Abstract
SchistoDB (http://schistoDB.net/) is a genomic database for the parasitic organism Schistosoma mansoni, one of the major causative agents of schistosomiasis worldwide. It currently incorporates sequences and annotation for S. mansoni in a single user-friendly database. Several genomic scale analyses are available as well as ESTs, oligonucleotides, metabolic pathways and drugs. In this article, we describe the data sets and its analyses, how to query the database and tools available in the website.
INTRODUCTION
The flatworm Schistosoma mansoni is one of the major etiological agents of human intestinal schistosomiasis mansoni. The disease affects over 200 million individuals in 74 developing countries and causes high morbidity in infected populations (1). Current strategies of disease control depend heavily on the use of the sole drug available for mass treatment, praziquantel (1). Treatment is effective in single dose and has resulted in decreased morbidity at endemic areas. However, it is highly desirable that control strategies include other countermeasures such as vaccines and new drugs. In addition, Praziquantel is not efficacious against all life cycle forms present in the human host and there is evidence that drug resistance may arise in schistosomes (2). The S. mansoni genome is ∼270 Mb contained in eight pairs of chromosomes (3). The present work focuses on the computational genome analysis of this parasitic species.
CONTENT OF THE CURRENT RELEASE
SchistoDB contains several different S. mansoni data sets and the results of different computational analyses. One highlight of the database is its integration to the metabolic pathway prediction generated using the SRI PathwayTools software (4). Pathway analysis allowed us to select putative drug target candidates. The database also contains all drugs available on KEGG drug database (5), thus enabling us to indicate enzymes known to be targeted in other organisms. Protein topology and cellular location predictions are important tools for the selection of vaccine candidates. We expect that SchistoDB will contribute to efforts towards the identification of drug and vaccine candidates in addition to a more comprehensive analysis of genes.
Data
SchistoDB provides access to the latest draft genome sequence and annotation of S. mansoni (6,7) (Puerto Rico strain) obtained from the Wellcome Trust Sanger Institute and the mitochondrial genome (8) (NMRI strain). The current database version (Release 2.0) also contains oligonucleotides (9) used in the Agilent 44 K element array widely used by the community and ESTs mapped to the genome. The database provides the results of computational analyses including open reading frames (ORFs) >50 aa and protein feature predictions such as signal peptides, transmembrane domains, hydrophobicity plots and InterPro domains (10), Gene Ontology (11) function predictions, EC Number assignment and BLAST similarities to the NCBI non-redundant protein database and Protein Data Bank database (12). We also loaded the OrthoMCL (13) group of genes from S. mansoni with orthologous genes from 86 other eukaryotic and prokaryotic genomes. In addition, drugs provided by KEGG (5) were loaded and their targets were associated to S. mansoni genes that have matching EC numbers. Users are able to visualize all data types in record pages and by queries using the query interface (see Data-mining tools section) (Table 1).
Table 1.
Data types and sources that have been integrated into SchistoDB and the number of genes that are impacted
| Data type | Data source | Gene number |
|---|---|---|
| Protein coding genes | Sanger | 13 339 |
| Orthologs | OrthoMCL | 9516 |
| GO—Gene Ontology Terms | InterProScan | 5667 |
| EC—Ezyme Commission Numbers | SchistoCyc | 712 |
| ESTs | GenBank | 9534 |
| PDB—Protein Data Bank | RSCB PDB | 2713 |
Database architecture
SchistoDB uses GUS 3.5 to systematically load data into an underlying Oracle database. The open source database schema (GUS—Genomics Unified Schema) uses controlled vocabularies and ontologies to provide wide relations between the different data types and analyses. Online access to SchistoDB occurs via the GUS WDK (Web Development Kit, www.gusdb.org/wdk) which facilitated the creation of the website. The use of GUS significantly facilitates the data loading and analysis process, enabling future and frequent release cycles. GUS and WDK have been used for the development of other databases such as PlasmoDB (14).
DATA-MINING TOOLS
SchistoDB currently provides approximately 30 different queries of the data and several tools for analyzing, retrieving or viewing the data such as BLAST, Pathway Tools and GMOD Genome Browser (15). Once the appropriate selection of data types to display has been achieved, users can integrate different search results using the ‘Query History’ page. Refining the original query iteratively until a narrow list of genes of interest is obtained, providing a manageable number of targets to validate, a time consuming and expensive process. The data can also be downloaded in flat file format for further analysis.
GBrowse genome browser (www.gmod.org) is used in SchistoDB to display gene models, EST alignments, BLAST results, etc. GBrowse enables visualization of the parasite genome and gene models, ORF identification, and facilitates downloading of data in various formats. Different tracks display each analyses or distinct data sets within the genome browser.
Schistosoma mansoni metabolic pathways are available through Pathway Tools web interface where several queries provide access to pathways, reactions, enzymes, compounds and other elements. The graphical overview allows the user to visualize the complete set of pathways and highlight specific reactions or perform organism comparison and expression analyses.
Mining for candidate drug and vaccine targets will benefit from many of the analyses available. SchistoDB integrates different datasets in a relational database that has permitted us to apply a technique known as genomic filtering (16). Genomic filtering allows the identification of gene products that might be of interest for drug targeting based on several criteria e.g. absence of alternative pathways that consume or produce a given compound, presence or similarity to the host molecule to avoid toxicity, EST evidence, cellular location, known drugs that target the same gene product in other organisms or 3D models of the protein. The presence of signal peptides and transmembrane domains will be important for the identification of vaccine candidates. EST evidence permits the verification if the putative target is expressed in the relevant life cycle stages. The identification of similar proteins with structure information permits homology modeling of S. mansoni proteins which will contribute to the design of new chemicals and the identification of exposed antigenic peptides. The user could perform complex operations with the results, such as use Boolean operators (AND, NOT, OR) to search for proteins that, for example, have a signal peptide, do not have transmembrane domains and are expressed in the schistosomula life cycle stage according to EST evidence, to identify secreted proteins.
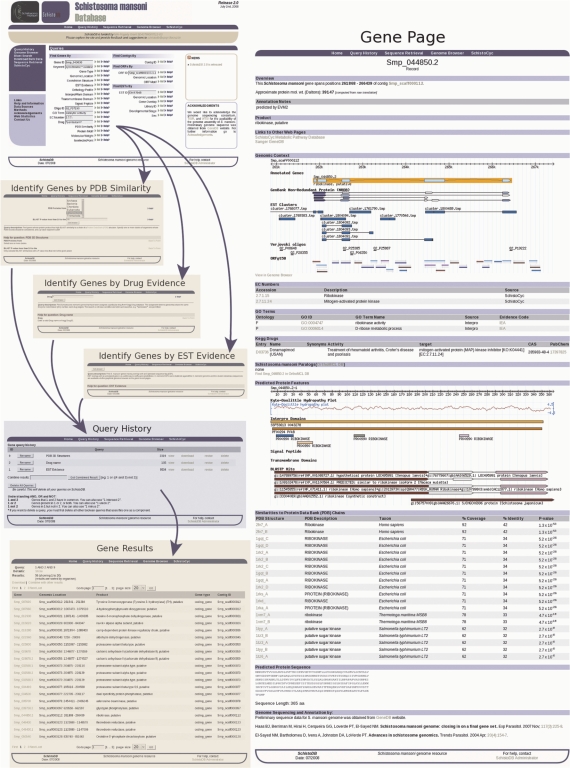
Figure 1 shows an example where the combination of the queries ‘Genes by PDB similarity’, ‘Genes by Drug Evidence’ and ‘Genes by EST Evidence’ generates a narrow list of 56 genes from a total of 13 339. That means, 56 genes have similar 3D structures in PDB database, drugs known to target the same gene product in other organims and also overlaping ESTs. Clicking on any of the gene identifiers opens a page with information on that gene. The search can be downloaded with user-selectable features.
Figure 1.
Screenshots from SchistoDB displaying the flow of a query. From the initial page users select from the various query choices for identifying genes, contigs, ORFs or ESTs. From each query a results page is displayed. The results may be downloaded, combined or the query revised. The query history page allows the user to manipulate previous results. Individual genes are displayed in the results page and it links to the gene page. In the example, the gene for ribokinase is displayed. The gene results page includes: annotation, links to SchistoCyc and GeneDB, the gene model, BLAST hits, EST clusters, microarray oligonucleotides, ORFs, EC, Gene Ontology, KEGG Drugs, Orthology, protein domains, the predicted protein, mRNA and coding sequences.
FUTURE DIRECTIONS
The current version contains only S. mansoni data, so the expansion of the database will start with the integration of data sets from other Schistosome species. We also expect to load and integrate other data types such as SNPs, microarray and SAGE. As new data are added, we will include additional queries and tools to view these data.
FUNDING
National Institutes of Health – Fogarty International Center (5D43TW007012-03 to A.Z., R.L.V.M. and A.J.D.). Funding for open access charge: National Institutes of Health – Fogarty International Center (5D43TW007012-03).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the genome sequencing consortium, TIGR and WTSI for the availability of the genome assembly and annotation of S. mansoni. Without their generous pre-publication contribution, this integrated database resource would not be possible. Special thanks to the GUS developers and to the EupathDB group, that provided essential support to accomplish this work.
REFERENCES
- 1.Chitsulo L, Engels D, Montresor A, Savioli L. The global status of schistosomiasis and its control. Acta Trop. 2000;77:41–51. doi: 10.1016/s0001-706x(00)00122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pica-Mattoccia L, Cioli D. Sex- and stage-related sensitivity of Schistosoma mansoni to in vivo and in vitro praziquantel treatment. Int. J. Parasitol. 2004;34:527–533. doi: 10.1016/j.ijpara.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Simpson AJG, Sher A, McCutchan TF. The genome of Schistosoma mansoni: isolation of DNA, its size, bases and repetitive sequences. Mol. Biochem. Parasitol. 1982;6:125–137. doi: 10.1016/0166-6851(82)90070-6. [DOI] [PubMed] [Google Scholar]
- 4.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 5.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Sayed NMA, Bartholomeu D, Ivens A, Johnston DA, LoVerde PT. Advances in schistosome genomics. Trends Parasitol. 2004;20:154–157. doi: 10.1016/j.pt.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Haas BJ, Berriman M, Hirai H, Cerqueira GG, Loverde PT, El-Sayed NM. Schistosoma mansoni genome: closing in on a final gene set. Exp. Parasitol. 2007;117:225–228. doi: 10.1016/j.exppara.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Le TH, Blair D, McManus DP. Mitochondrial DNA sequences of human schistosomes: the current status. Int. J. Parasitol. 2000;30:283–290. doi: 10.1016/s0020-7519(99)00204-0. [DOI] [PubMed] [Google Scholar]
- 9.Verjovski-Almeida S, Venancio TM, Oliveira KCP, Almeida GT, DeMarco R. Use of a 44k oligoarray to explore the transcriptome of Schistosoma mansoni adult worms. Exp. Parasitol. 2007;117:236–245. doi: 10.1016/j.exppara.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, Copley R, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahl A, Brunk B, Crabtree J, Fraunholz MJ, Gajria B, Grant GR, Ginsburg H, Gupta D, Kissinger JC, Labo P, et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarter JP. Genomic filtering: an approach to discovering novel antiparasitics. Trends Parasitol. 2004;20:462–468. doi: 10.1016/j.pt.2004.07.008. [DOI] [PubMed] [Google Scholar]