Abstract
Understanding the consequences on host physiology induced by viral infection requires complete understanding of the perturbations caused by virus proteins on the cellular protein interaction network. The VirusMINT database (http://mint.bio.uniroma2.it/virusmint/) aims at collecting all protein interactions between viral and human proteins reported in the literature. VirusMINT currently stores over 5000 interactions involving more than 490 unique viral proteins from more than 110 different viral strains. The whole data set can be easily queried through the search pages and the results can be displayed with a graphical viewer. The curation effort has focused on manuscripts reporting interactions between human proteins and proteins encoded by some of the most medically relevant viruses: papilloma viruses, human immunodeficiency virus 1, Epstein–Barr virus, hepatitis B virus, hepatitis C virus, herpes viruses and Simian virus 40.
INTRODUCTION
Viruses interfere with fundamental cellular processes, such as gene expression, cell growth and differentiation, by perturbing the cellular regulatory networks. The molecular mechanisms underlying this subversion of cell physiology mediated by viral infection can be understood only by uncovering how viral proteins perturb cellular protein interaction networks.
Elucidating mechanisms of viral action may thus be better achieved within an interpretative framework relying not on individual genes, but rather on entire biological pathways and networks. Although cellular protein interaction maps already exist for a few model organisms, and recent efforts have been made in order to compile from public databases viral interactions maps (1), there is currently no resource archiving and publicly providing exhaustive and detailed interaction maps between viral and host proteins, with the possible exception of the HIV-1 Human Protein Interactions Database (http://www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions) and of the PIG database (http://pig.vbi.vt.edu).
VirusMINT fills this gap by collecting and annotating, in a structured format, all interactions reported in the scientific literature between viral and host proteins (mainly human).
Although several automation proposals have been put forward to increase the efficiency and accuracy of curation (2), manual curation is, to date, the best way to populate databases with high-quality data.
The curation efforts to date has concentrated mainly on viruses known to be associated with infectious diseases and oncogenesis in humans, such as adenovirus, Simian virus 40 (SV40), human papilloma viruses, Epstein–Barr virus (EBV), hepatitis B virus (HBV), hepatitis C virus (HCV) and herpes viruses. Future plans include regular update of the data set and its extension to new viruses.
DATA CURATION
Protein–protein interactions were manually curated from the literature or imported from other databases: MINT (3), IntAct (4) and HIV-1 Human Protein Interactions Database. Data uploaded from MINT and Intact did not require any extra curation effort, as these databases fully describe the interaction details in their entries, reporting relevant information including interaction detection method, experimental role of interactors and participant identification method. Furthermore these two databases already have adopted PSI-MI standards (5), greatly facilitating data management. The HIV-1 Human Protein Interactions Database does not conform to PSI-MI standards and does not provide a full description of experimental details, only the functional relationship between the interaction partners. Thus, only a subset of interactions reported in the HIV-1 Human Protein Interactions Database data set could be imported, namely those representing enzymatic reactions, physical associations and co-localization. These interactions were automatically remapped to the most appropriate term in the PSI-MI controlled vocabulary (Table 1).
Table 1.
Remapping from HIV database terminology to PSI-MI controlled vocabulary
| HIV database | PSI-MI |
|---|---|
| Acetylated by | Acetylation reaction |
| Acetylates | Acetylation reaction |
| Cleaved by | Cleavage reaction |
| Co-localizes with | Colocalization |
| Fractionates with | Colocalization |
| Deglycosylates | Deglycosylation reaction |
| Glycosylated by | Glycosylation reaction |
| Methylated by | Methylation reaction |
| Myristoylated by | Myristoylation reaction |
| Palmitoylated by | Palmitoylation reaction |
| Phosphorylated by | Phosphorylation reaction |
| Phosphorylates | Phosphorylation reaction |
| Binds | Physical interaction |
| Associates with | Physical interaction |
| Complexes with | Physical interaction |
| Ubiquitinated by | Ubiquitination reaction |
Interaction data derived from articles curated according to the IMEx manual were first uploaded in MINT and then reimported in VirusMINT while, in order to rapidly populate VirusMINT with new viral interactions, we also applied a quick curation strategy, conforming to MIMIx standards (6) but without reporting the full experimental details substantiating each interaction. Each interaction is thus described by the experimental method, the interaction type and the experimental roles of the interactors, as defined by the PSI-MI ontology. Interacting proteins were remapped from NCBI identifiers to UniprotKB identifiers (7) wherever possible using the PICR service (8).
To distinguish between viral proteins generated from the same precursor (and which therefore point to the same Uniprot KB accession number), we took advantage of a new term, polyprotein fragment, recently introduced in the PSI-MI ontology. Distinct polyprotein fragments are annotated with their name, the range of the protein with respect to the polyprotein precursor and the ‘polyprotein fragment’ ontology term. Each fragment is considered and displayed in VirusMINT as a distinct molecule.
DATA SELECTION
To select relevant articles from the literature we developed a simple text mining script. The implemented parser, based on ‘context free grammar’, identifies sentences containing interaction information (9). We first searched PubMed for abstracts containing virus names. Each sentence in the selected abstracts was then individually examined for presence of interaction keywords, which were largely based on the list of Temkin and Gilder (9). To further increase efficiency of the parser this list was enriched with new tags, based mainly on the methods most commonly used for the identification of protein–protein interactions, and with the name list of the viral proteins of interest.
DATA SEARCH
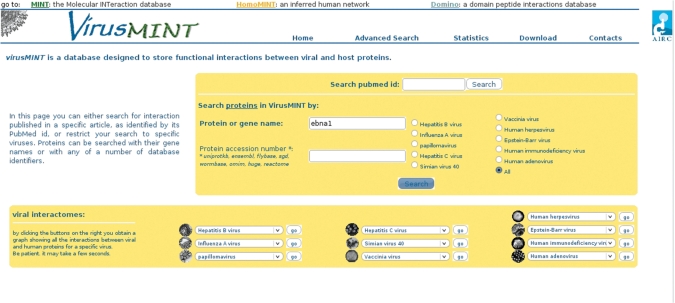
Whereas it is possible to perform a quick search based on protein or gene name, or based on an identifier from external databases from the VirusMINT home page, the ‘Advanced Search page’ allows for more flexible queries based on criteria such as the viral data set of interest (Figure 1) or a publication reference (PubMed ID number or DOI identifier). The search returns a list of proteins, and by clicking on the protein name the browser will present in the left frame a summary of the Uniprot Knowledgebase record for the selected protein, and in the right frame the list of interacting partners. Interactions involving proteins obtained from the processing of the same viral polyprotein are considered as distinct in VirusMINT. Each interaction is also assigned a confidence score (10). A summary of the reported interactions with related experimental details can be accessed by clicking on the number in the ‘interactions’ column.
Figure 1.
The Advanced Search page showing search options. VirusMINT can be queried for protein or gene name and for various database identifiers. Queries can be restricted to a strain of interest by clicking the corresponding radio button. The bottom half of the page lists all viruses represented in the database, and clicking on the corresponding virus name returns a graph of all interactions between viral and human proteins.
VISUALIZATION
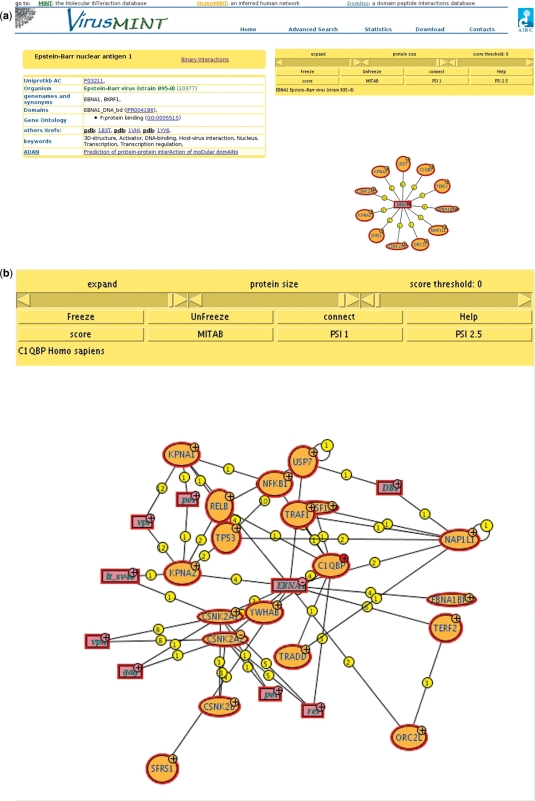
The ‘VirusMINT viewer’ button launches a Java applet that shows a graph of all interaction partners for that protein (Figure 2a). Node size is proportional to the molecular weight of the protein and node color is used to distinguish different species. Proteins linked to OMIM (11) diseases are highlighted in red. Edges are weighted according the number of supporting experimental evidences. The graph displayed by the VirusMINT viewer can be expanded (left click on ‘+’), or edited interactively by moving or deleting nodes (right click). The ‘score’ scroll bar is used to filter interactions according to a user-defined confidence threshold. In the interactome viewer, the confidence score takes into account the interactions involving proteins in all the strains of a particular virus. Finally, the ‘connect’ button interrogates the MINT database to add all the interactions between the proteins displayed in the graph (Figure 2b).
Figure 2.
The VirusMINT viewer. (a) By clicking on the VirusMINT viewer button all the interactions of the protein of interest will be displayed as a graph. (b) The ‘connect’ button interrogates the MINT database for all interactions between the proteins displayed in the graph.
VirusMINT also provides an innovative graphic display to visualize the full interactome of a given virus. This function is available both from the Homepage and from the Advanced Search Page, where it is also possible to restrict the query to a single viral strain. If no strain is specified, ortholog proteins from each strain are grouped to provide a ‘collapsed’ unique interactome for all available viral data. The interactome viewer displays both virus–virus and virus–host interactions. For smaller interactomes, the MINT database is queried for non-viral proteins to provide additional connections in the virus–host graph. The interactome viewer launches as a compact interface, where proteins are represented by dots rather than circles, and where all viral proteins are easily identifiable in red font. It is possible to switch to the classic viewer representation by scrolling the appropriate bar. A mouse click on a virus node triggers the display on the left frame of all strains in which orthologs of the protein are represented in VirusMINT. Clicking the edges displays a summary of the experimental evidences of the selected interaction. For both the ‘?’ button will open a pop-up window with detailed information about the selected protein.
‘Extension buttons’ found in the MINT viewer have not been implemented in the VirusMINT viewer, since VirusMINT viewer already displays all available data about the interactome of the selected virus.
DATA SUBMISSION
Authors of publications reporting protein interactions involving viral proteins are encouraged to submit the interaction data directly to VirusMINT. From the download page it is possible to obtain a preformatted spreadsheet file containing instructions for the compilation of the different fields.
STATISTICS
VirusMINT contains interaction data for 557 proteins encoded by 149 different viral strains, corresponding to 2007 unique interactions supported by 5483 experimental evidences derived from more than 1690 articles. Currently, 477 articles describing 1415 unique interactions supported by 2635 experimental evidences were manually curated in addition to the imported interactions (Table 2).
Table 2.
Detailed summary of interactions stored in VirusMINT
| VirusMINT |
Intact |
HIV |
Total |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pmids | Virus interactions | Hosts interactions | Evidences | Pmids | Virus interactions | Hosts interactions | Evidences | Pmids | Virus interactions | Hosts interactions | Evidences | Pmids | Virus interactions | Hosts interactions | Evidences | |
| EBV | 17 | 1 | 179 | 438 | 4 | 5 | 18 | 21 | 1 | 184 | 456 | |||||
| HBV | 5 | 11 | 19 | 5 | 8 | 11 | 19 | |||||||||
| HCV | 3 | 8 | 43 | 151 | 1 | 1 | 8 | 32 | 44 | 159 | ||||||
| Human adenovirus | 57 | 9 | 112 | 243 | 4 | 10 | 17 | 61 | 9 | 122 | 254 | |||||
| Human herpesvirus | 45 | 297 | 83 | 491 | 45 | 297 | 83 | 491 | ||||||||
| HIV | 29 | 7 | 48 | 120 | 1206 | 570 | 2744 | 1235 | 7 | 618 | 2864 | |||||
| Influenza A virus | 8 | 5 | 17 | 24 | 8 | 5 | 17 | 24 | ||||||||
| Papillomavirus | 121 | 10 | 282 | 610 | 2 | 1 | 5 | 9 | 123 | 11 | 287 | 619 | ||||
| SV 40 | 63 | 5 | 57 | 149 | 63 | 5 | 57 | 149 | ||||||||
| Vacciniavirus | 30 | 74 | 44 | 165 | 30 | 74 | 44 | 165 | ||||||||
| Others | 71 | 43 | 80 | 225 | 71 | 43 | 80 | 225 | ||||||||
For each virus is reported the source database, the number of unique virus–virus interactions (virus interactions), the number of unique virus–host interactions (host interactions) and the number of experimental evidences. The publications have been imported into VirusMINT in the following order: (i) import of missing interactions from Intact (IMEx curation rules) and (ii) import of HIV missing interactions from the HIV-1 Human Protein Interactions Database.
DATA DOWNLOAD
The VirusMINT data set is freely available and can be obtained by clicking the ‘Download’ link on the VirusMINT homepage. It is released in two different formats: flat files and PSI-2.5 XML files.
FUNDING
Associazione Italiana per la Ricerca sul Cancro; ENFIN Network of Excellence (LSHG-CT-2005-518254); Dana-Farber Cancer Institute Strategic Initiative; National Human Genome Research Institute (P50-HG004233); and National Institute of Environmental Health Science (R01-ES015728). Funding for open access charge: XXX.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ceol A, Chatr-Aryamontri A, Licata L, Cesareni G. Linking entries in protein interaction database to structured text: the FEBS Letters experiment. FEBS Lett. 2008;582:1171–1177. doi: 10.1016/j.febslet.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 3.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stumpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 7.Uniprot Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Côté RG, Jones P, Martens L, Kerrien S, Reisinger F, Lin Q, Leinonen R, Apweiler R, Hermjakob H. The protein identifier cross-referencing (PICR) service: reconciling protein identifiers across multiple source databases. BMC Bioinformatics. 2007;8:401. doi: 10.1186/1471-2105-8-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temkin JM, Gilder MR. Extraction of protein interaction information from unstructured text using a context-free grammar. Bioinformatics. 2003;19:2046–2053. doi: 10.1093/bioinformatics/btg279. [DOI] [PubMed] [Google Scholar]
- 10.Chatr-Aryamontri A, Ceol A, Licata L, Cesareni G. Protein interactions: integration leads to belief. Trends Biochem. Sci. 2008;33:241–242. doi: 10.1016/j.tibs.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 11.McKusick VA. A Catalog of Human Genes and Genetic Disorders. Baltimore: Johns Hopkins University Press; 1998. Mendelian Inheritance in Man. [Google Scholar]