re-curated many models for major domain families, and has also inherited redundancy that is intrinsic to the imported collections.
A domain annotation resource may represent a set of homologous sequence fragments as two or more separate models, for various reasons. The molecular functions within that set may be quite diverse, for example, or a single model may be ineffective in database search applications when the sequence fragments are too dissimilar. Two or more redundant models may match overlapping regions on a query sequence, and the annotation derived from those models may be in conflict. In many such cases, CDD will now provide annotation with the name and description of a conserved domain superfamily, where the latter is defined as a set of evolutionarily related single-domain models.
Before domain models can be clustered into superfamilies, a subset of models must be flagged as multi-domain models and exempted from clustering. Multi-domain models are defined as those whose footprints overlap with two or more sequential single-domain footprints, so that they might merge the corresponding single domains into a single cluster. With multi-domain models excluded, single domain models are subjected to single-linkage clustering, where two models are considered related if they annotate a set of protein sequences with diverse taxonomic origins in significantly overlapping intervals. The RPS-BLAST E-value threshold for clustering is set to 1E-05, and sequences must be from three or more diverse taxonomy nodes. The resulting single-domain clusters contain a mixture of models from various sources.
The cluster names and descriptions are generated automatically, by picking a representative model and copying its name and description. If the cluster contains an NCBI-curated domain or domain hierarchy, the model or the hierarchy's parent model is selected as the superfamily representative. If the cluster contains more than one NCBI-curated hierarchy, the hierarchy with the highest coverage of a nonredundant set of proteins is selected. If the cluster contains no NCBI-curated model, a model imported from the Pfam collection is selected (the model with highest coverage, if more than one). If the cluster does not contain a model imported from Pfam either, a model imported from the SMART collection is selected, and so on.
Not all superfamily cluster names are generated computationally. A small batch of superfamilies has been reviewed, and in some cases names and descriptions have been modified by CDD curators. Larger superfamily clusters have also been reviewed for putative errors in the clustering procedure, and some of the clusters have been split up accordingly. To this end, we maintain a black-list for clustering, which specifies pairs of models that are not supposed to end up in the same cluster, but have been observed to co-cluster occasionally, such as when one type of domain occurs as an insert in another type of domain, and when the RPS-BLAST alignment tool happens to over-extend N- or C-terminal partial alignments into the inserted domain for several sequences in the Entrez protein database. The type II fibronectin domains (cd00062) and zinc metallopeptidases (cd00203) would be one example.
Curation of superfamily descriptions and content will be an ongoing activity in CDD curation. Representing groups of sequences or sequence fragments related by common ancestry as superfamilies is a frequent practice in many protein classification resources, of course, and the results of such classification efforts will not always coincide. In general, superfamily clusters in CDD will coincide with ‘clans’ in the Pfam resource and with superfamilies in the SCOP classification (8), for example, although we have not attempted to quantify the agreement.
Superfamilies are recorded as explicit CD models, which do not contain a multiple sequence alignment but rather a list of single-domain accessions. Most of the conserved domain superfamilies contain only a single model, and often represent domains found in relatively narrow taxonomic lineages. Only superfamilies that represent two or more individual models have been indexed in the Entrez/CDD database. The accessions assigned to superfamily models start with ‘cl’, so that they can be distinguished from regular single-domain models, whose accessions start with ‘cd’, ‘pfam’, ‘smart’, ‘COG’, ‘PRK’, etc., depending on the source database. With each CDD release, single domains will be clustered anew, and the results of that clustering will vary as they depend on the set of single-domain models tracked by the database as well as on the content of NCBI's Entrez/protein database. Superfamily accessions will be preserved if the composition of the associated cluster does not change by 50% or more.
SPECIFIC DOMAIN MATCHES
In previous versions of CDD, the default domain annotation reported for any protein with matches to CDD would show the top ranked hit for any particular interval of the protein query. Source databases were treated differently; hits to NCBI-curated domain models were listed ahead of other models unless the RPS-BLAST E-value associated with the match remained above a threshold of 1E-05. Annotation with NCBI-curated models was emphasized, as their content and behavior in database searches were better understood.
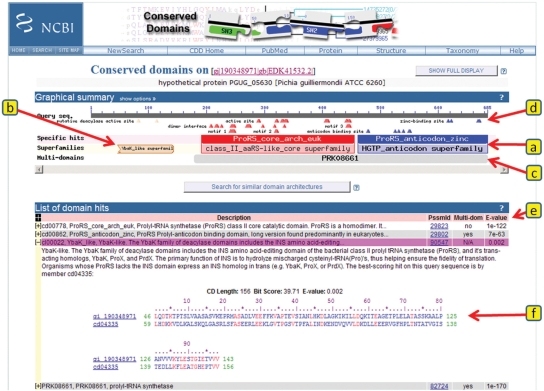
CDD now behaves a bit differently. If a region on a user query matches an NCBI-curated model, and if that match is the top-ranked match according to previously defined criteria, the alignment's bit-score must also exceed a model-specific threshold in order to qualify this specific match for annotation on the query protein sequence. Such matches are labeled ‘Specific hits’ on the CD-Search results pages, and they are visually separated from other matches, labeled ‘Non-specific hits’. The default concise display of CD-Search results summarizes all overlapping hits to models from the same superfamily. It also explicitly shows ‘Specific hits’, when present, but does not display ‘Non-specific hits’. The latter are shown only in the full display. A typical concise display is shown in Figure 1, see the figure legend for further explanations.
Figure 1.
CDD-based annotation on a recently predicted protein sequence. This summary is the default concise version of the annotation view as generated by CD-Search, using precalculated alignment information. The view is divided into two panels, a graphical summary (items a through d) and a table detailing the matches (items e and f). The query sequence is represented as a gray bar in the top portion of the graphical summary, with a ruler indicating sequence length and coordinates. (a) ‘Specific hits’ to NCBI-curated domain models are indicated in a separate area below the query sequence, and the corresponding balloons are rendered in bright colors. The extent of the hits also defines annotations with conserved domain ‘Superfamilies’, which are indicated in the area below the ‘Specific hits’, and enclosed in boxes to indicate superfamily relationships. If the full display is selected, an area summarizing ‘Non-specific hits’ will be shown as well, and the boxes will be drawn to resolve superfamily relationships, where highest ranked match for each superfamily defines the extents of each corresponding box. ‘Non-specific hits’ and ‘Superfamilies’ balloons are rendered in pastel colors, with each homologous superfamily being assigned a separate color. (b) If a region of the query has no ‘Specific hits’, only the ‘Superfamilies’ annotation is shown in the concise default display. If a match to a conserved domain model is incomplete, as in this case, the balloon is rendered with a jagged edge to indicate a missing region. (c) In the default concise display, matches to multi-domain models are rendered as gray balloons in a separate area of the summary graph. Only the best-ranked nonoverlapping multi-domain models are shown. (d) Functional sites as annotated on NCBI-curated domain models are mapped to the query sequence. Sites are mapped from the highest ranked model only, and they are colored to correspond to their source model. When no ‘Specific hits’ are available, such as in (b), sites may still be mapped if they have been annotated on the parent model of a hierarchy that gave a ‘Non-specific hit’. Both conserved domain balloons and site annotations are hot-linked so that moving the mouse over the objects displays pop-ups with additional information, and so that clicking on the objects generates summary pages for the particular domain model, embedding the user query sequence in the alignment for further analysis, if applicable. (e) A table view summarizes what the graphical view indicates as well, listing E-values, multi-domain status and various identifiers for the conserved domain models identified as matches. The table rows can be expanded (f) to display detailed sequence alignment information between the query and the domain model's consensus sequence. An alignment of all sequences comprising a domain model, with or without the query sequence embedded, is accessible by clicking on the domain's balloon representation in the graphical summary or its unique numerical identifier (PSSM-Id) in the tabular summary, respectively.
The model-specific score thresholds for specific hits are set as the minimum bit-scores observed for models matched to the very sequences which have been used in their construction. In other words, when a match to a user query sequence is labeled ‘Specific hit’, it scores at least as well as one or more sequences that are part of that NCBI-curated domain model. This is a stringent threshold, but it allows us to assign domain matches with high confidence.
Another element that lends confidence to the assignment of domain matches is the use of domain models that are based on the alignment of similarly sized sequence regions from proteins believed to be bona fide members of the family being profiled. In contrast, the presence of short sequence fragments in an alignment and the inclusion of false positives (sequence, which may not actually be related by common descent), could weaken a model-specific threshold score, which in turn might lead to erroneous ‘specific-hit’ assignments on query proteins. Because we have control over the NCBI curation process, which is specifically designed to only include member sequences with the desired attributes, we feel confident about applying model-specific threshold scores with the NCBI-curated subset of CDD, and only hits to that subset appear as ‘specific hits’ at this time. Further study is needed on the other data subsets in CDD to determine if and what kind of model-specific thresholds can be applied with the same level of confidence.
All matches to imported domain models (not curated by NCBI as part of CDD) are therefore labeled ‘Non-specific hits’ at this point, as their content and behavior in database searches are less well understood.
NCBI-curated domain hierarchies may also split up a family of homologous sequence fragments into many specific subfamilies, which may also carry very specific functional annotation. A query sequence that matches one of these particular subfamilies should only be annotated with that function, if the match is strong enough, and not if that particular subfamily happens to provide the best scoring match by coincidence. If no ‘Specific hit’ can be detected, the system defaults to annotation based on the superfamily description, which tends to be rather generic. The development of the model-specific score threshold and an evaluation of its effect on the accuracy of annotation can be found in (9). A simple simulation indicates that the potential for misclassification due to sub-families not represented in curated CD hierarchies has been sharply reduced, from about 50% to about 6%, with the introduction of model specific score thresholds.
ANNOTATION OF FUNCTIONAL SITES
CDD curators have recorded functionally conserved sites on protein domain models in cases where they can provide evidence for the presence of such sites. Evidence may come from a published paper, or it may come from a 3D structure that exemplifies certain aspects of the domain's function. Previously, such functional site annotation was visible on the conserved domain summary reports generated by CDD's web services, and in visualization of domain models using the helper application Cn3D (10). More recently, CDD-based annotation of functional sites is available for proteins in NCBI's Entrez query and retrieval system. In addition, the CD-Search web-service, which visualizes domain annotation on protein sequences, whether precalculated or obtained from live database searches, now labels such inferred sites on the protein sequence. An example is provided in Figure 1.
Just as domain functional annotation may be provided as ‘Specific hit’ annotation in cases where the score of a match to an NCBI-curated model exceeds a model-specific threshold, CDD maps functional sites from a specific model to a query protein sequence only if the match exceeds that threshold score. If the match to an NCBI-curated model scores below the model-specific threshold, CD-search will only show functional sites that have been recorded on the parent model of the associated domain hierarchy and can be mapped from that parent model to the user query sequence. This mapping of superfamily ‘generic’ sites is made possible by NCBI-curated domain hierarchies following a strictly validated alignment model that permits such computation and has been discussed previously (11).
NEW VERSION OF CDTree/Cn3D
CDTree/Cn3D is a helper application which enables users of the CDD resource to examine NCBI-curated domain hierarchies in great detail. CDTree/Cn3D can be set up so that user query sequences can be embedded in previously defined hierarchical classifications, and so that the assignment of a user query to any particular sub-family can be challenged or confirmed (12). CDTree/Cn3D is also the software tool used by CDD curators, and can be used to build up multiple sequence alignment representations of proteins and protein domains from scratch or starting from existing legacy data. It provides a convenient interface to searching NCBI's protein sequence databases via PSI-BLAST (13), where the search models are constructed from user-edited multiple sequence alignments.
A new version of CDTree/Cn3D has been made available recently, which now supports Mac OSX as well as Windows operating systems and extends the number of features.
CDD CONTENTS AND AVAILABILITY
CDD is a database in NCBI's Entrez query and retrieval system, and can be searched by keyword. CDD is linked to other resources in Entrez, and explicit ‘Conserved Domains’ links are available for the majority of protein sequences tracked by Entrez, which point directly to visualization of the domain annotation. The pre-calculated annotation is updated several times a day, as the protein sequence database continues to grow.
CDD can also be searched with a protein query sequence through the CD-Search tool, described earlier (6), which uses the RPS-BLAST algorithm to compare query sequences against position-specific scoring matrices derived from the model collection in CDD. A single CD-Search with an average sized protein (several hundred amino acids) against the default search set should not take longer than a few seconds, where most of the time is spent formatting the output. Users of the service can choose between the default search set, which collates NCBI-curated domain models and those imported from SMART, Pfam, COGs and NCBI's Protein Clusters database, and individual search sets, such as all of the above and the KOGs collection (3), which is not part of the default set.
When users submit protein query sequences to NCBI's protein-BLAST service, CD-Search is run in parallel, and its results are displayed on intermediate and final protein-BLAST results pages.
To facilitate analysis of large numbers of proteins, RPS-BLAST and CDD data sets can be downloaded by FTP and installed locally. A standalone version of RPS-BLAST is packaged with other BLAST executables, available at ftp://ftp.ncbi.nih.gov/blast/executables/LATEST/, and is also available as part of the NCBI toolkit distribution (see ftp://ftp.ncbi.nih.gov/toolbox). Preformatted search databases, ready for use with RPS-BLAST, are available on the CDD FTP-site at ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd. The FTP site also contains individual search models and instructions on how to customize search databases. Database searches run locally will, in general, run significantly faster than database searches submitted and retrieved via the CD-Search web service.
Precomputed domain annotations for all protein sequences in the Entrez protein database are available for download from the CDART FTP site at ftp://ftp.ncbi.nih.gov/pub/mmdb/cdart/, and are meant to support large-scale analysis of conserved domain assignments and domain architecture.
CDD itself is updated several times a year. The current version, v2.14 contains a total of 26 660 models. A total of 2395 of these are superfamily ‘clusters’, which explain relationships between homologous and redundant models from various collections. The 3368 models are explicit alignment models curated as part of the CDD project. Version v2.15 is scheduled for release at the end of September 2008, and version v2.16, which will mirror the Pfam release 23, is scheduled for release in late 2008.
With CDD release 2.14, the help documentation has been thoroughly revised and now reflects all recent changes. Find URLs and FTP addresses in the table below:
| CDD | Database home page | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
| CDD help | CDD help documentation | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml |
| CDD FTP | CD models, prebuilt search databases | ftp://ftp.ncbi.nih.gov/pub/ mmdb/cdd |
| CD-Search | Live and precomputed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| CDTree/Cn3D | Domain hierarchy viewer and editor | http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml |
| rpsblast | Stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution | ftp://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: http://www.ncbi.nlm.nih.gov/BLAST/download.shtml |
FUNDING
Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Funding for open access charge: Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the authors of Pfam, SMART, COGs and NCBI's Protein Clusters database; and Paul Thiessen and the NCBI Information Engineering Branch for continuing assistance with software development. We also thank Kira Makarova and Michael Galperin for providing updates to COGs.
REFERENCES
- 1.Finn RD, Tate J, Mistry I, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: and updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheeler DL, Barret T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2005;32(Web Server Issue):W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreeva A, Howorth D, Chandonia J.-M, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong JH, Marchler-Bauer A. Protein subfamily assignment using the conserved domain database. BMC Research Notes. 2008 doi: 10.1186/1756-0500-1-114. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2003;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]