Abstract
ExplorEnz is the MySQL database that is used for the curation and dissemination of the International Union of Biochemistry and Molecular Biology (IUBMB) Enzyme Nomenclature. A simple web-based query interface is provided, along with an advanced search engine for more complex Boolean queries. The WWW front-end is accessible at http://www.enzyme-database.org, from where downloads of the database as SQL and XML are also available. An associated form-based curatorial application has been developed to facilitate the curation of enzyme data as well as the internal and public review processes that occur before an enzyme entry is made official. Suggestions for new enzyme entries, or modifications to existing ones, can be made using the forms provided at http://www.enzyme-database.org/forms.php.
INTRODUCTION
The IUBMB Enzyme List, ‘Nomenclature and Classification of Enzymes by the Reactions they Catalyse’, is a functional classification system, whereby enzymes are classified on the basis of the overall reaction catalysed (1). The IUBMB Nomenclature Committee (NC-IUBMB), in association with the IUPAC-IUBMB Joint Commission on Biochemical Nomenclature (JCBN), has overall responsibility for the maintenance and development of the Enzyme List. The assignment of EC numbers and preparation of descriptive entries for each enzyme is coordinated by our group at Trinity College Dublin. In this article, we describe the development of ExplorEnz (2), which is now the primary source of IUBMB enzyme data. The enzyme data, including associated literature references, are stored in MySQL databases that can be accessed through a web application located at http://www.enzyme-database.org.
ENZYME CLASSIFICATION AND THE ENZYME LIST
Enzymes are classified according to the reactions they catalyse. Each enzyme, or group of enzymes that catalyse the same reaction, is given a four-part EC number, each part of which provides information about the reaction(s) catalysed. The structure of the catalytic molecule is not taken into account in the classification process so enzymes with different structures that catalyse the same reaction will have the same EC number. Further details of the rules used for enzyme classification have been published elsewhere (1,3,4) as well at the ExplorEnz website.
In addition to the EC number, an ‘accepted name’ is assigned. This is usually the most commonly used name for the enzyme, provided that it is not misleading or ambiguous. Other information provided for each enzyme includes: the reaction(s) catalysed, other names by which the enzyme is/has been known, the systematic name, comments, references and links to other relevant databases (Table 1). Where appropriate, glossary entries provide further information about less well known biochemical terms. In many instances, there are also diagrams of individual reactions, of the reaction mechanism or of the related metabolic pathways. These are supplied as raster images in GIF format and clicking on an EC number in a diagram provides a link back to the relevant enzyme entry in the database.
Table 1.
A description of the fields in the Enzyme List
| Field | Description |
|---|---|
| EC number | A four-component identifier, which classifies an enzyme according to class, subclass, sub-subclass, the final component being a serial number within that sub-subclass. |
| Accepted name | Usually the name by which the enzyme is most commonly known so long as it is not ambiguous or misleading. |
| Reaction | The reaction catalysed; this field may sometimes hold two or more sequential reactions; in some cases, alternative reactions are given. |
| Glossarya | This field provides explanatory information on the chemical names given in the reaction, along with links to further details on the IUBMB/IUPAC websites. |
| Other name(s)a | A list of alternative names by which the enzyme has been known. Ambiguous, or obsolete names, where given, are flagged as such. |
| Systematic namea | A formal, unambiguous, name that is composed of two parts: (i) the name of the substrate, or a list of substrates separated by colons; (ii) a term ending in –ase, which describes the type of reaction involved. The second part of the name is sometimes qualified by a further term given in parentheses: e.g. ‘(ADP-forming)’. |
| Commentsa | Extra information on the nature of the reaction catalysed, metal-ion requirements and links to associated enzymes. |
| Links to other databases | A list of some other databases that hold data on the enzyme. |
| References | References to the primary literature, citations of which are given as evidence of the reaction catalysed, the properties and the function of the enzyme. |
aDo not occur in all enzyme entries.
The official Enzyme List has been available for many years in the form of a series of flat files at http://www.chem.qmul.ac.uk/iubmb/enzyme/. These files, being small in size, are advantageous in areas with poor download speeds, but they also have a number of drawbacks. Some of these drawbacks, which have been addressed in ExplorEnz, are the lack of a comprehensive search facility, of a log detailing changes that have been made to the data and, perhaps most importantly, an automated means of providing individual users and database providers with the most up-to-date version of the data in downloadable form. Many databases, including BRENDA (5), ExPASy (6), GO (7) and KEGG (8), incorporate the IUBMB Enzyme List data into their datasets. Prior to the development of ExplorEnz, they had no easy way of knowing which enzyme entries had had minor modifications made to them or what those changes were. This problem has been addressed in ExplorEnz by providing a daily update of the database, in SQL and XML format, for download. The replication facility provided by MySQL allows curators of other databases to have real-time updates of the data. In addition to reducing the workload of database providers in obtaining the Enzyme List data, the download facilities of ExplorEnz are an important means of improving consistency among the databases by ensuring that all have access to the most up-to-date version.
DATABASE CONSTRUCTION
Prior to construction of the database, the HTML files of the Enzyme Nomenclature website at http://www.chem.qmul.ac.uk/iubmb/enzyme/ were converted to plain-text files. A set of Perl scripts was written to convert this plain-text version of the Enzyme List into a tabular format that could be used as input to MySQL. A pattern-matching system based on a library of regular expressions was developed in order to regenerate automatically the formatting of chemical names using HTML mark-up. The reason for having a plain-ASCII and an HTML version of the data is to provide the user with the correctly formatted output, while allowing them to search using plain text, as they would with other databases. Access to the database is provided by a set of web applications written in PHP. The data can also be exported in the style employed by the IUBMB website at http://www.chem.qmul.ac.uk/iubmb/enzyme/newenz.html, which prevents duplication of effort and ensures consistency among the datasets whenever new/modified data are added to ExplorEnz.
At the time of writing, ExplorEnz contained 1119 entries in class 1 (oxidoreductases), 1179 entries in class 2 (transferases), 1127 entries in class 3 (hydrolases); 371 entries in class 4 (lyases); 165 entries in class 5 (isomerases) and 141 entries in class 6 (ligases), giving a total of 4102 current enzyme entries. EC numbers no longer in use are listed as being either deleted or transferred, of which there are 796 instances. Table 2 shows a more complete set of these data, broken down by class.
Table 2.
Tallies of the EC numbers held in ExplorEnz
| Class 1 (Oxidoreductases) | Class 2 (Transferases) | Class 3 (Hydrolases) | Class 4 (Lyases) | Class 5 (Isomerases) | Class 6 (Ligases) | All classes | |
|---|---|---|---|---|---|---|---|
| Current | 1119 | 1179 | 1127 | 371 | 165 | 141 | 4102 |
| Transferred | 146 | 51 | 276 | 64 | 3 | 2 | 542 |
| Deleted | 63 | 59 | 98 | 23 | 7 | 4 | 254 |
| Total | 1328 | 1289 | 1501 | 458 | 175 | 147 | 4898 |
The most recent version of these data can be viewed at http://www.enzyme-database.org/stats.php
WEB INTERFACE AND SEARCH RESULTS
The search interface allows searching of all of the fields, or any selected subset thereof, as shown in Figure 1a. Text fields within the database are set as case-insensitive and an asterisk (*) may be used as a wildcard character. By default, all of the fields in the enzyme entry are displayed in the results output, but the user has the option to limit the output displayed to fields of their choosing; for example, it is possible to search for an enzyme name within the fields Accepted name and Other names but to display only the Reaction field in any records matching the search pattern. All of the entries that match the search criteria are displayed on a single page by default, but the user can specify the number of entries to be displayed on each page (up to 250, in predefined increments). Figure 1 shows the basic search form and sample query results.
Figure 1.
(a) The default search interface of ExplorEnz, which shows an example of a search within the EC-number field for sub-subclass 1.3.3.−. (b) An example of ExplorEnz output, showing the first two enzyme entries returned for the query shown in (a).
ExplorEnz makes use of the regular-expression pattern-matching facility of MySQL. A brief guide to regular expressions is provided at http://www.enzyme-database.org/regex.php. The most basic use of this feature would be for case-sensitive searches. An ‘Advanced search’ option allows the user to search for up to four different text patterns simultaneously, using Boolean algebra to include or exclude terms from the results. Search terms are matched against a plain-ASCII version of the data but HTML-encoded results are returned.
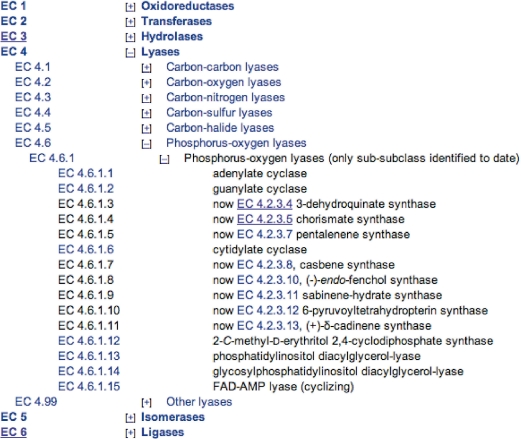
Searches that are restricted to the EC-number field will match EC numbers exactly. Any part of the EC number can be replaced by a wildcard; hence, an EC-only search for ‘1.1.1.*’ will return all enzyme entries with EC numbers in sub-subclass 1.1.1. An alternative way to search for an EC number is to use the dynamically generated table of contents of the Enzyme List, which is available from the homepage. As shown in Figure 2, the table of contents displays the class, subclass, sub-subclass and accepted names of each whole or partial EC number, which, when clicked upon, will return the relevant enzyme entry, or set of entries. Clicking on the class or subclass title (e.g. ‘Oxidoreductases’) will open a separate window with information describing the contents of that class. The data on a specific enzyme entry can be obtained using the general URL: http://www.enzyme-database.org/query.php?ec=x.y.z.w (for EC x.y.z.w).
Figure 2.
EC table of contents. Enzyme classes, subclasses and sub-subclasses can be opened or closed by clicking on the ‘+’ or ‘−’ symbols. Clicking on a partial EC number selects the range of EC numbers it contains while clicking on a full EC number shows the full entry for that enzyme. The class and sub-class headings are linked to descriptions of their contents.
Text searches may use complete or partial terms, e.g. ‘N-form’ will return ‘N-formyl-’, ‘N-formimido-’, etc. Terms entered using the asterisk (*) as a wildcard are also accepted, such as ‘N-*yl-CoA.’ An additional feature is that the search term can be highlighted in the results page, making it easier for the user to locate relevant information in the results.
The enzyme database maintains a FULLTEXT index, which permits the use of MySQL's natural language full-text search functions, and provides an alternative way to query the database using Boolean logic, in addition to the ‘advanced query’ method described above. Full-text searches can be enabled on the simple search page by checking the ‘Full-text search’ button. The search words entered by the user are then matched against the full-text index and ranked in order of relevance. Unlike substring searching, this search mode does not expand the query term automatically with wildcard characters, although these may be added by the user; thus, ‘glucos* sugar’ would match entries that contain ‘glucose’ or ‘glucosamine’ (etc.) or ‘sugar’ in the selected fields. Terms prepended with a ‘+’ are required to appear in the results; terms with ‘−’ prepended will be excluded from the results. The characters + and − thus simulate the Boolean operators AND and NOT, respectively. For example, ‘+glucose +oxidase’ will require that both terms appear among the selected fields of each record, whereas ‘glucose oxidase’ will return those entries matching either, or both, of the two search words.
As new information on enzymes becomes available, it sometimes becomes necessary to delete existing entries or to transfer an entry to a different category. Deleted and transferred entries are included in the ExplorEnz output, which will be of help to those wishing to trace the fate of specific entries. The last field in an enzyme entry provides a history of the enzyme since its creation, e.g. ‘EC 1.14.19.3 created 1986 as EC 1.14.99.25, transferred 2000 to EC 1.14.19.3’.
The results of a search can be converted into a format more suitable for printing, without loss of the formatting, by clicking on the printable version button on the results page. In the printable version, the font size is reduced and rendered in black and white only, the ‘Links to other databases’ field is omitted and underlining of links is suppressed. If desired, the search results, in either the default or printable view, can be translated to a PDF using third-party software. In addition to printable output, ExplorEnz also provides readymade PDF files of all enzyme entries, with one file for each class (http://www.enzyme-database.org/downloads.php). These PDFs are produced by pdfLaTeX from the HTML data that are held in the database, after a processing step that converts the hypertext markup into LaTeX. Each PDF has a table of contents, a bibliography and an index of the accepted names. The LaTeX package, hyperref, has been used to provide each file with hyperlinks, both internal and external, to link to pages within each document as well as to link the EC numbers to the corresponding ExplorEnz web page.
CURATION
Tools
A second, developmental, database was also created. In addition to the enzymes of the official Enzyme List, this includes enzymes that are in the process of being classified, as well as revised versions of existing entries. A curatorial interface to this database was constructed, called CurEnz, which permits modification of existing entries as well as the addition of new candidate enzymes. It allows the curator to add unformatted text, which is then automatically rendered into the correctly formatted HTML using the regular expression-based system referred to earlier. This feature reduces the time required to input the data, and ensures consistent formatting of chemical terms throughout the database. PubMed identifiers (PMIDs) can be used to import correctly formatted references from PubMed into the database automatically. Changes to the curatorial database are logged to a separate table, so that the history of each enzyme entry is preserved. Global changes in terminology, field name or nomenclature that are applied to all entries at once are not logged; such changes are recorded separately, along with an explanation of the reason for the change, at http://www.enzyme-database.org/changes.php.
New submissions
Novel enzymes for inclusion in the Enzyme List, as well as proposed revisions to existing enzyme entries, can be submitted through web forms available on the ExplorEnz website.
Since enzyme classification is based on the reaction(s) catalysed, there must be published chemical/biochemical evidence that a particular enzyme is responsible for a particular reaction before classification can be considered. Amino acid sequence data, or the existence of an apparent gap in a biochemical pathway, are not, in themselves, sufficient for classification purposes. Before assigning an EC number to a new enzyme, it must first be established that it catalyses a reaction that has not been classified previously. This is easier to determine if the substrate specificity of the enzyme is high, i.e. if the enzyme uses only one compound (or a small number of compounds) as the substrate. Many enzymes, such as alcohol dehydrogenase (EC 1.1.1.1), have broad substrate specificity, which means that they can use many substrates to give many products. In order to assign a new EC number to an alcohol dehydrogenase, there would need to be published evidence that the substrate(s) that the new enzyme uses differ(s) in some significant way from the substrate specificities of alcohol dehydrogenases already present in the Enzyme List. The same enzyme from different species does not receive a separate EC number. This information is now available at the ExplorEnz website and, in more detail, elsewhere (3).
If sufficient published data are available to warrant a new enzyme entry, a draft entry is created in CurEnz, where it undergoes internal review by a panel of experts before starting the 1-month public review process at http://www.enzyme-database.org/newenz.php. If at the end of the public review process no evidence has arisen to challenge the validity of the entry, the enzyme's status becomes official and it is added to the main Enzyme List and modified entries replace the pre-existing version.
CONCLUSIONS
ExplorEnz is the primary source of new EC numbers, from which all other databases containing the Enzyme Nomenclature data can be updated. It preserves the formatting of chemical names: the unformatted names form part of the searchable data, while the formatted versions are stored within the database as HTML. It offers a comprehensive range of search options, from a simple search for a single EC number to more advanced search options using regular expressions. The user has control over the output, being able to limit it to a subset of the available fields. Daily updates of the data as SQL and XML files can be obtained from the main website. MySQL replication is another option that is available to interested parties, upon application to the corresponding author.
Future developments of the enzyme database will include the separation of primary and secondary (alternative) reactions, the continued expansion of glossary and synonyms fields (with, for example, pharmacological terms), and the inclusion of information on reaction thermodynamics. In addition, we hope to provide vector graphic versions of the reaction diagrams, in a format that will make them searchable and more suitable for printing.
AVAILABILITY
The database is publicly accessible at http://www.enzyme-database.org as HTML, XML or SQL. The data are copyrighted by the IUBMB, but are freely available for use in other applications provided that the original source of the data is acknowledged.
FUNDING
Funding for open access charge: Science Foundation Ireland (07/IN.1/B930-Tipton).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Prof. Toni Kazic (University of Missouri) for assistance in writing the Perl scripts that were used during the cleaning and parsing of the original data.
REFERENCES
- 1.IUBMB. Enzyme Nomenclature: Recommendations (1992) of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. San Diego, CA: Academic Press; 1992. Accessible at: http://www.chem.qmul.ac.uk/iubmb/enzyme/ (2 September 2008, date last accessed) [Google Scholar]
- 2.McDonald AG, Boyce S, Moss GP, Dixon HBF, Tipton KF. ExplorEnz: a MySQL database of the IUBMB enzyme nomenclature. BMC Biochem. 2007;8:14. doi: 10.1186/1471-2091-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tipton KF, Boyce S. In: Nature Encyclopedia of Life Sciences. London: Nature Publishing Group; 2000. Enzyme Classification and Nomenclature. [Google Scholar]
- 4.Boyce S, Tipton KF. History of the enzyme nomenclature system. Bioinformatics. 2000;16:34–40. doi: 10.1093/bioinformatics/16.1.34. [DOI] [PubMed] [Google Scholar]
- 5.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32 doi: 10.1093/nar/gkh036. D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]