Abstract
The Genomes TO Protein Structures and Functions (GTOP) database (http://spock.genes.nig.ac.jp/~genome/gtop.html) freely provides an extensive collection of information on protein structures and functions obtained by application of various computational tools to the amino acid sequences of entirely sequenced genomes. GTOP contains annotations of 3D structures, protein families, functions, and other useful data of a protein of interest in user-friendly ways to give a deep insight into the protein structure. From the initial 1999 version, GTOP has been continually updated to reap the fruits of genome projects and augmented to supply novel information, in particular intrinsically disordered regions. As intrinsically disordered regions constitute a considerable fraction of proteins and often play crucial roles especially in eukaryotes, their assignments give important additional clues to the functionality of proteins. Additionally, we have incorporated the following features into GTOP: a platform independent structural viewer, results of HMM searches against SCOP and Pfam, secondary structure predictions, color display of exon boundaries in eukaryotic proteins, assignments of gene ontology terms, search tools, and master files.
INTRODUCTION
Proteins encoded by genomes generally function after adopting proper 3D structures. A rapid increase in the number of entirely sequenced genomes led to an unprecedented growth in the number of hypothetical proteins resulting from genome annotation. Protein structures and functions can be inferred from amino acid sequences by using advanced computer programs. There is no doubt in the importance of structural and functional annotations of hypothetical proteins. The GTOP project was started in 1999 as reported (1) and was taken over by the DNA Data Bank of Japan (2) in 2007, under which the database has been continuously updated. GTOP is a database that provides protein annotation of 3D structures and functions based on similarity searches against PDB (3), SCOP (4), and Swiss-Prot (5), 2D structure predictions, Pfam (6) protein families, PROSITE (7) functional motifs, prediction of trans-membrane regions, and others.
There are several databases of the 3D structures of all the genome-encoded proteins. For example, SUPERFAMILY (http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/) (8) provides SCOP domain assignments to proteins encoded by completely sequenced genomes. A collection of comparative protein 3D structure models is available at Modbase (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi) (9) in some entirely sequenced genomes. Gene3D (http://gene3d.biochem.ucl.ac.uk/Gene3D/) (10) makes public CATH-based domain assignments and functional annotations to proteins in more than 500 genomes. Functional and domain assignments including intrinsically disordered (ID) regions can be found at PEDANT (http://pedant.gsf.de/) (11).
From the previous report, we have added a large body of data and tools to GTOP, for example ID region assignments, exon information on eukaryotic proteins, an efficient mechanism to search within a user-specified set of genomes, and tools for phylogenetic profile search. Since its inception, GTOP has employed a user-friendly interface to let the user grasp features of a query protein at a glance. The interface has been improved with the addition of new information. A GTOP user can readily obtain comprehensive structural and functional data of all the proteins encoded by entirely sequenced genomes.
UPDATE IN GTOP THAT CONTRIBUTED TO IMPROVED STRUCTURAL ASSIGNMENTS
A list of the genomes stored in GTOP is available at http://spock.genes.nig.ac.jp/~genome/org.html, together with the abbreviations of organism names used in the database. In the 2002 paper, we reported that GTOP contained protein data of 41 genomes (1). The database has grown to cover a total of 797 genomes, with 41, 466, 114 and 176 genomes of archaea, eubacteria, eukaryota and bacteriophages, respectively. The following data are subject to regular renewal: (i) amino acid sequences encoded by genomes newly sequenced after the previous update, (ii) amino acid sequences that existed in the previous version but were subsequently modified and (iii) reference databases such as PDB, SCOP, Swiss-Prot, Prosite, and Pfam whose new versions were released. The sequences fallen in category (ii) were recalculated to keep annotations up-to-date. Update category (iii) is crucial to keep annotations up-to-date, because most annotations in GTOP are obtained by homology search programs or those based on homology search.
The main focus of GTOP is structural annotations made by homology searches against the PDB and SCOP databases. Although GTOP used PSI-BLAST (12) in the previous report, it now employs reverse-PSI-BLAST (13), as this method gives comparable results in drastically reduced computation time. HMM searches using the SUPERFAMILY profiles (8) of SCOP domains were additionally conducted, as they are particularly effective in identifying small domains such as DNA binding domains.
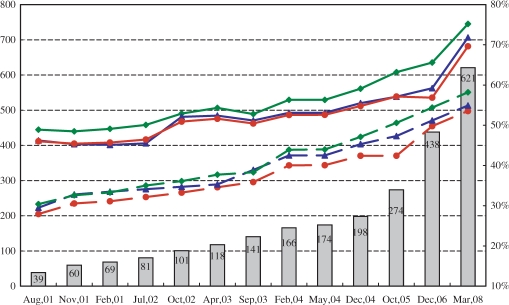
Figure 1 presents a time course of the number of the genomes stored and the average fractions of proteins with 3D annotations made by BLAST and reverse-PSI-BLAST. The fraction of sequences with alignments to PDB shows a steadily increasing trend, reflecting the growth of the PDB database. The fraction aligned by reverse-PSI-BLAST exceeds that by BLAST, reflecting the higher sensitivity of the former method. However, one should note that in this statistics a sequence is considered to be annotated if it has at least one PDB hit by BLAST or reverse PSI-BLAST and it may have large tracts of structurally undetermined regions. When statistics is evaluated residue-wise, the fractions of regions aligned to PDB sequences in the latest version in human and Escherichia coli proteins are 47% and 64%, respectively.
Figure 1.
The time courses of the number of genomes included and the fraction of the sequences with homologs in the PDB. The line graphs represent the ratios of the sequences with homologs in the PDB, while the column graph stands for the number of genomes in GTOP. The scales for the fraction and the number of genomes are shown at the right and left ends, respectively. The blue, green, and red lines correspond to fruit fly, E. coli, and the overall average, respectively. The solid and dotted lines respectively show the ratios obtained using reverse PSI-BLAST, and those using BLAST. The exact numbers of genomes are displayed near the top of the rectangles.
ID REGIONS
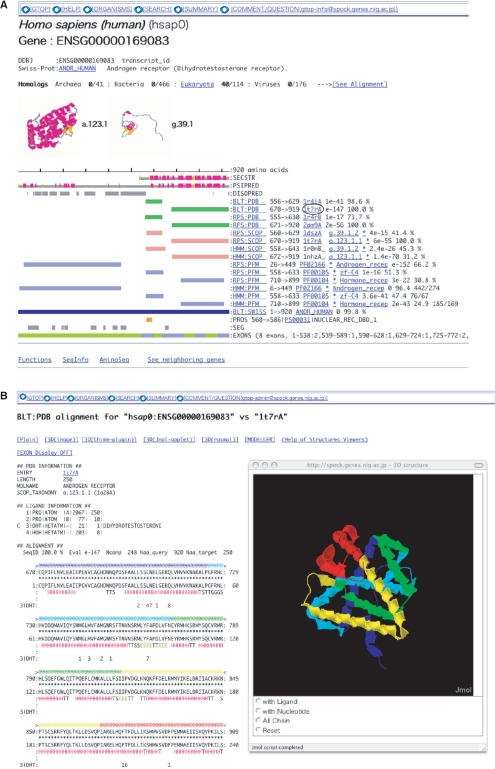
As most proteins do not entirely consist of structural domains, the fraction of residues with structural assignments will not reach unity; outside of globular domains there exist ID regions that assume no specific 3D structures by themselves, and tend to contain active regions in proteins involved in crucial biological processes such as signal transduction and transcriptional regulation (14–16). Recent research revealed that ID regions exist predominantly on the cytoplasmic side of eukaryotic proteins (17), play important roles in cell signaling, transcriptional control (18). We predicted ID regions in proteins stored in GTOP by the DISOPRED2 (19) program and presented them. Figure 2A shows a GTOP screen shot of human androgen receptor, a typical protein with long ID regions. As this example illustrates, GTOP graphically displays complex domain architectures of eukaryotic proteins composed of structural domains and ID regions.
Figure 2.
GTOP view examples. (A) The domain assignments of the human androgen receptor are presented in color bars to facilitate intuitive grasp of molecular architecture of the protein. This is a typical protein with long ID regions: the N-terminal half of the protein consists mainly of ID regions (18,22), consistent with the ID regions predicted by DISOPRED2 (gray bars on the line marked by DISOPRED). (B) A structurally aligned region of the same protein is shown in the exon view. This page can be obtained by clicking on the characters ‘1t7rA’ circled in Figure 2A, and by clicking on the EXON Display and 3D (Jmol-applet) buttons in the top section of the pop-up screen. The 3D structure is shown in five colors. By the 3D viewer, the sequence alignment is displayed with the exons represented in the same colors.
EXON BOUNDARIES IN EUKARYOTIC PROTEINS
The existence of introns and exons is a unique feature of eukaryotic genes and the location of exon boundaries in the corresponding protein structure is of interest (20). We thus developed tools to display exon boundaries on amino acid sequences and 3D structures. Figure 2B shows an example of the exon boundary view. The exons are presented in 5 colors both in the 3D structure and the sequence displays, from which the boundaries can be clearly seen. We developed a 3D viewing system incorporating Jmol applet (http://www.jmol.org/) so that the user can view 3D structures in the browser without installing additional software. Alternatively Rasmol (21) or Chime (http://www.mdl.com/) can be used. Exon information is also presented in green and blue stripes (near the bottom of Figure 2A).
SEARCH TOOLS
GTOP strives to keep precomputed annotations of all the amino acid sequences of proteins derived from all the completely sequenced genomes. One clear benefit of having precomputed annotations beside the rapidity of supplying information is to make inter-genomic comparative analyses possible. Phylogenetic profile search is one analytical tool that exploits this advantage: a user-specified search produces the presence and absence pattern of features such as SCOP folds, superfamilies, and families, Pfam domains, PROSITE motifs, and the number of trans-membrane helices. The user can conduct a search for a specific feature that are present in certain species and/or absent in others; for example, a search for a SCOP domain present in all the eubacterial species and absent in all the eukaryotic species in GTOP. The summary section of GTOP also offers comparative statistics, which has the ratio of 3D annotations in each genome, the frequencies of SCOP folds, superfamilies, and families, Pfam domains and PROSITE motifs.
Expansion of the database resulted in increased search time. The tools for keyword, homology, and text searches in GTOP were thus modified so that the user can reduce search time through selection of the genomes in which to conduct a search. The user can easily specify organisms with the use of check boxes placed next to organism names.
MASTER FILES
An annotation summary of each protein, consisting of abbreviated one-line descriptions, is saved in a master file. Master file information for each protein is displayed below a GTOP diagram of the type shown in Figure 2A. All the available data of each genome have been compiled in one file, freely downloadable from ftp://spock.genes.nig.ac.jp/pub/gtop/. Explanations of the meanings for each HEADER can be found at http://spock.genes.nig.ac.jp/~genome/mas-doc.html.
FUTURE DIRECTIONS
Despite the wealth of currently available structural data and use of sensitive programs, considerable fractions of most proteins have neither structural domains nor ID regions assigned. We are currently developing a system to accurately classify the fraction into structural domains and ID regions. Excitingly this will result in reliable identification of structural domains whose 3D structures remain undetermined. We expect that the installation of this system will provide further insights into the protein structure. We are also considering incorporation of protein–protein interaction data to enrich GTOP further.
FUNDING
The GTOP database is supported in part by the Target Protein Research Program from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and in part by the Bioinformatics Research and Development Project from the Japan Science and Technology Agency. Funding for open access publication charge: the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Kawabata T, Fukuchi S, Homma K, Ota M, Araki J, Ito T, Ichiyoshi N, Nishikawa K. GTOP: a database of protein structures predicted from genome sequences. Nucleic Acids Res. 2002;30:294–298. doi: 10.1093/nar/30.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugawara H, Ogasawara O, Okubo K, Gojobori T, Tateno Y. DDBJ with new system and face. Nucleic Acids Res. 2008;36:D22–D24. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, Flippen-Anderson JL, Ionides J, Kamada C, Krissinel E, et al. Remediation of the protein data bank archive. Nucleic Acids Res. 2008;36:D426–D433. doi: 10.1093/nar/gkm937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UniProt_Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;35:D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeats C, Lees J, Reid A, Kellam P, Martin N, Liu X, Orengo C. Gene3D: comprehensive structural and functional annotation of genomes. Nucleic Acids Res. 2008;36:D414–D418. doi: 10.1093/nar/gkm1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riley ML, Schmidt T, Artamonova, Wagner C, Volz A, Heumann K, Mewes HW, Frishman D. PEDANT genome database: 10 years online. Nucleic Acids Res. 2007;35:D354–D357. doi: 10.1093/nar/gkl1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 15.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 16.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 17.Minezaki Y, Homma K, Nishikawa K. Intrinsically disordered regions of human plasma membrane proteins preferentially occur in the cytoplasmic segment. J. Mol. Biol. 2007;368:902–913. doi: 10.1016/j.jmb.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 18.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J. Mol. Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Homma K, Kikuno RF, Nagase T, Ohara O, Nishikawa K. Alternative splice variants encoding unstable protein domains exist in the human brain. J. Mol. Biol. 2004;343:1207–1220. doi: 10.1016/j.jmb.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 21.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 22.Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43:3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]