Abstract
The Carbohydrate-Active Enzyme (CAZy) database is a knowledge-based resource specialized in the enzymes that build and breakdown complex carbohydrates and glycoconjugates. As of September 2008, the database describes the present knowledge on 113 glycoside hydrolase, 91 glycosyltransferase, 19 polysaccharide lyase, 15 carbohydrate esterase and 52 carbohydrate-binding module families. These families are created based on experimentally characterized proteins and are populated by sequences from public databases with significant similarity. Protein biochemical information is continuously curated based on the available literature and structural information. Over 6400 proteins have assigned EC numbers and 700 proteins have a PDB structure. The classification (i) reflects the structural features of these enzymes better than their sole substrate specificity, (ii) helps to reveal the evolutionary relationships between these enzymes and (iii) provides a convenient framework to understand mechanistic properties. This resource has been available for over 10 years to the scientific community, contributing to information dissemination and providing a transversal nomenclature to glycobiologists. More recently, this resource has been used to improve the quality of functional predictions of a number genome projects by providing expert annotation. The CAZy resource resides at URL: http://www.cazy.org/.
INTRODUCTION
Due to the extreme variety of monosaccharide structures, to the variety intersugar linkages and to the fact that virtually all types of molecules can be glycosylated (from sugars themselves, to proteins, lipids, nucleic acids, antibiotics, etc.), the large variety of enzymes acting on these glycoconjugates, oligo- and polysaccharides probably constitute one of the most structurally diverse set of substrates on Earth. Collectively designated as Carbohydrate-Active enZymes (CAZymes), these enzymes build and breakdown complex carbohydrates and glycoconjugates for a large body of biological roles (collectively studied under the term of Glycobiology). Therefore, CAZymes have to perform their function usually with high specificity. Because carbohydrate diversity (1) exceeds by far the number of protein folds, CAZymes have evolved from a limited number of progenitors by acquiring novel specificities at substrate and product level. Such a dizzying array of substrates and enzymes makes CAZymes a particularly challenging subject for experimental characterization and for functional annotation in genomes.
Nearly 20 years ago, the first foundation for a family classification of CAZymes was seen in an effort that classified cellulases into several distinct families based on amino-acid sequence similarity (2). Soon after, the family classification system based on protein sequence and structure similarities, was extended to all known glycoside hydrolases (2–4), and subsequently extended to all CAZymes involved in the synthesis, degradation and modification of glycoconjugates. The classification of CAZymes has been made available on the web since September 1998. Because based on amino-acid sequence similarities, these classifications correlate with enzyme mechanisms and protein fold more than enzyme specificity. Consequently, these families are used to conservatively classify proteins of uncharacterized function whose only known feature is sequence similarity to an experimentally characterized enzyme, avoiding overprediction of enzyme activities.
At present, CAZy covers approximately 300 protein families in the following classes of enzyme activities:
Glycoside hydrolases (GHs), including glycosidases and transglycosidases (3–5). These enzymes constitute 113 protein families that are responsible for the hydrolysis and/or transglycosylation of glycosidic bonds. GH-coding genes are abundant and present in the vast majority of genomes corresponding to almost half—presently about 47%—of the enzymes classified in CAZy. Because of their widespread importance for biotechnological and biomedical applications, GHs constitute so far the best biochemically characterized set of enzymes present in the CAZy database.
Glycosyltransferases (GTs). These are the enzymes responsible for the biosynthesis of glycosidic bonds from phospho-activated sugar donors (6–8). They form over 90 sequence-based families and present in virtually every single organism and represent about 41% of CAZy at present.
Polysaccharide lyases (PLs) cleave the glycosidic bonds of uronic acid-containing polysaccharides by a β-elimination mechanism (6). They are presently found in 19 families in CAZy (7), corresponding to only about 1.5% of CAZy content. Many PLs have biotechnological and biomedical applications and, despite their small overall number, they are among the CAZymes with the highest proportion of biochemically characterized examples present in the database.
Carbohydrate esterases (CEs). They remove ester-based modifications present in mono-, oligo- and polysaccharides and thereby facilitate the action of GHs on complex polysaccharides. Presently described in 15 families (7), CEs represent roughly 5% of CAZy entries. As the specificity barrier between carbohydrate esterases and other esterase activities is low, it is likely that the sequence-based classification incorporates some enzymes that may act on non-carbohydrate esters.
Carbohydrate-binding modules (CBMs). These are autonomously folding and functioning protein fragments that have no enzymatic activity per se but are known to potentiate the activity of many enzyme activities described above by targeting to and promoting a prolonged interaction with the substrate. CBMs are most often associated to the other carbohydrate-active enzyme catalytic modules in the same polypeptide and can target different substrate forms depending on different structural characteristics (9,10). However, occasionally they can be present in isolated or tandem forms not coupled with an enzyme. Roughly 7% of CAZy entries contain at least one CBM module. CBMs are presently classified in 52 families in CAZy (7).
In addition to protein families that are well curated by the CAZy database, CAZymes are known to contain domains not acting on carbohydrates, including other enzymes—such as proteases, myosin motors or phosphatases, etc.—and a variety of protein–protein or protein–cell wall binding domains—cohesins, SLHs, TPR, etc.
The CAZy family classification system covers all taxonomic groups, and provides the ground for common nomenclature for CAZymes across different glycobiologists (11,12) generally specialized only in some specific groups of organisms. Day-to-day inspection of new enzyme characterizations reported in the literature regularly led and continues to lead to the definition of new enzyme families. Significantly, the CAZy families, originally created following hydrophobic cluster analysis in the 1990s from very limited number of sequences available (2–6) and later complemented by BLAST- and HMMer-based sequence similarity approaches, are globally surviving the challenge of time in spite of a hundred-fold increase in the number of sequences.
DATABASE CONTENT
The CAZy database contains information from (i) sequence annotations from publicly available sources, namely the NCBI, including taxonomical, sequence and reference information, (ii) family classification and (iii) known functional information. This data allow the exploration of an enzyme (CAZyme), all CAZymes in an organism or a CAZy protein family. The addition of new family members and the incorporation of biochemical information extracted from the literature are updated regularly, following careful inspection. Newly released three-dimensional (3D) structures and genomes are analyzed as they are released by public databases. Daily update releases from GenBank form the bulk of sequence additions to the database (8) are complemented by weekly PDB releases (13). Presently only genome released through these GenBank releases are analyzed regularly, whereas other genomes protein predictions are analyzed upon request as part of collaborative efforts (vide infra).
Another feature of CAZy is that the number of families, the family-associated information and content are continuously updated. When new families are created, old previously released genomes and sequence in public databases are reanalyzed to take the additional new family into account to ensure completeness in sequence description. Internally, curators include and maintain all referenced biochemical and other characterization data from the literature and the analysis of full sets of protein sequences present in a single genome. Because of this continuous effort of data addition, new families are frequently added and reflect the advances in experimental characterization of CAZymes. New families are exclusively created based on the availability of at least one biochemically-characterized member for which a sequence is available and the information published in peer-reviewed scientific literature. This sequence then serves as a seed for the family that is gradually extended with sequences that share statistically significant similarity.
Only functional assignments based on experimental data are included in the CAZy database by the association of EC numbers to protein sequences. Therefore inferred functional assignments are not included. Experimental data are ideally a direct enzyme analysis, but also could include indirect evidence such as gene knockout experiments with extensive characterization. Because there is a shortage of EC numbers, relative to the number of functions characterized experimentally, some incomplete EC numbers such as 3.2.1.-, 2.4.1.-, 2.4.2.- and 2.4.99.- are also included in the database. In addition, as the described functions in CAZy are only of enzymatic nature, additional and complementary binding and inhibitory functions known to be associated with several CAZy proteins will be curated and explored in the near future.
SEMI-AUTOMATIC MODULAR ASSIGNMENT
Carbohydrate-active enzymes, can exhibit a modular structure (Figure 1), where a module can be defined as a structural and functional unit (7,14). Each family in CAZy is dependent on the definition of a common segment in each full sequence that ultimately contains the catalytic or binding module. The definition of the limits within the sequence of the composing modules depends on available information derived from a combination of different approaches:
protein 3D structures,
reported deletion studies and
protein-sequence analysis and comparisons.
Different sequence comparison tools are used to define enzyme families, particularly gapped BLAST (9) and HMMER (10) using hidden markov models (HMMs) made from each family. All the sequences corresponding to the catalytic and binding of carbohydrate-active enzymes are excised from the full protein sequence and grouped in a BLAST library. Positive hits against this ‘high quality’ library, are entered into the database by trained curators following manual check on a daily basis with a small number of sequences with high identity (>85%) ungapped alignments to previously examined sequences being entered automatically.
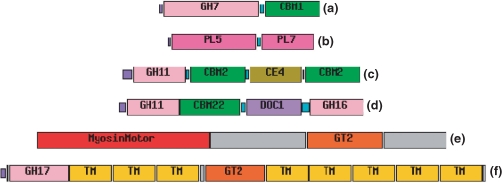
Figure 1.
Examples of modular carbohydrate-active enzymes. (a) Cellobiohydrolase I from Hypocrea jecorina (SP P00725); (b) alginate lyase from Sphingomonas sp. A1 (GB BAB03312.1); (c) xylanase from Cellulomonas fimi (GB CAA54145.1); (d) xylanase D/lichenase from Ruminococcus flavefaciens (GB CAB51934.1); (e) chitin synthase from Emericella nidulans (GB BAA21714.1); (f) cyclicβ-1-3-glucan synthase from Bradyrhizobium japonicum (GB AAC62210.1).
A new layer dealing with the analysis of whole protein sets issued from genomes has been introduced recently. Modular annotation has been in fact applied to genome data released by the NCBI, with over 750 genomes analyzed. Approximately 1–3% of the proteins encoded by a typical genome correspond to CAZymes (10,11). In addition to publicly released sequences, annotation of proteins in recently sequenced genomes prior to full release are regularly performed by the CAZy team in collaboration with scientists from all over the globe.
MANUAL FUNCTIONAL ANALYSIS
All too often, functional annotation methods employed during whole genome annotation are erroneous and lack consistent language (12,15). While sequence similarity to genes annotated by GO or best BLAST hits can be a good-starting point to assignment to pathways or possible general functions, such as serine/theonine kinase, many automatic functional assignments are unfortunately much more specific. This is particularly true in the case of CAZymes, since related families of the latter group together enzymes of widely differing specificity.
The CAZy database employs practices that aim to eliminate the problems with automatic annotation. Biochemical characterization of new proteins from the literature is used to create new protein families, to annotate their referring entries and to update family descriptions (6). These biochemical assignments are also employed to help the manual curator estimate the likely general functions and add descriptions that indicate which enzymatically characterized proteins are related to new sequences. Inclusion of reference data compiled by communities centered on model organisms is considered for the future. Bibliographic references are included in CAZy by a specific layer that includes over 16 000 different bibliographic references. These references were extracted automatically from individual accessions using ProFal (16) and about one-third was entered manually.
When functional predictions are made, they arise from manual curation by examination of closely related sequences and when biochemical information is not available, such as the case for many genome projects, very general functional tags are used to convey general functions of a family. Recently, we have begun further breaking down families into subfamilies in the hope of grouping proteins by specificity using sequence similarity. This would allow us to give more insights into possible functions. This new classification can also give insights into conserved active sites and active site specificity, when comparing biochemically characterized enzymes. Currently subfamily assignments are available publicly only for GH13 (14), GH1, GH2 and GH5 (released with this publication). This effort will be continued in the future with many more subfamilies being incorporated into the CAZy knowledge base in the future. Subfamilies identify subgroups of sequences that are more homogeneous in their functional properties. Most identified subfamilies are monospecific. If polyspecific, the functional variability is low and typically limited to two or three EC activities. There, often the known subfamily functions often share a substrate or product. Furthermore, rational enzyme engineering may be used to switch the functions for several cases (data not shown). Subfamilies also open the door for further enzymatic characterization—a few subfamilies as still no known activity—or for the identification of meaningful targets for structural characterization.
LARGE-SCALE ANALYSIS AND COLLABORATION
Internal CAZy tools, such as our semi-automatic modular assignment presently allow the analysis of a larger number of sequences than a few years ago, making it possible to perform large-scale analyses, such as the annotation of CAZyme systems in genomes and metagenomic investigations of the breakdown of complex carbohydrates. A typical genome analysis begins with the assignment of protein models to one or several CAZy families (depending on the number of CAZy modules present within the sequence). This family assignment is then followed by the prediction of general functional classes using a manual examination of alignments to closely related sequences, taking care to identify the retention of active-site residues. Once a genome is categorized by family and functional classes, gene content analysis is utilized to give insights into how newly sequenced organisms might be similar or different from closely related species. Differences in genome content, i.e. relative family size, might reflect the relative diversity or complexity of the inherent biological processes (17) and therefore, the biology of the compared species. For example, differences suggesting a more pronounced pectin metabolism in ‘dicot’ Arabidopsis versus ‘monocot’ rice have been noted (17) as well as expected differences in cell-wall metabolism between short-lived annual Arabidopsis versus long-lived poplar tree have been suggested (18). With the advent of a variety of post-genomic techniques, a new vision of the CAZymes as significant components of carbohydrate-based systems now emerges. Examples include: N- and O-glycosylation of proteins, starch metabolism, biosynthesis of the cell-wall and its subcomponents. Geisler-Lee et al. (19) have combined bioinformatics and transcriptome analysis of various poplar and Arabidopsis tissues and organs and have shown that CAZyme transcripts are particularly abundant in wood tissues.
NEW FEATURES
In addition to a website facelift, the new CAZy website comes with a host of new features. Primarily, we are now offering users the ability to search the CAZy site for information by GenBank protein accession number, family or organism rather than navigate long static pages as prior to 12/31/2008 (Figure 2). To the new site we are also including, pages to describe new releases, new genomes and other new features. In addition, tools developed in the lab are available for interactive use.
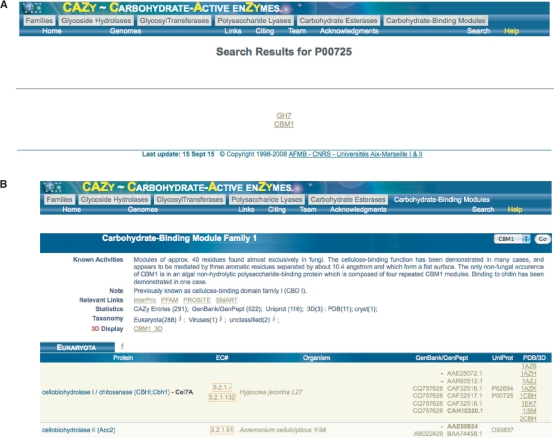
Figure 2.
(A) Once a search is performed, such as for a protein accession (P00275), the resulting page indicates the modular families that compose that protein. (B) Upon clicking the resulting links provided in A, users are directed to a page about the family and gives a listing of all annotated members.
FUTURE TRENDS
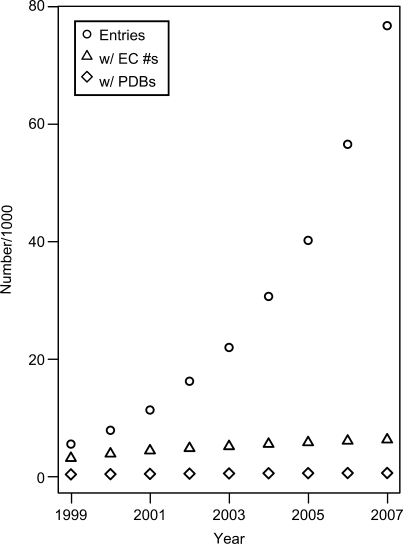
The CAZy database is a fluid database always changing and growing as additional data becomes available. In the last 2 years, the number of sequences in CAZY has nearly doubled and the number of available genomes is over 750. We believe this trend will continue in the coming years. Unfortunately, while sequencing is forever more rapid, progress in structural information and biochemical characterization is much slower. The number of biochemical data has grown only by 8% over the last 2 years (Figure 3). This means that the gap is widening between available sequences and biochemically characterized enzymes, making better methods for high-throughput biochemical characterization advantageous.
Figure 3.
The number of protein containing CAZy modules were noted in December of the years 1999–2007. Within this set (Open circle), the number of enzymatically characterized proteins (triangle) and those with solved structures (open diamond) were also counted. In December 2007, <10% of proteins in CAZy were characterized enzymatically and <1% had a solved structure. In 8 years, the number of sequences has increased 14-fold, while the number of enzymatic and structural characterization has mearly doubled. Therefore, the porportion of proteins with functional and stuctural information is decreasing rapidly unless high throughput functional efforts are made in this category of enzymes.
As started previously, we are actively pursuing the classification of subfamilies within each family. This further level of classification is important for instance to identify key residues or motifs important to define specificity. Finally, we hope to offer soon a page to submit sequences for a sequence similarity search and keyword search on our website.
AVAILABILITY ON THE WEB
The CAZy database is available at www.cazy.org. Information about selected families is available through the website and at www.cazypedia.org. Software from the group is available at www.cazy.org/tools.
FUNDING
The authors wish to thank the Departement des Sciences de la Vie of CNRS for a 2-year funding grant to B.L.C. and Novozymes for a contract supporting V.L.
Conflict of interest statement. P.M.C. is affiliated to Université de Provence (Aix-Marseille I) and B.H. and C.R. are members of CNRS.
REFERENCES
- 1.Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 10(12) structures for a reducing hexasaccharide: the Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994;4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- 2.Henrissat B, Claeyssens M, Tomme P, Lemesle L, Mornon JP. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989;81:83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- 3.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1993;293(Pt 3):781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 1996;316(Pt 2):695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yip VL, Withers SG. Breakdown of oligosaccharides by the process of elimination. Curr. Opin. Chem. Biol. 2006;10:147–155. doi: 10.1016/j.cbpa.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies G, Henrissat H, Svensson B, editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Res. 2004;32:D23–D26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy SR. Multiple alignment using hidden Markov models. Proc. Intl Conf. Intel. Syst. Molec. Biol. ISMB. 1995;3:114–120. [PubMed] [Google Scholar]
- 11.Davies GJ, Gloster TM, Henrissat B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr. Opin. Struct. Biol. 2005;15:637–645. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Doerks T, Bairoch A, Bork P. Protein annotation: detective work for function prediction. Trends Genet. 1998;14:248–250. doi: 10.1016/s0168-9525(98)01486-3. [DOI] [PubMed] [Google Scholar]
- 13.Bourne PE, Addess KJ, Bluhm WF, Chen L, Deshpande N, Feng Z, Fleri W, Green R, Merino-Ott JC, Townsend-Merino W, et al. The distribution and query systems of the RCSB Protein Data Bank. Nucleic Acids Res. 2004;32:D223–D225. doi: 10.1093/nar/gkh096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stam MR, Danchin EG, Rancurel C, Coutinho PM, Henrissat B. Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng. Des. Sel. 2006;19:555–562. doi: 10.1093/protein/gzl044. [DOI] [PubMed] [Google Scholar]
- 15.Gilks WR, Audit B, De Angelis D, Tsoka S, Ouzounis CA. Modeling the percolation of annotation errors in a database of protein sequences. Bioinformatics (Oxford, England) 2002;18:1641–1649. doi: 10.1093/bioinformatics/18.12.1641. [DOI] [PubMed] [Google Scholar]
- 16.Couto FM, Silva JM, Coutinho PM. ProFAL: PROtein Functional Annotation through Literature. In: Pimentel E, Brisaboa NR, Gomez J, editors. Proceedings of the 8th Conference on Software Engineering and Databases. Spain: Alicante; 2003. pp. 747–756. [Google Scholar]
- 17.Yokoyama R, Rose JK, Nishitani K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004;134:1088–1099. doi: 10.1104/pp.103.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science (New York, NY) 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 19.Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunneras S, et al. Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 2006;140:946–962. doi: 10.1104/pp.105.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]