Abstract
The UniPROBE (Universal PBM Resource for Oligonucleotide Binding Evaluation) database hosts data generated by universal protein binding microarray (PBM) technology on the in vitro DNA-binding specificities of proteins. This initial release of the UniPROBE database provides a centralized resource for accessing comprehensive PBM data on the preferences of proteins for all possible sequence variants (‘words’) of length k (‘k-mers’), as well as position weight matrix (PWM) and graphical sequence logo representations of the k-mer data. In total, the database hosts DNA-binding data for over 175 nonredundant proteins from a diverse collection of organisms, including the prokaryote Vibrio harveyi, the eukaryotic malarial parasite Plasmodium falciparum, the parasitic Apicomplexan Cryptosporidium parvum, the yeast Saccharomyces cerevisiae, the worm Caenorhabditis elegans, mouse and human. Current web tools include a text-based search, a function for assessing motif similarity between user-entered data and database PWMs, and a function for locating putative binding sites along user-entered nucleotide sequences. The UniPROBE database is available at http://thebrain.bwh.harvard.edu/uniprobe/.
INTRODUCTION
The characterization of transcription factors’ (TFs’) DNA-binding specificities represents a critical step towards understanding the regulation of gene expression and elucidating the biophysical properties governing protein–DNA interactions. The study of DNA-binding specificities therefore has profound implications for the analysis and prediction of the regulatory networks that govern intracellular function, responses to external stimuli, differentiation and development in an organism. Despite recent advances in this field, the vast majority of TFs in most major model organisms and pathogens remain either uncharacterized or poorly described (1).
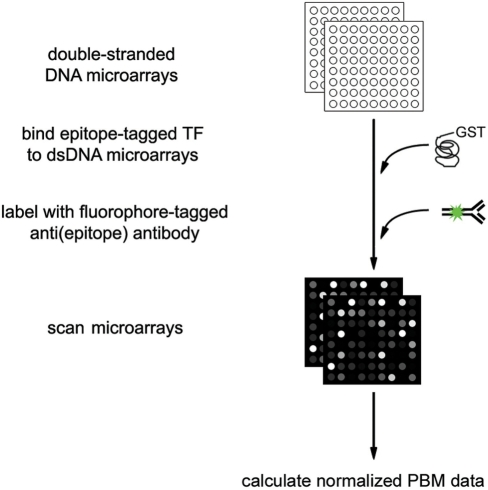
The development of universal (2) protein binding microarray (PBM) technology (3) (Figure 1) offers a new avenue for the exploration of protein–DNA binding specificity. Universal PBMs provide an efficient and comprehensive method for in vitro interrogation of DNA-binding preferences. PBM technology complements other currently available technologies, such as chromatin immunoprecipitation coupled with either microarray readout (4–7) or high-throughput sequencing (8–10) that identify genomic regions bound in vivo.
Figure 1.
Universal PBM schema. Universal PBMs containing all possible 10-mers within 60-mer probes are first synthesized as single-stranded oligonucleotide arrays, to which a common primer is annealed and extended in order to biochemically convert the single-stranded array to a double-stranded DNA (dsDNA) array (these steps are not shown in the figure) (2). The dsDNA array is then bound by protein, stained with a fluorophore-conjugated antibody, and scanned; the quantified array data are then normalized by the relative amounts of DNA in each spot, and used to calculate k-mer binding data (2). PWMs can be calculated either from the k-mer binding data using our Seed-and-Wobble algorithm (2) or from the 60-mer probe data using other motif finding algorithms (34).
Universal PBMs achieve comprehensive, high-resolution determination of proteins’ DNA-binding preferences by measuring the binding preferences of a protein over all possible k-mers of a given length (2,11). Currently employed custom array designs contain a set of 60-bp DNA probes that encompass all possible permutations of either 9 (Bulyk Lab, unpublished data) or 10 bp (12), depending upon the microarray design (2,12). In addition to covering all contiguous 9-mers or 10-mers, these array designs also offer an extensive set of gapped permutations that provide coverage of binding sites of greater length. Together, these data can be synthesized to produce high-confidence measurements of the relative preferences of a protein for all possible sequence variants belonging to a wide range of k-mer patterns typically found in TF binding site motifs (2,12). PBM enrichment scores from the PBM signal intensity data are typically calculated for each of the more than 2.3 million 8-mers (i.e. binding site ‘words’ with eight informative nucleotide positions, including all contiguous 8-mers and a large collection of gapped 8-mers). These 8-mers encompass the full affinity range of DNA binding preferences, from the most preferentially bound k-mers to low-affinity k-mers and nonspecifically associated sequences (2).
The TRANSFAC (13) and JASPAR (14) databases contain hundreds of matrices constructed from DNA binding site data from various data types; indeed, a given position weight matrix (PWM) in TRANSFAC frequently is derived from binding sequence data compiled from multiple experimental methods, which include lower throughput approaches such as gel retardation (i.e. electrophoretic mobility shift assays), DNase I footprinting, immunoprecipitation, supershift assays and methylation protection and higher throughput approaches such as in vitro selection (15) (SELEX). The PAZAR database (16) is a meta-database that contains TF binding site data. It contains PWMs from the JASPAR core database and in vivo TF binding site data and cis-regulatory module information from various sources, including other databases. A review of databases of cis-regulatory modules is beyond the scope of this article.
The universal PBM technology has several key advantages over in vitro selection approaches, such as SAGE–SELEX (17). SELEX does have the capability to interrogate sequences spanning a wide range of affinities, but it requires a significant increase in cost and labor to achieve the necessary depth of sequencing. Moreover, SELEX data have limited sensitivity because one cannot distinguish DNA binding site sequence variants missing from the collected data from those that are truly not bound by the given TF. In a survey of all SELEX datasets in the 2006 JASPAR database, we found that the median total number of binding site sequences in a JASPAR SELEX dataset is just 28, while the median number of nonredundant binding site sequences is just 11. In contrast, the DNA binding profiles obtained from our PBM experiments provide information on the direct binding preferences of a given protein over all k-mer DNA binding sequence variants, measured in vitro using the universal PBM technology; the number of sequence variants examined is limited only by the number of features on the microarray. In addition to the k-mer binding profiles, these procedures also provide DNA binding sequence PWMs derived from the k-mer data using our Seed-and-Wobble algorithm (2).
The UniPROBE database hosts the high-resolution DNA binding profiles obtained from PBM experiments on known and predicted TFs (2,3,18–21). The database currently contains DNA binding profiles for many proteins not included in similar databases such as JASPAR (14) and TRANSFAC (13), and it offers several tools for searching the database and analyzing user-defined binding profiles or DNA sequences. The resources and analysis tools offered by the UniPROBE database promise to facilitate previously untenable, downstream genomic analyses, and we anticipate that it will represent an important genomic resource as additional PBM data are compiled.
DATABASE CONSTRUCTION
The UniPROBE database is managed by a MySQL relational database that provides the back-end for user queries and facilitates the data retrieval necessary for the site's analysis tools. All HTML pages are dynamically generated by PHP scripts hosted on an Apache server, and several JavaScript libraries provide interactive interfaces that facilitate site navigation and form accessibility. The Apache server also hosts all downloadable data, available by HTTP connection.
DATABASE CONTENT
The UniPROBE database hosts the results of PBM experiments, subsequent computational analyses performed on these data, and protein annotations. The site currently hosts PBM data for over 175 nonredundant proteins from a wide range of organisms, including the prokaryote Vibrio harveyi, the eukaryotic malarial parasite Plasmodium falciparum, the parasitic Apicomplexan Cryptosporidium parvum, the yeast Saccharomyces cerevisiae, the worm Caenorhabditis elegans, mouse and human (2,18,20,21). These data already encompass the majority of mouse homeodomain TFs and will soon include more than 100 additional mouse proteins (labs of Bulyk, M.L. and Hughes, T.R., unpublished results), nearly 90 additional S. cerevisiae proteins (labs of Bulyk, M.L. and LaBaer, J., unpublished results), and over 20 additional C. elegans proteins (labs of Bulyk, M.L. and Walhout, A.J., unpublished results). The UniPROBE database will additionally host data for Drosophila melanogaster TFs from ongoing projects in the Bulyk laboratory, and we anticipate the addition of several datasets from other laboratories using this microarray technology.
For each DNA-binding protein, the server holds several different data types, including: (i) unprocessed 60-mer probe signal intensity data; (ii) normalized probe intensities; (iii) TF-binding DNA profile representations and (iv) publication-specific data. The unprocessed (or ‘raw’) Agilent array data include information on probe position, 60-mer probe sequence, and Cy3 (DNA, from incorporated Cy3-dUTP) and Alexa 488 (protein, from Alexa 488 conjugated anti-GST antibody) signal intensities, the latter of which are necessary for accurately assessing relative DNA binding (2). The normalized probe intensities are derived from the raw data after adjusting for relative DNA concentrations at each spot and for spatial nonuniformities within the microarray. The in-house software used for normalization and subsequent binding profile generation will soon be available for download as the Universal PBM Analysis Suite (11).
The database offers several different binding profile representations for assessing TF specificity and affinity. First, we use our Seed-and-Wobble algorithm (2) to generate PWM motifs, which represent the observed probability of finding a given nucleotide in a given position within a TF's DNA target site. PWMs currently serve as one of the primary methods for quantitative representation of DNA binding site motifs (13,14,22), and they are useful for creating graphical sequence logos (23,24) and for performing sequence analysis using any of several published software tools (22,25). Graphical sequence logos of the Seed-and-Wobble motifs, generated using the algorithms defined by enoLOGOS (24), are also present in the UniPROBE database.
Second, we provide two k-mer-based DNA binding profiles for each TF. The universal PBM designs facilitate k-mer binding profile construction because they allow for full coverage of all 8-mers of width 12 or less. The k-mer profiles have several advantages over the traditional PWM model. For example, comprehensive coverage of ungapped and gapped 8-bp sequence variants can provide insight into nucleotide interdependence within DNA binding site sequences; whereas, mononucleotide independence is implicit in traditional PWMs (26,27). The database's first k-mer-based binding profile consists of the median signal intensities and PBM enrichment scores associated with each contiguous 8-mer, where enrichment scores are calculated using a variant of the Wilcoxon–Mann–Whitney statistic and range from 0.5 for the most favored k-mers to −0.5 for the most disfavored k-mers. The second k-mer-based binding profile includes the top-scoring gapped 8-mer patterns (up to 10 positions) as determined by a 0.25 enrichment score cutoff; we used this threshold to avoid excessive file size for the gapped pattern profile and note that the Universal PBM Analysis Suite (11) can be used to generate full profiles for all gapped 8-mer patterns up to 12-nt positions in length.
In addition to these PBM data files, the UniPROBE database also provides relevant experimental information and factor-specific annotations. Experimental features include protein expression method, sequence and construct information (i.e. full-length protein or DNA-binding domain only). Factor annotations include functional descriptions and links to a variety of protein and gene reference databases. The website structure and interface section subsequently discusses these external annotations in further detail.
WEBSITE STRUCTURE AND INTERFACE
The Browse page of the UniPROBE database site presents a table containing each hosted TF, that protein's structural class, and the publication with which the protein's PBM data are associated. The entries are accompanied by brief descriptions of protein function retrieved from IHOP (28) or from a species-specific database such as SGD (29) or WormBase (30). The table then presents a link to a zipped file containing all factor-associated PBM data and a link to a Details page containing further factor annotations and a display of relevant features from the PBM experiments.
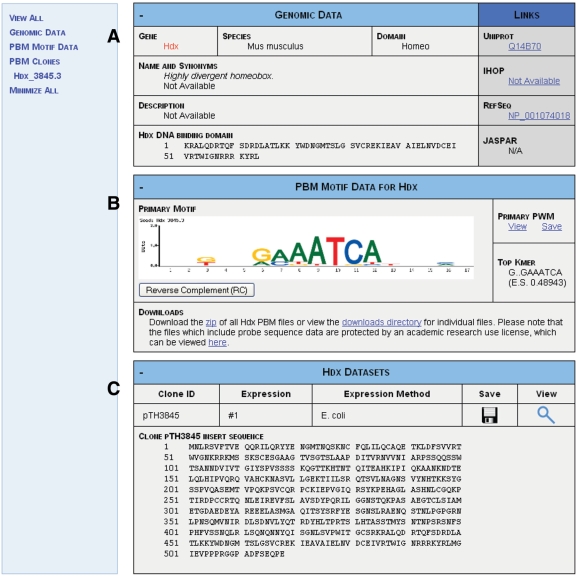
The Details page (Figure 2) described above has several components. The first section (Figure 2A) provides additional annotations for the factor of interest, including unique gene or protein accession numbers (if available as provided by the species-specific database), gene synonyms, DNA-binding domain amino acid sequence (if available) and links to databases such as IHOP (28), RefSeq (31), UniProt (32) and JASPAR (14). The second section (Figure 2B) displays the PWM and the matrix motif logo derived from Seed-and-Wobble analysis of PBM k-mer data. Although it does not directly display k-mer data, links to the data files containing complete k-mer data, normalized 60-mer probe data, and raw probe data are provided below the motif logo. The final section of the Details page (Figure 2C) displays a table that presents the experimental conditions and protein sequences used to produce each PBM dataset for the TF of interest.
Figure 2.
Details Page for the Mus musculus TF Hdx. This page includes (A) gene and protein annotations for Hdx, (B) PBM-derived motif data for the factor and (C) PBM experimental information for the Hdx data.
In addition to the Browse and Details pages, several other pages facilitate the download of specific materials. The Downloads page distributes ZIP files containing all instances of specific data types (i.e. all PWMs or all raw data) in the entire database, along with ZIP files of the data associated with each given publication. The Downloads page also provides links to the core SQL tables used by the database and documentation for these tables. As an alternative method of accessing UniPROBE files, one can use the Apache HTTP Server index to browse for files or use the Explore page to view the directory structure in a Microsoft Explorer style interface.
Most pages in the database website also provide forms for performing text searches on the database and for interrogating PBM-derived binding profiles. The following section describes these tools in detail.
SEARCH AND ANALYSIS TOOLS
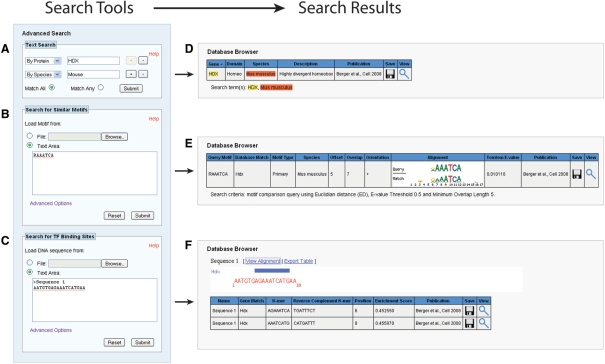
Several tools for conducting database searches and performing analyses on TF k-mer binding profiles (Figure 3) enhance the database's utility. A simple search field in the top right corner of the site's horizontal navigation bar provides a modified full-text MySQL search across species names, gene names, synonyms and annotations. Under the Advanced Search navigation bar, a customizable text search allows the user to enter multiple key words within specific database fields for a higher precision query. These terms may be linked by AND or OR Boolean operators by selecting the Match All or Match Any radio buttons, respectively (Figure 3A). The Browse page presents the match results (Figure 3B) for both search methods in the same table format used by the default Browse display, which provides basic annotations for each matching protein and links to download or view the associated PBM data.
Figure 3.
Database search tools and formatted query results. Search options include (A) a text-based search, (B) a tool for comparing standard motif representations against PBM-derived motifs in the database using the Tomtom program (33) from the Meta-MEME suite (22) and (C) a tool for scanning FASTA-formatted nucleotide sequences for matches to TF 8-mer binding profiles in the UniPROBE database. The Database Browser table formats the search results for viewing by (D) highlighting text search term matches, (E) presenting a graphical view of motif alignment and (F) illustrating 8-mer binding site matches along the input sequence (x-axis).
The Advanced Search bar also contains two analysis tools available for online use. The first tool, which uses the Tomtom program (33) from the Meta-MEME suite (22), provides a platform for comparing standard motif representations against the PBM-derived PWMs in the database (Figure 3C). The user may enter or upload up to 20 binding site representations in one of the following formats: frequency matrix, count matrix, Meta-MEME 3.x motif or IUPAC motif. As additional options, the user can restrict the query to PWMs from a particular species, specify a maximum similarity threshold, set a minimum matrix overlap or choose the comparison algorithm. Available algorithms include Euclidean distance, Pearson correlation, Kullback–Liebler Divergence and the Sandelin–Wasserman function, which are described in detail in the documentation for the Tomtom program (33). Upon query submission, the tool returns a table of statistics describing the best scoring alignment between a given pair of motifs, a Tomtom E-value (33) that quantifies their similarity and a graphical alignment of the two motifs’ logos (Figure 3D). This table also provides links to each matching factor's Details page for further investigation of the PBM data.
The second analysis tool uses contiguous 8-mer PBM enrichment score data to scan user-supplied input DNA sequences for putative TF binding sites (Figure 3E). To perform this DNA scan, the user must first upload or enter a FASTA file containing up to 30 DNA sequences, each of a 10-kb maximum permissible length. The user then specifies the species of interest and an enrichment score threshold, and the tool scans the input DNA sequence using an 8-bp sliding window to detect whether any TFs from the species of interest have enrichment scores greater than the user's threshold for that particular sequence. The website displays the results of the scan both as an HTML table available for plain-text download and as a simple graphic indicating binding site position and TF identity (Figure 3F). This tool may be useful not only for generating hypotheses about putative regulatory interactions but also for minimizing the unintentional creation of new binding sites for unrelated factors when designing site-directed mutagenesis experiments.
FUTURE DIRECTIONS
The upcoming publication of several large PBM datasets of yeast, fly and mouse TFs will contribute significantly to the breadth of coverage in the database. We encourage users of the database to register at http://thebrain.bwh.harvard.edu/uniprobe/register.php to receive updates concerning the addition of new datasets and changes to the database interface or analysis tools. We also encourage other labs generating universal PBM data to contact us by email if they wish to add their data to the UniPROBE database following the acceptance of their data for publication. The development of several additional tools may also enhance the website, and they may include a download manager, a local BLASTP function for identifying matches in our database to a user-specified query protein, and a DNA-binding preference prediction tool for user-specified query proteins.
AVAILABILITY AND LICENSE
All data hosted by the UniPROBE database are freely available for distribution at the database website. The sequences of the 60-mer DNA probes synthesized on our custom-designed universal arrays are available under the terms of the academic research use license described at http://thebrain.bwh.harvard.edu/uniprobe/academic-license.php. All pages have been tested under Firefox 2.0, Firefox 3.0 and Internet Explorer 7.
FUNDING
National Institutes of Health (R01 HG003985 to M.L.B.). Funding for open access charge: National Institutes of Health (R01 HG003985 to M.L.B.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank A. Philippakis and F. S. He for the survey of SELEX datasets in the 2006 JASPAR database, I. Adzhubey for technical assistance, M. Berger for helpful discussions and S. Gisselbrecht, R. P. McCord, A. Gehrke, and A. Aboukhalil for critical reading of the article.
REFERENCES
- 1.Bulyk ML. DNA microarray technologies for measuring protein–DNA interactions. Curr. Opin. Biotechnol. 2006;17:422–430. doi: 10.1016/j.copbio.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep P.W., 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 5.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 6.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 7.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 8.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 9.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 10.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 11.Berger M, Bulyk M. Nat. Protoc. Universal protein binding microarrays for the comprehensive characterization of the DNA binding specificities of transcription factors. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philippakis AA, Qureshi AM, Berger MF, Bulyk ML. Design of compact, universal DNA Microarrays for protein binding microarray experiments. (Presented at RECOMB 2007 conference) J. Comput. Biol. 2008;15:655–665. doi: 10.1089/cmb.2007.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliphant AR, Brandl CJ, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol. Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portales-Casamar E, Kirov S, Lim J, Lithwick S, Swanson MI, Ticoll A, Snoddy J, Wasserman WW. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8:R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P. High-throughput SELEX SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 18.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi Y, Qin Y, Berger MF, Ballow DJ, Bulyk ML, Rajkovic A. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol. Reprod. 2007;77:312–319. doi: 10.1095/biolreprod.107.060459. [DOI] [PubMed] [Google Scholar]
- 20.De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, Llinas M. Specific DNA-binding by Apicomplexan AP2 transcription factors. Proc. Natl Acad. Sci. USA. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol. Microbiol. 2008;70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundy WN, Bailey TL, Elkan CP, Baker ME. Meta-MEME: motif-based hidden Markov models of protein families. Comput. Appl. Biosci. 1997;13:397–406. doi: 10.1093/bioinformatics/13.4.397. [DOI] [PubMed] [Google Scholar]
- 23.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs' target genes and functions. Nat. Methods. 2008;5:347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulyk ML, Johnson PL, Church GM. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–1261. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Man TK, Stormo GD. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001;29:2471–2478. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat. Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 29.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers A, Antoshechkin I, Bieri T, Blasiar D, Bastiani C, Canaran P, Chan J, Chen WJ, Davis P, Fernandes J, et al. WormBase 2007. Nucleic Acids Res. 2008;36:D612–D617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber BR, Bulyk ML. Meta-analysis discovery of tissue-specific DNA sequence motifs from mammalian gene expression data. BMC Bioinformatics. 2006;7:229. doi: 10.1186/1471-2105-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]