Abstract
We have developed ExDom, a unique database for the comparative analysis of the exon–intron structures of 96 680 protein domains from seven eukaryotic organisms (Homo sapiens, Mus musculus, Bos taurus, Rattus norvegicus, Danio rerio, Gallus gallus and Arabidopsis thaliana). ExDom provides integrated access to exon-domain data through a sophisticated web interface which has the following analytical capabilities: (i) intergenomic and intragenomic comparative analysis of exon–intron structure of domains; (ii) color-coded graphical display of the domain architecture of proteins correlated with their corresponding exon-intron structures; (iii) graphical analysis of multiple sequence alignments of amino acid and coding nucleotide sequences of homologous protein domains from seven organisms; (iv) comparative graphical display of exon distributions within the tertiary structures of protein domains; and (v) visualization of exon–intron structures of alternative transcripts of a gene correlated to variations in the domain architecture of corresponding protein isoforms. These novel analytical features are highly suited for detailed investigations on the exon–intron structure of domains and make ExDom a powerful tool for exploring several key questions concerning the function, origin and evolution of genes and proteins. ExDom database is freely accessible at: http://66.170.16.154/ExDom/.
INTRODUCTION
Domains are the fundamental structural components of proteins and perform vital biological functions. Homologous domains with highly similar structure and function, but lacking extensive sequence similarity, are abundantly found in proteins from different eukaryotic organisms (1,2). Eukaryotic genes coding for a protein with multiple domains may contain several introns within the coding sequences of each domain. Since the origin and evolution of the proteins are tightly linked to those of its coding genes, modern research has increasingly focused on analyzing the relationship of the domain structure of eukaryotic proteins to the exon–intron structure of their coding split genes (3–7). Software tools to analyze the interrelated structures of domains, proteins, exons and genes will be extremely useful to modern research in genomics and proteomics, and could enable new insights and discoveries concerning the origin and evolution of genes and proteins.
There is currently a lack of integrated resources for comparative analysis of the exon–intron structure of genes mapped to the domain structure of proteins across different organisms. Domain annotation resources such as Pfam (8), ProDom (9) and InterPro (10) provide detailed information on protein domains, and genome annotation resources like RefSeq (11) and Ensembl (12) provide extensive gene structure annotations. But integrated access to the exon–intron structure of protein domains is not available in both types of resources. SEDB (13) and XdomView (14) are two tools that map protein structure data from RCSB Protein Data Bank (PDB) (15) to gene structures annotated in GenBank (16). However, the BLAST (17) based mapping methodologies used by these tools tend to retrieve several gene structure matches for a single queried protein sequence, and hence may have limitations in identifying reliable and exact one-to-one mapping of a queried protein to its corresponding gene entry. These tools also lack facilities for comparative analysis of the gene structure of a user-specified domain among different protein hits, or for analyzing variations in protein domain architecture. Several resources for in-depth analysis of domain features such as the domain architecture in proteomes (18), the domain homology in different transcriptomes (19) and the role of alternative splicing in protein structure (20) are currently available. However, these resources lack comparative analytical capabilities for investigating exon–intron structure variations among the domains of multiple proteins.
We have developed a unique, publicly available database called ExDom (http://66.170.16.154/ExDom/) that provides integrated access to the exon–intron structure of protein domains from seven eukaryotic genomes (Homo sapiens, Mus musculus, Bos taurus, Rattus norvegicus, Gallus gallus, Danio rerio and Arabidopsis thaliana). ExDom is the first resource that provides reliable one-to-one mapping of domain data from the Pfam database to exon–intron structure annotations in the RefSeq database. It efficiently integrates 96 680 Pfam annotated domains in 40 390 UniProtKB/Swiss-Prot (21) proteins to their corresponding exon–intron structures in the 40 390 RefSeq genes that code for these proteins. ExDom's sophisticated web-based graphical user interface allows efficient intergenomic and intragenomic comparative analysis of exon–intron structures of common, shared and unique domains among different proteins. Users can also analyze the exon–intron structure variations of alternatively spliced genes, which produce domain architecture variations in multiple protein isoforms. The reliable data content and the extensive analytical facilities for exploring the data make ExDom a vital utility for the comprehensive and integrated analysis of gene structures of domains across similar and divergent protein groups.
DATABASE CONTENTS
ExDom database contains an extensive collection of exon–intron structures of genes mapped to protein domains, which can be visualized and analyzed with its user-friendly web interface. Table 1 gives the current data content in ExDom and reveals the diversity in domain and gene structure features among the seven eukaryotic organisms currently available in ExDom. We find that a significant fraction (40.6%) of ExDom proteins contain multiple domains. Most of the domains in one organism tend to have homologous domains in several other organisms. A given domain can also occur in several proteins within the same organism. We also find that domain architecture variations among protein isoforms are very common in all seven organisms. Another interesting observation is the abundance of introns within the coding regions of domains. In ExDom, 67.7% of domains have introns and the average number of introns per domain is 3.3.
Table 1.
Summary of ExDom contents
| Organism | Unique Pfam domains | Total Pfam Domains | Proteins mapped to their gene structures | Proteins with multiple domains | Proteins with isoforms | Domain architecture variations among protein isoforms | Domains with introns | Average number of introns in domains | Intra-genomic common domainsa | Inter-genomic common domainsb |
|---|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | 3298 | 39 927 | 13 220 | 5860 (44.3%) | 1366 (10.4%) | 1072 (78.5%) | 25 824 (64.7%) | 3.5 | 2057 (62.4%) | 3154 (95.6%) |
| Mus musculus | 3089 | 24 596 | 10 465 | 4259 (40.7%) | 321 (3.1%) | 247 (77%) | 17 343 (70.5%) | 3.6 | 1739 (56.3%) | 3060 (99%) |
| Bos taurus | 1739 | 6145 | 3275 | 1140 (34.8%) | 2 (0.06%) | 1 (50%) | 4651 (75.7%) | 3.4 | 727 (41.8%) | 1734 (99.7%) |
| Rattusnorvegicus | 2107 | 10 218 | 4844 | 1904 (39.3%) | 35 (0.7%) | 27 (77.1%) | 7549 (73.9%) | 3.5 | 972 (46.1%) | 2099 (99.6%) |
| Gallus gallus | 892 | 3008 | 1379 | 573 (41.6%) | 3 (0.2%) | 3 (100%) | 2291 (76.2%) | 3.3 | 301 (33.7%) | 886 (99.3%) |
| Danio rerio | 824 | 2393 | 1293 | 410 (31.7%) | – | – | 1723 (72%) | 3.4 | 272 (33%) | 812 (98.5%) |
| Arabidopsis thaliana | 1353 | 10 393 | 5914 | 2258 (38.2%) | 203 (3.5%) | 155 (76.4%) | 6065 (58.4%) | 2.7 | 856 (63.3%) | 1067 (78.9%) |
aCommon domains found in different proteins of the same organism.
bCommon domains found in proteins of at least one other organism.
DESIGN AND IMPLEMENTATION
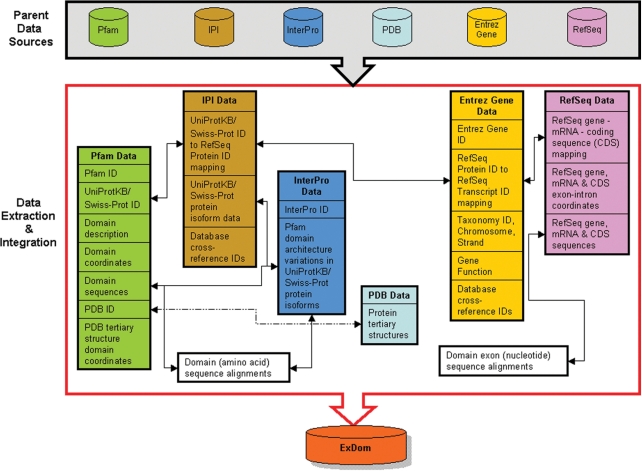
The ExDom database was developed by extracting and seamlessly integrating domain data from Pfam-A (the section of Pfam database with curated and well-characterized domain annotations) and InterPro, protein data from International Protein Index (IPI) (22), tertiary structure data from PDB and gene data from RefSeq and Entrez Gene. This data were then stored in a MySQL 4.1 relational database along with cross-references to gene and protein annotations from several other resources. Figure 1 gives an overview of the ExDom database development. The MySQL database was normalized and indexed to ensure efficient data retrieval through the query options available in ExDom's web interface. We will perform regular updates to the ExDom database to ensure that its contents are up-to-date with the periodic updates in its six parent data sources. Update schedules of ExDom will be synchronized with the update schedules of the Pfam database.
Figure 1.
Schematic overview of the data extraction and integration involved in ExDom database development.
The ExDom web interface developed in Perl CGI (http://www.perl.org/), runs on an Apache 2.0.53 (http://www.apache.org/) web server and utilizes the Perl DBI (http://www.cpan.org/) module to query and fetch data from the back-end MySQL database (http://www.mysql.com/). The graphical display of the protein-domains mapped onto the exon–intron structures of genes were implemented with Flash MX 6.0 vector graphics programming (http://www.adobe.com/). Multiple sequence alignments of protein and exon sequences of specific domains were performed using MultAlin (23) and the alignments were graphically displayed using the Jalview alignment editor tool (24). The graphical visualization of the tertiary structures of domain-specific exonic regions was implemented using the Jmol 3D structure viewer (25). Additional details on the methodologies used for creating the ExDom database and web interface are provided in the ‘Documentation’ section in the ExDom website.
ANALYTICAL CAPABILITIES
Graphical visualization of the exon–intron structure of protein domains
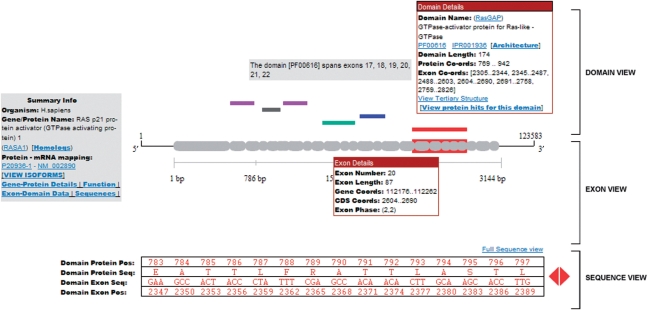
ExDom provides a two-dimensional graphical display tool called the ‘ExDom Plot’ (Figure 2) for visualization and analysis of domains of a protein and their corresponding exon–intron structures in their coding genes. The ExDom Plot also has a summary information box with details such as the gene/protein name, the name of the organism to which the protein belongs, a link to homologous gene information in the HomoloGene database (26), RefSeq mRNA ID to UniProtKB/Swiss-Prot protein ID mapping and links to several information pages that provide additional protein and gene information.
Figure 2.
ExDom Plot for RASA1 protein.
Each ExDom Plot has the following graphical elements:
‘Domain View’: This provides a graphical display of the domain architecture of a given protein. Domains are represented with uniquely color-coded rectangular boxes. Information on the domain such as domain name, Pfam ID, Pfam Accession, InterPro Accession, domain length, domain start and end positions in the protein sequence, its nucleotide positions in the coding gene sequence and a link to the tertiary structure view of the domain (whenever the tertiary structure data of a protein domain is available in PDB) are also available in the Domain View.
‘Exon View’: This gives the exon–intron structure of the gene that codes for the protein displayed in Domain View. Exon–intron structures are shown with gray rectangular rounded boxes representing exons and gaps between them representing introns. Exon View also provides details on the number, length and phase of the exons, as well as the coordinates of the exons in the gene and in the spliced coding sequence.
‘Sequence View’: This provides a scrollable view of the amino acid sequence of each domain alongside the corresponding nucleotide sequence (grouped into codons) which codes for the domain. Users can also further explore the domain-exon sequences by clicking on the ‘Full Sequence View’ link. This provides a comprehensive view of the exon sequences mapped on the entire domain in a single display page, enabling users to efficiently analyze sequence details of large domains.
It is the synchronized display of these three graphical elements in the ExDom Plots that enables users to efficiently analyze the exon–intron structure of domains. When a user moves the cursor over a domain of a given protein displayed in the Domain View area, this triggers the highlighting of the corresponding exon–intron structures of the domain in the Exon View area. Simultaneously, the associated amino acid and nucleotide sequences of the domain are displayed in the Sequence View area.
Comparative display and analysis using ExDom Plots
The comparative display features in ExDom allow users to visualize and analyze the domain architecture and domain-specific exon–intron structure variations across multiple proteins. Users can specifically search for the prevalence of a given domain in the seven organisms in ExDom, retrieve all proteins that possess this domain and view the ExDom Plots of these protein hits. The domain-specific color coding in the ExDom Plots is a particularly useful feature for comparative analysis, with common domains displayed in uniform color across the multiple protein hits. This allows easy visual comparisons of the various features of common domains, domains shared across subsets of proteins, and domains unique to one protein, among the different proteins displayed in a single web page (Supplementary Figure 1).
Multiple Sequence Alignment Viewers
ExDom's Sequence Alignment Viewers allow users to efficiently analyze the sequence variations among common domains found in different proteins. The Protein Alignment Viewer gives a graphical display of the multiple sequence alignment of the amino acid sequences of a common domain across different protein hits and the Exon Alignment Viewer provides a graphical multiple sequence alignment display of the spliced exons that code for the common domain in the different proteins. The alignments were generated using MultAlin, and the graphical display was implemented in Jalview. The alignment viewers provide useful analytical facilities for exploring the degree of conservation of domain sequences across proteins and also for analyzing the distribution of exonic sequences within the aligned coding nucleotide sequences of domains. Several other advanced features to analyze and edit the alignments are available in both Alignment Viewers. Users can also download the alignment files in text format.
Analysis of domain–gene structure variations in protein isoforms
Alternative transcription and alternative splicing can produce significant variations in the domain architecture of their protein isoforms. In ExDom, we have mapped the domain architecture variations in UniProtKB/Swiss-Prot protein isoforms to their specific gene structure variations in their respective RefSeq mRNA isoforms (mapping procedure details are available in the ‘Documentation’ page in the ExDom web interface). Users can analyze these variations using the comparative isoform display feature in ExDom which displays the ExDom Plots of the protein isoforms next to each other, thus allowing easy analysis and identification of the exon–intron structure variations that may be responsible for the changes in domain architecture in the protein isoforms.
Domain Tertiary Structure Viewer
Out of the 40 390 proteins in ExDom, 18 010 (44.6%) had tertiary structure data available in the PDB database. However, only 2309 (5.7%) PDB proteins had complete tertiary structure annotations for their Pfam domains. In ExDom, we integrated this Pfam tertiary structure data to gene structure data, and developed an advanced graphical viewing tool implemented in Jmol for analyzing the exon distributions within the tertiary structure of domains. A comparative analysis facility which allows users to simultaneously view the exon-domain tertiary structures of any two proteins is also available and is very useful for analyzing variations in exon locations in the tertiary structures of a common domain found in two different proteins.
Integrated access to gene and protein information
Gene and protein annotations from several database resources are integrated into ExDom. The Summary Info Box in the ExDom Plots of proteins provides access to the following information pages (Supplementary Figure 2): (i) Gene–Protein Details: provides information such as Entrez Gene ID, Gene Symbol, Gene Name, Gene Synonyms, Strand, Map Location, Contig Details, Organism Name, Taxonomy ID and UniProtKB/Swiss-Prot ID (ii) Functions: provides functional annotations for the gene/protein from GO (27) database and also a hyperlink to PubMed (26) citations on gene/protein function; (iii) Exon-Domain Data: users can view the gene structure data correlated to domain data in tabular format and download this information; (iv) Sequences: users can view gene, mRNA, CDS and protein sequences and download them; and (5) Database cross-references: provides link integration to 30 different resources including, EuSplice (28), UniProtKB/Swiss-Prot, SUPERFAMILY (29), PRINTS (30) and Ensembl.
QUERYING EXDOM
The web interface of ExDom has the following intelligent search options that allow users to effortlessly retrieve and analyze the exon–intron structure of domains.
Domain-centric search
Users can submit searches for a domain of interest and retrieve ExDom Plots of proteins that contain this domain. Domain-centric searches can be performed using the domain name or with specific domain identifiers (Pfam Accession or InterPro Accession). Users can also search with keywords that describe the features of a domain or its functionality. For example, a keyword search for the term ‘zinc’ retrieves 220 unique Pfam domains in the ExDom hit page. All of these domains have biological roles associated with the metal zinc. The hit page also provides details of each domain such as the Pfam Accession, the number of proteins that have the domain, the domain name and the domain description.
Gene/protein-centric search
These searches can be used to retrieve the exon–intron structures of domains from an individual gene or protein entry. Users can search based on Entrez Gene ID, gene name, gene symbol, UniProtKB/Swiss-Prot ID, RefSeq Protein ID or RefSeq Transcript ID. For example, a search for the Entrez Gene ID ‘4968’ of the ‘8-oxoguanine DNA glycoxylase’ gene, retrieves the ExDom Plots of its eight protein isoforms, which display the variations in domain architecture and exon–intron structure among these isoforms.
Drill-down domain searches
ExDom allows users to drill down its domain-centric search through the protein hits displayed in the ExDom result page. For example, a search submitted for ATP dependent DNA ligase domain (DNA_ligase_A_M) in ExDom using its Pfam Accession ‘PF01068’, retrieves eight protein hits that contain this domain. Looking at the ExDom Plots, one can see that there are several additional domains in the protein hits. The ExDom Plot of second protein hit, lig4 (M. musculus) shows that this protein has three additional domains—DNA_ligase_A_N, DNA_ligase_A_C and BRCT. To retrieve all proteins in ExDom that contain the BRCT domain, one can simply click on the drill-down search option—‘View protein hits for this domain’ which is found in the Domain View pop-up box. This will retrieve 47 protein hits that have the BRCT domain. Once again, each of these BRCT domain-containing protein hits has several additional domains, and, if interested, users can again use the drill-down search to retrieve proteins hits for any of these domains.
EXPLORING THE UTILITY OF THE DATABASE WITH A SAMPLE QUERY
The utility and the features of the ExDom database are demonstrated here with a sample query for the Sterol transfer family (SCP2) domain, which is involved in binding and intracellular transport of cholesterol and other lipids. Submitting a ‘Domain Keyword’ search in ExDom for the term ‘SCP2’ retrieves the result page with ExDom Plots for the fourteen proteins that have the SCP2 domain. The ExDom Plot of each protein provides detailed information on the domain architecture and the exon–intron structures of domains.
The ‘Summary Info’ boxes (on the left) show that the 14 proteins come from six different organisms—H. sapiens (human), M. musculus (mouse), R. norvegicus (rat), B. taurus (cow), G. gallus (chicken) and D. rerio (zebrafish). The protein hits are arranged in the result page based on the gene/protein names as follows: (i) Stomatin–like 1 in human (STOML1) and mouse (Stoml1); (ii) Sterol carrier protein 2 in chicken (SCP2), human (SCP2), mouse (Scp2) and rat (Scp2); (iii) hydroxysteroid dehydrogenase like 2 in zebrafish (hsdl2), mouse (Hsdl2) and rat (Hsdl2); (iv) hydroxysteroid (17-beta) dehydrogenase 4 in human (HSD17B4), mouse (Hsd17b4) and rat (Hsd17b4) and (v) chromosome 20 open reading frame 79 (C20orf79) in human and chromosome 20 open reading frame 79 ortholog (C13H20orf79) in cow. This arrangement based on gene/protein name similarity ensures that potentially homologous proteins are clustered together in the result pages. One can check whether these proteins, which are grouped together based on their names, are known homologs of each other by analyzing their homolog annotations in HomoloGene database (accessible through the ‘Homologs’ link in the Summary Info box). This reveals that each of the five name-based groups displayed in the ExDom result page falls into specific homologous groups annotated in HomoloGene.
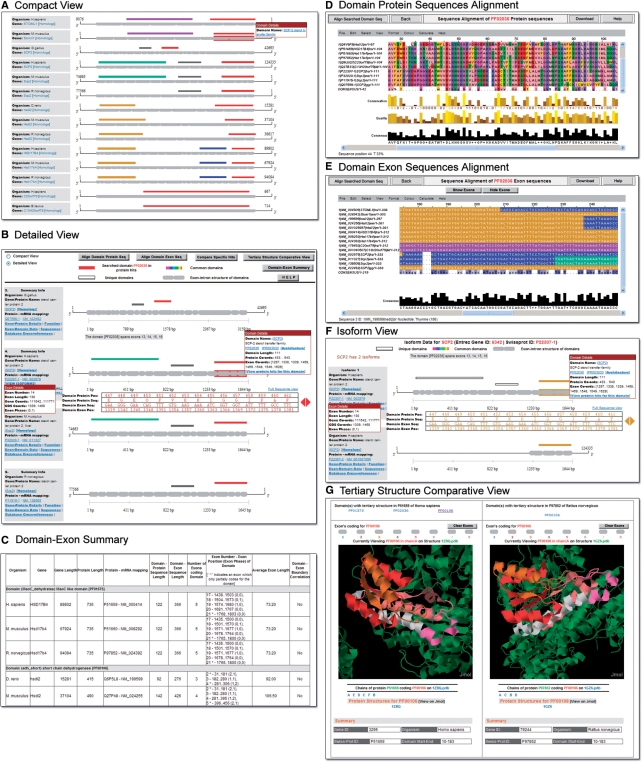
To perform a comprehensive comparison of the distribution of various domains in the protein hits, users can select the ‘Compact View’ radio button from the top menu bar. This displays a comprehensive overview of the domain architecture in all 14 protein hits that can be efficiently compared and analyzed in a single view (Figure 3A). One can see that the red rectangular boxes that represent the SCP2 domains are found in all 14 protein hits. Additional domains seen in the protein hits are: (1) Band_7 (violet box)—an integral membrane protein involved in regulation of cation conductance; (2) adh_short (brown box)—an NAD or NADP-dependent oxidoreductase domain; (3) MaoC_dehydratas (blue box)—a domain involved in the synthesis of monamine oxidase; (4) Thiolase_N (green box) and (5) Thiolase_C (gray box) domains are both involved in degradative pathways such as fatty acid beta-oxidation.
Figure 3.
Results of the sample query for the protein domain ‘SCP2’ in ExDom. (A) Compact View of the ExDom Plots of 14 protein hits. (B) Detailed View of the ExDom Plots of the four SCP2 proteins—SCP2 (chicken), SCP2 (human), Scp2 (mouse) and Scp2 (rat). (C) Domain-Exon Summary page. (D) Protein Sequence Alignment Viewer with multiple amino acid sequence alignment of the SCP2 domain in 14 protein hits. (E) Nucleotide Sequence Alignment Viewer with spliced coding nucleotide sequence alignment of the SCP2 domain in 14 protein hits. (F) Isoform display of human SCP2 protein. (G) Tertiary Structure Comparative Viewer displaying adh_short domain in HSD17B4 (human) and rat Hsd17b4 (rat).
One can see that Band_7, MaoC_dehydratas, Thiolase_N and Thiolase_C are found only within their specific homologous protein groups and are absent in nonhomologs. However, SCP2 occurs in all fourteen proteins and adh_short is found in both the HSDl2 and the HSD17B4 homologs. One can also see that the domain architecture tends to be uniform within any given homologous protein group, with all homologs having the same set of domains, arranged in the same order. The only exception to this is the SCP2 homolog group, where SCP2 (human) and Scp2 (mouse) proteins have Thiolase_N, Thiolase_C and SCP2 domains, but SCP2 (chicken) and Scp2 (rat) have Thiolase_C and SCP2 domains, but lack the Thiolase_N domain.
Users can analyze and compare the exon–intron structure of domains from different proteins in detail by switching back to the default ‘Detailed View’ option in the result page. Figure 3B shows the ‘Detailed View’ of the ExDom Plots of the four homologous SCP2 proteins. An analysis of the domain lengths (displayed in the ‘Domain Details’ pop-up) of the four homologous SCP2 proteins reveals that the lengths of SCP2 (111 amino acids), Thiolase_C (78 amino acids) and Thiolase_N (230 amino acids), domains remain unchanged across these proteins.
Additional details on the domain-exon structure of the 14 protein hits can be accessed by clicking on the ‘Domain-Exon Summary’ link in the top menu bar in the result page (Figure 3C). Using the information available in this page, one can perform several interesting comparative analyses of domain features in the protein hits. For example, one can clearly see that the length of the domain-coding exons and the number of introns intervening exons, vary among the four homologous SCP2 proteins. The human Thiolase_N spans nine exons, separated by eight introns and has an average exon length of 76.67 bases, while mouse Thiolase_N spans seven exons intervened by six introns and has an average exon length of 98.57 bases. However, the exon–intron structure of Thiolase_C remains the same in all four homologs with two introns separating the three coding exons which have an average exon length of 78 bases. The SCP2 domain has a more variable exon-intron structure. Human SCP2 domain spans four exons (average exon length: 83.25 bases), the mouse and rat homologs of this domain each have five exons (average exon length: 66.60 bases). The chicken homolog has six exons and an average exon length of 55.50 bases.
Analysis of the correlation of exon ends to domain ends is a useful feature for investigating the role of exon shuffling (31) in domain evolution. We analyzed this correlation using the ExDom Plots of the four SCP2 proteins. In all four proteins, none of the domains have ends which coincide with their exons’ ends. In all cases, there was a minimum distance of at least 15 bases between the domain ends and exon ends. We also analyzed the position at which an intron intervenes the coding sequence (intron location) using the exon phase information in the ExDom Plots. Several domain-coding exons in the SCP2 proteins have symmetrical exon phases of [(0, 0), (1, 1) or (2, 2)].
Widening our analysis to include all 14 protein hits, we found that the variations in exon–intron structure, domain length and other features of the SCP2 domain are not limited to the four SCP2 proteins, but occur in all protein hits (Supplementary Table 1).
To compare the sequence similarities in the SCP2 domain among all protein hits, one can click on the ‘Align Domain Protein Seq’ button available in the top menu bar of the result page and view the multiple sequence alignment of the amino acid sequence of this domain from all 14 hits (Figure 3D). The Jalview applet provides a detailed graphical display of the alignment with specific color-coding based on the physicochemical properties (hydrophilic, hydrophobic, aromatic, etc.) of amino acids—a feature that allows users to easily identify sequence similarities. The nucleotide sequence alignment of the spliced coding sequences of the SCP2 domains from the 14 proteins can be accessed by clicking on the ‘Align Domain Exon Seq’ button (Figure 3E). These results are graphically displayed with specific color coding based on percentage nucleotide identity. Users can also use the ‘Show Exons’ option in the alignment viewer to highlight exons in contrasting colors for easy identification of exon boundaries. These features allow users to explore the similarities and differences in the nucleotide sequences of the SCP2 domains in detail.
In cases where one to one mapping of a protein isoform to its specific mRNA isoform is available in ExDom, one can analyze variations in the domain architecture and in the exon–intron structure of the multiple protein isoform using the isoform display page. In our example, only the human SCP2 protein has isoform data available and users can access this data by clicking on the ‘View Isoforms’ link in the Summary Info box of this protein. This retrieves the comparative isoform display page with ExDom Plots of the two isoforms of human SCP2 protein (Figure 3F). While isoform 1 (P22307-1) is mapped to the mRNA isoform NM_002979 and has three domains (Thiolase_N, Thiolase_C and SCP2), isoform 2 (P22307-2) which is mapped to the mRNA isoform NM_001007099 has only the SCP2 domain. Looking at the coding sequence of these isoforms in the ExDom Plots, we can see that isoform 2 does not have the first 11 exons and has only a portion of the 12th exon. However, the remaining four exons are exactly the same in both isoform 1 and isoform 2. To analyze this further, one can access the RefSeq record for NM_001007099 through the link in the Summary Info box, and view the detailed annotations for this mRNA. This reveals that the large truncation in mRNA isoform NM_001007099 that codes for protein isoform 2 is caused by the alternative transcription initiation by a downstream promoter which results in the omission of 1212 nucleotide region that code for both Thiolase_N and Thiolase_C domains.
Out of the 14 protein hits, two (human HSD17B4 and rat Hsd17b4) have solved tertiary structures. Users can view the tertiary structure of domains within these two proteins by clicking on the ‘View Tertiary Structure’ link provided in the ‘Domain Details’ pop-up. One can see that rat Hsd17b4 has tertiary structure for only the adh_short domain. However, human HSD17B4 has tertiary structure data for all three domains. Clicking on the link to the tertiary view of the SCP2 domain of this protein, one can view the SCP2 domain sequence displayed in 3D with red color. Exon icons are given in gray above the viewer and indicate that the exons 22, 23 and 24 code for the SCP2 domain. Clicking on each exon icon, one can identify the location of individual exons in the tertiary structure of the domain by the change of color in the exonic region. Each exon is colored differently from its neighboring exons, allowing users to easily view the exon boundaries in the domain. Users can zoom in, rotate the structure or utilize any of the several additional viewing options in Jmol to analyze the exon locations in the tertiary structure of SCP2 domain. ExDom also has an advanced tertiary structure view option which allows users to compare the tertiary structure of two protein domains against each other. This feature can be accessed through the ‘Tertiary Structure Comparative View’ button in the result page menu bar. For example, one can simultaneously view the adh_short domain which has solved PDB structure for both human HSD17B4 and rat Hsd17b4 proteins (Figure 3G) and compare the exon distributions in their domain structures.
FUTURE DEVELOPMENT
Our aim in developing ExDom was to create an integrated platform for the comparative analysis of exon-domain relationships in diverse organisms for investigating the origin and evolution of gene structures of protein domains. In the future, we plan to enhance the analytical capabilities in ExDom by including the domain-exon structure mapping of several additional organisms of varying evolutionary divergence. This will enable efficient phylogenetic analysis of exon-domain relationships in eukaryotes and in-depth analysis of the evolutionary history of protein domains. We also plan to add several tools for analyzing exon-domain relationships based on protein function and protein interaction networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Genome International Corporation. Partial funding for open access charge: Genome International Cooperation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Anup Viswanathan for his valuable contributions to ExDom web interface design and Vipin T Sreedharan, Brajendra Kumar, Sharath K.S. and Vinoth Sankaran for providing valuable technical support.
REFERENCES
- 1.Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 2.Marsden RL, Lee D, Maibaum M, Yeats C, Orengo CA. Comprehensive genome analysis of 203 genomes provides structural genomics with new insights into protein family space. Nucleic Acids Res. 2006;34:1066–1080. doi: 10.1093/nar/gkj494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Roos AD. Conserved intron positions in ancient protein modules. Biol. Direct. 2007;2:7. doi: 10.1186/1745-6150-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M, Walch H, Wu S, Grigoriev A. Significant expansion of exon-bordering protein domains during animal proteome evolution. Nucleic Acids Res. 2005;33:95–105. doi: 10.1093/nar/gki152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt EE, Davies CJ. The origins of polypeptide domains. Bioessays. 2007;29:262–270. doi: 10.1002/bies.20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy SW, Gilbert W. Complex early genes. Proc. Natl Acad. Sci. USA. 2005;102:1986–1991. doi: 10.1073/pnas.0408355101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 8.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catherine B, Emmanuel C, Sébastien C, Yoann B, Sandrine D, Daniel K. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns B, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flicek P, Aken BL, Beal K, Ballester BM, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leslin CM, Abyzov A, Ilyin VA. Structural exon database, SEDB, mapping exon boundaries on multiple protein structures. Bioinformatics. 2004;20:1801–1803. doi: 10.1093/bioinformatics/bth150. [DOI] [PubMed] [Google Scholar]
- 14.Vivek G, Tan TW, Ranganathan S. XdomView: protein domain and exon position visualization. Bioinformatics. 2003;19:159–160. doi: 10.1093/bioinformatics/19.1.159. [DOI] [PubMed] [Google Scholar]
- 15.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Lee B, Lee D. DAhunter: a web-based server that identifies homologous proteins by comparing domain architecture. Nucleic Acids Res. 2008;36:W60–W64. doi: 10.1093/nar/gkn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kutchma A, Quayum N, Jensen J. GeneSpeed: protein domain organization of the transcriptome. Nucleic Acids Res. 2007;35:D674–D679. doi: 10.1093/nar/gkl990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birzele F, Küffner R, Meier F, Oefinger F, Potthast C, Zimmer R. ProSAS: a database for analyzing alternative splicing in the context of protein structures. Nucleic Acids Res. 2008;35:D63–D68. doi: 10.1093/nar/gkm793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot: the manually annotated section of the UniProt KnowledgeBase. Methods Mol. Biol. 2007;406:89–112. [Google Scholar]
- 22.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 23.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 25.Herráez A. Biomolecules in the Computer: Jmol to the rescue. Biochem. Educ. 2006;34:255–261. doi: 10.1002/bmb.2006.494034042644. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2008;32:D13–D21. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhasi A, Pandey RV, Utharasamy SP, Senapathy P. EuSplice: a unified resource for the analysis of splice signals and alternative splicing in eukaryotic genes. Bioinformatics. 2007;23:1815–1823. doi: 10.1093/bioinformatics/btm084. [DOI] [PubMed] [Google Scholar]
- 29.Wilson D, Madera M, Vogel C, Chothia C, Gough J. The SUPERFAMILY database in 2007: families and functions. Nucleic Acids Res. 2007;35:D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al. PRINTS and its automatic supplement, preprints. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]