Abstract
The datasets on gene expression are the valuable source of information about the functional state of an organism. Recently, we have acquired the large dataset on expression of segmentation genes in the Drosophila blastoderm. To provide efficient access to the data, we have developed the FlyEx database (http://urchin.spbcas.ru/flyex). FlyEx contains 4716 images of 14 segmentation gene expression patterns obtained from 1579 embryos and 9 500 000 quantitative data records. Reference data are available for all segmentation genes in cycles 11–13 and all temporal classes of cycle 14A. FlyEx supports operations on images of gene expression patterns. The database can be used to examine the quality of data, analyze the dynamics of formation of segmentation gene expression domains, as well as to estimate the variability of gene expression patterns. Currently, a user is able to monitor and analyze the dynamics of formation of segmentation gene expression domains over the whole period of segment determination, that amounts to 1.5 h of development. FlyEx supports the data downloads and construction of personal reference datasets, that makes it possible to more effectively use and analyze data.
INTRODUCTION
The availability of accurate quantitative datasets is of critical importance for the success of systems biology studies. The datasets on gene expression are the valuable source of information about the functional state of the organism. In order to draw meaningful inferences from gene expression data, it is important that each gene is surveyed in a spatiotemporal context, preferably in situ or in a living organism.
In the course of investigation of segment determination in Drosophila we have acquired the large dataset on the expression of segmentation genes (1,2). This dataset has cellular resolution in space and 6.5 min resolution in time (3). To provide efficient access to the data we developed the FlyEx database (4), designed as a spatiotemporal atlas on gene expression. Besides the quantitative data, it contains confocal images on gene expression patterns used to extract the data, as well as a set of reference images and data representing the most typical expression pattern for a given developmental time.
The expression localization data has been gathered by the dedicated model species databases like BDGP and FlyBase (5,6), MEPD (7), ZFIN (8), GDX and EMAGE (9,10). These resources contain rich collections of the images of gene expression patterns annotated with regard to space and developmental time. The important feature that sets FlyEx apart from other spatiotemporal atlases is the availability of highly accurate quantitative gene expression data. This characteristics attracts attention of many scientific groups, which widely use FlyEx to study the mechanism of pattern formation, infer regulatory interactions in the segmentation genetic network and develop new mathematical models (http://urchin.spbcas.ru/flyex/refs.jsp).
Here, we describe the contents of FlyEx and its user interface.
DATABASE CONTENTS
Dataset on segmentation gene expression
The expression of segmentation genes plays a crucial role in the establishment of the Drosophila body plan. The dataset on segmentation gene expression describes the dynamics of segment determination over 1.5 h of development, from cleavage cycle 10 to cleavage cycle 14A. At this time, the embryo is a syncytium and composed of about 5000 nuclei surrounded by islands of cytoplasm and located below the plasma membrane (11). The invagination of membranes, which seal nuclei into distinct cells, starts at about 15 min from the onset of cycle 14A and is completed by the end of this cycle, when gastrulation begins. Cleavage cycles from 10 to 13 are relatively short (10 min at an average), while cleavage cycle 14A lasts about 50 min (12).
The dataset includes quantitative data on expression of 14 segmentation genes controlling the determination of segments (13). These genes are maternal coordinate genes bicoid (bcd) and caudal (cad), gap genes Kruppel (Kr), knirps (kni), giant (gt), hunchback (hb) and tailless (tll), pair-rule genes even-skipped (eve), fushi-tarazu (ftz), hairy (h), runt (run), odd-skipped (odd), paired (prd) and sloppy-paired (slp). The quantitative data was obtained in 1580 wild-type (OregonR) Drosophila melanogaster embryos.
The quantitative gene expression data was acquired from images of gene expression patterns obtained by confocal scanning microscopy of fixed embryos immunostained for segmentation proteins, as described in (14,15). The flies were kept at room temperature. Approximately half of the embryos was additionally stained with an anti-histone H1-4 antibody (Chemicon International Inc., USA) to mark the nuclei. Most of embryos were scanned in Differential Interference Contrast optics to collect data on the blastoderm morphology for embryo staging. All the images were acquired in 8-bit format. The microscope at our disposal has four channels and thus allows to only scan four fluorescent labels at a time. Each embryo is scanned for the expression of three genes, and each gene was detected in a single channel. As segmentation proteins are transcription factors and the expression of segmentation genes is largely a function of position along the anterior–posterior (A–P) axis of the embryo (16), only two optical sections through the embryo nuclei separated by two micrometers were recorded. The resultant raw images were averaged, cropped and rotated to yield the embryo image, which displays the expression pattern of a single gene.
We have developed a data pipeline to extract quantitative data on segmentation gene expression from confocal images (17). This pipeline consists of image segmentation, background removal, temporal characterization of an embryo, data registration and data averaging, described below.
Image segmentation recognizes nuclei from background and converts three images of gene expression patterns obtained in one embryo into a table. Each raw of this table contains data for one nucleus. Each nucleus is characterized by a unique identification number, the x- and y-coordinates of its centroid, and the average fluorescence levels of three proteins scanned in the embryo. The x- and y-axes are chosen such that they are tangent to the anterior and ventral sides, respectively, of an embryo image and hence, the x-axis corresponds to the anteroposterior axis of the embryo and the y-axis to the dorsalventral axis. In the segmented data files, x- and y-coordinates are expressed as percent of the maximum size of the embryo in the x and y directions with 0% at the anterior pole and most ventral position, respectively. This compensates for size differences from embryo to embryo. When necessary, physical coordinates can be regenerated from stored data.
To segment images a nuclear mask is constructed, using either the image stained for histones, if any, or the pixel maximum image of all the available channel images. This mask is superimposed on each channel image to compute the coordinates of nuclei centroids and mean fluorescence intensities over each nucleus (15). The mask is a binary image, in which all the pixels located within a nucleus are white (pixel value 255), while the rest of pixels is black (pixel value 0).
Data normalization removes a background noise (18). The signal is approximated by the nearly symmetrical 2D paraboloid and removed by a linear mapping of intensity that transforms fluorescence at or below the background level to zero and transforms the maximum fluorescence (255) to itself.
Temporal characterization is performed to determine the developmental age of each embryo and reconstruct the temporal dynamics of gene expression. The calculation of nuclei number is used to stage embryos prior to cycle 14A. However, cleavage cycle 14A is about 50 min long and therefore, during this cycle, other morphological markers are to be used for staging embryos. A generally used method is the examination of the blastoderm morphology and the measurement of degree of membrane invagination in terms of the membrane/cortex ratio. The cortex is a surface layer of the blastoderm rich in microtubules, microfilaments, spectrin, myosin, actin-binding proteins, intermediate filaments and depleted of yolk. From these measurements, the embryo age can be inferred using the standard curve that gives membrane invagination as a function of developmental time (19). The method requires to obtain the images of the blastoderm morphology in each embryo, measure the membrane/cortex ratio and is time-consuming for this reason. To automate embryo staging, we have developed a method to predict the embryo age from its gene expression pattern (20). The method is based on analysis of the highly dynamic expression pattern of the eve gene, which was scanned in all embryos, and standardization of these expression patterns against a small training set of embryos with a known developmental age. As a prediction method the support vector regression is used.
Besides, thorough visual inspection of the expression patterns of the eve gene was applied to classify the embryos belonging to cycle 14A by eight temporal classes (2). The embryos were scanned without regard to age, and all the classes are approximately equally populated, hence we expect our dataset to be uniformly distributed in time and each class represents about 6.5 min of development.
The background removal and temporal characterization are applied to improve quality and exhaustively characterize the quantitative data obtained in individual embryos. However, the precise estimation of the degree of natural and experimental variation in data, as well as the construction of a spatiotemporal atlas necessitate the acquisition of reference, or typical data. To obtain such a data two additional processing steps are introduced.
Registration eliminates small individual differences in sizes among embryos of a given temporal class and brings individual embryos to the common coordinate system. Two registration methods, the spline or SpA method and the FRDWT or wavelet method, are used. These methods differ in procedure applied to extract Ground Control Points (2,21).
Data averaging is used to construct reference data for each segmentation gene and each time interval. We call such a data as integrated data.
The database
Currently, the database contains data for 1579 embryos and the addition of data to FlyEx is ongoing. We store 4716 images, each displaying the expression pattern of one of 14 segmentation genes. The nuclear mask is available for 441 embryos. All embryos contained in FlyEx belong to the blastoderm cleavage cycles from 10 to 14A (12). At present FlyEx stores about 9 500 000 quantitative data records.
The data on embryo sizes are available for 1565 embryos. The info files produced by confocal microscope are present for 1572 embryos. These files contain accessory information, which describes the acquisition system (optical transfer function, spectral properties, noise characteristics, etc.), configuration parameters, imaging mode, as well as basic information about a user, date and so on. This contextual information is indispensable for image processing and analysis.
The experimentally measured ages in terms of minutes from the onset of cycle 14A are stored for 120 embryos. The validity of this data is supported by images of the blastoderm morphology and information on the membrane/cortex ratio used as input to read the developmental time from the standard curve. In addition, the images of blastoderm morphology are available for 121 embryos, however, information on the membrane/cortex ratio has not yet been obtained from these images. The predicted ages in terms of minutes from the onset of cycle 14A are available for 656 embryos.
FlyEx stores the coefficients of normalization and registration to generate data without background and registered data from quantitative gene expression data on-the-fly. Six normalization coefficients are stored for each image, displaying the expression pattern of one gene. Two registration coefficients are available for each registration method and each embryo, belonging to temporal classes from 2 to 8 of cleavage cycle 14A.
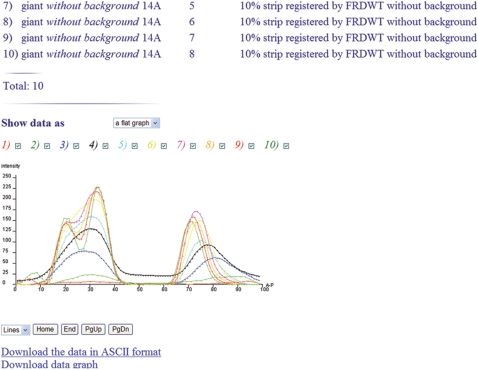
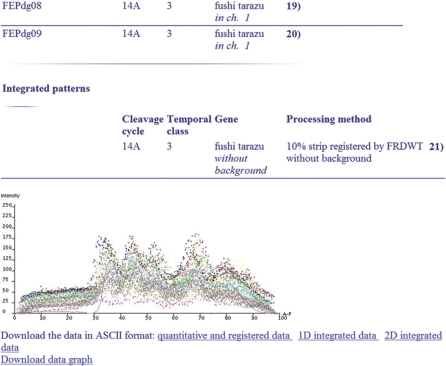
FlyEx contains reference data of two types: one-dimensional integrated data for the central 10% strip along the midline of an 2D lateral projection of an embryo and data for the projection as a whole, called the 2D integrated patterns (2,21). Both types of integrated data are constructed form individual gene expression data by the database itself and published by a database administrator. The one-dimensional integrated data are available for cycles 11–13 and all temporal classes of cycle 14A, while the integrated patterns are only constructed from normalized data registered by FRDWT and for temporal classes from 2 to 8 of cycle 14A. In total 530 sets of one-dimensional integrated data and 94 sets of integrated patterns are published in FlyEx. The total number of quantitative data records for one-dimensional integrated data is about 270 000. Besides publicly available integrated data, FlyEx can store integrated data constructed by the user and protected by a password from deleting.
DATABASE ACCESS
The information access is the most critical issue in a database design. To adapt the FlyEx interface to professional experience of various users we provide several ways to access the database.
Both sequential browsing and search forms can be used. However, in the current FlyEx version only search forms provide access to extended information about embryos, such as the images of the blastoderm morphology, embryo group name, embryo sizes, membrane/cortex ratio, measured and predicted ages from the onset of cycle 14A.
Three types of search forms are provided. The first form allows a user to retrieve data by embryo name, the second one permits to select the data by metadata information (e.g. gene name(s), cleavage cycle, time class, data types, etc.). In this case an embryo list is dynamically generated, and from this list the data for individual embryos can be easily accessed.
The third form, the most convenient one, called the Natural language queries, makes it possible to formulate and execute a query in natural language. The text of the query is entered in the QUERY text field. The QUERY EXAMPLES list contains a set of predefined standard queries for convenience. On selection of a query from the list this query is automatically displayed in the QUERY field and may be edited before execution. By pressing the SEND QUERY button, the query will be executed and a result will appear in a new browser window. In the upper part of this window, the query in natural language is displayed, in which words used to retrieve the information from the database are shown in red. Below the query result is displayed as a table. The detailed description of the Natural language interface to FlyEx is given in (22).
To support the work of a beginner, the conceptual schema of information on the expression of segmentation genes in Drosophila is constructed. This schema presents some basic concepts in the domain, as well as the relations between them as a graph. The conceptual schema can be used as a guide to clarify the structure of information and the meaning of terms, as well as to visually construct queries by selection of one or several concepts of interest as described in (4,22). When a query is visually constructed, the user selects a concept of interest by a mouse click and submits the query by pressing the SEND QUERY button. After submission the user is supplied with a list of predefined queries sorted according to their relevance. These queries can be used to refine a term meaning and make completely explicit the structure and content of the information.
The images of gene expression patterns retrieved from the database can be scaled, subjected to contrast enhancement or can be filtered by intensity. Cuts of rectangular area are also supported. These operations improve the visualization of images.
The quantitative and processed data on gene expression in individual embryos are retrieved from FlyEx in the form of dynamically generated tables, 3D graphs or reconstructed images. The graphs display the spatial variation in the level of expression of scanned genes, while the reconstructed image shows the expression patterns of these genes. Special options are available for interactive modification and adjustment of graphical displays, namely, the selection of a gene expression pattern for visualization, cuts of a strip along the A–P axis of an embryo, intensity filtering and zoom in the x direction.
The integrated data for 10% strip can be displayed as a table or flat graph. At most 14 diagrams of reference data can be displayed as one graph or a table.
The integrated patterns can be presented to a user as a table, flat graph, 3D graph or as a reconstructed image, in which not more than three patterns can be visualized on the reference embryo. Special option allows the user to interactively display each of these patterns in different colors. This facilitates the comparison of relative positions of segmentation gene expression domains in the reference embryo.
A novel aspect of data visualization is a possibility to display the retrieved data as an ASCII text file or as a graph in image format and, in that way, to save the data for further analysis and usage.
The dynamics of gene expression in the reference 10% strip during cycles 11–14A is visualized as a flat graph. Two additional methods are used to display the dynamics of segmentation gene expression in the reference embryo during cycle 14A, namely 3D graph and reconstructed image.
DATABASE USE
FlyEx was designed to accomplish two main functions. First, this database encourage a user to analyze the dynamics of formation of segmentation gene expression domains. Second, FlyEx supports data download and construction of personal reference datasets, that makes it possible to more effectively use and analyze data.
To analyze the dynamics of segmentation, the Analysis tools form provides access to a variety of operations on images of gene expression patterns and data. These operations enable the user to perform the comparative analysis of data and images and furnish a straightforward tool to answer questions about (i) the relative localization of the domains of different segmentation genes at different developmental times, (ii) the quality of gene expression data, (iii) the dynamics of formation of the expression domains of a given gene (Figure 1), as well as about (iv) the level of variability of a given gene expression (4). Two major modifications introduced since 2004, when the first paper, describing FlyEx, was published, substantially enhance the comparative analysis. First, in a list generated following a query submission, each embryo is supplied with a thumbnail image, which displays gene expression pattern or quantitative data as a diagram. Such images provide insight into inherent characteristics and quality of the data, and, hence, can be used as guides in data selection. Second, to make the comparative analysis more efficient, we increase the maximum possible number of diagrams of quantitative data that can be displayed as one graph. If the number of diagrams does not exceed eight, they are shown in different colors, and display can be interactively adjusted to select a diagram for visualization. In case of analysis of the larger number of diagrams (Figure 2), the possibility to interactively adjust the display of diagrams is not provided yet.
Figure 1.
The dynamics of formation of gt expression domains over cleavage cycles 12–14A.
Figure 2.
One-dimensional quantitative data on expression of ftz in temporal class 3 extracted from 20 individual embryos are plotted as dots. The reference data are shown with solid line. The one-dimensional quantitative data are extracted from the central 10% strip along the midline of an embryo in the A–P direction.
All public information stored in FlyEx is free to download. To do it the user has to activate either Download data: Data from individual embryos or Download data: Integrated data link from the FlyEx main menu and specify data types, mode of data sorting, output file formats (ASCII, XML, or HTML for data and JPEG, GIF, TIFF or BMP for images), as well as e-mail address for confirmation of the completion of download.
The reference gene expression data are the most typical data on a given gene expression over a given time interval. This data not only gives comprehensive information about the dynamics of gene expression, but allows the user to estimate the level of natural variation in gene expression as well. The reference data published in FlyEx are constructed by averaging the expression of a given gene over all the embryos attributed to a given time interval. In view of critical importance of reference data, FlyEx supports the construction of personal integrated datasets protected by a password from deleting (Figure 3). The construction of both personal and published integrated data is carried out by the database itself, that decreases the possibility of errors and preserves metainformation, such as construction time and embryo list, for each dataset.
Figure 3.
The construction of reference data in FlyEx.
To construct a reference dataset the user selects the Construction of integrated patterns link from the FlyEx main menu, enters a password and chooses a type of data to be constructed (integrated data for 10% strip or integrated patterns). These actions present the user with a search form, in which gene name, embryo age and data processing method should be defined. The user is able to browse the list of selected embryos and choose embryos for data averaging subject to thumbnail images each displaying the diagram of quantitative data. The menu option Show patterns allows the user to visualize the constructed data and compare it with both the data published in the database and generated by other users. The user can also delete her/his personal data by selecting the Delete patterns option.
CONCLUSIONS AND FURTHER WORK
This article describes the design of FlyEx as the quantitative atlas on segmentation gene expression at cellular resolution. Proceeding from the observation that the expression of segmentation genes is largely a function of position along the A–P axis of the embryo body, we have constructed two spatiotemporal atlases. The first atlas is built from integrated data for 10% strip. The second atlas is composed from the integrated patterns of segmentation genes. Our strategic goal is to store, along with the reference data, the data derived from individual embryos, such as images of gene expression patterns, quantitative gene expression data and other types of processed data underlying averaged information. Besides its scientific significance, this information enables other researchers to scrutinize the quality of methods employed to obtain reference information on gene expression.
FlyEx is designed to answer user's; questions about the dynamics of formation of segmentation gene expression domains and support data dissemination. Currently, a user is able to monitor and analyze the dynamics of formation of segmentation gene expression domains over the whole time of segment determination, that amounts to 1.5 h of development. The dissemination of data in FlyEx is a subject of our special attention. All the data stored in the database are free to download both in batch mode and individually. This feature accounts for the wide use of the FlyEx data in current biological research.
In future, we plan to increase the contents of FlyEx by insertion of data on segmentation gene expression at the level of RNA, as well as data on the expression of segmentation genes in mutants. The capability to analyze data will also be extended to allow the user to process and analyze personal data from individual embryos.
FUNDING
The National Institutes of Health (RR07801); the Grant Assistant Program of the Civilian Research & Development Foundation (RUB1-1578-ST-05); the Netherlands Organization for Scientific Research (NWO) and the Russian Foundation for Basic Research (047.011.2004.013); the Russian Foundation for Basic Research (08-01-00315-a, 08-04-00712-a).
Conflict of Interest statement. None declared.
ACKNOWLEDGEMENTS
We thank E. Myasnikova and S. Surkova for preparation of data and helpful comments. The valuable advices of A. Samsonov are kindly acknowledged.
REFERENCES
- 1.Kosman D, Reinitz J, Sharp DH. Automated assay of gene expression at cellular resolution. In: Altman R, Dunker K, Hunter L, Klein T, editors. Proceedings of the 1998 Pacific Symposium on Biocomputing. Singapore: World Scientific Press; 1997. pp. 6–17. http://www.smi.stanford.edu/projects/helix/psb98/kosman.pdf (last accessed date August 10, 2008) [PubMed] [Google Scholar]
- 2.Myasnikova E, Samsonova A, Kozlov K, Samsonova M, Reinitz J. Registration of the expression patterns of drosophila segmentation genes by two independent methods. Bioinformatics. 2001;17:3–12. doi: 10.1093/bioinformatics/17.1.3. [DOI] [PubMed] [Google Scholar]
- 3.Surkova S, Kosman D, Kozlov KN, Manu Myasnikova E, Samsonova A, Spirov A, Vanario-Alons CE, Samsonova M, Reinitz J. Characterization of the Drosophila segment determination morphome. Dev. Biol. 2007;313:844–862. doi: 10.1016/j.ydbio.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poustelnikova E, Pisarev A, Blagov M, Samsonova M, Reinitz J. A database for management of gene expression data in situ. Bioinformatics. 2004;20:2212–2221. doi: 10.1093/bioinformatics/bth222. [DOI] [PubMed] [Google Scholar]
- 5.Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu SQ, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. Systematic determination of patterns of gene expression during drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. research088.1–0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grumbling G, Strelets V. Flybase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrich T, Ramialison M, Wittbrodt B, Assouline B, Bourrat F, Berger A, Himmelbauer H, Sasaki T, Shimizu N, Westerfield M, et al. Mepd: a resource for medaka gene expression patterns. Bioinformatics. 2005;21:3195–3197. doi: 10.1093/bioinformatics/bti478. [DOI] [PubMed] [Google Scholar]
- 8.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen JH, Yang Y, Venkataraman S, Richardson L, Stevenson P, Burton N, Baldock RA, Davidson DR. Emage: a spatial database of gene expression patterns during mouse embryo development. Nucleic Acids Res. 2006;34:D637–D641. doi: 10.1093/nar/gkj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson J, Ringwald M. The mouse gene expression database (gxd): 2007 update. Nucleic Acids Res. 2007;35:D618–D623. doi: 10.1093/nar/gkl1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campos-Ortega JA, Hartenstein V. Germany: Springer, Heidelberg; 1985. The Embryonic Development of Drosophila melanogaster. [Google Scholar]
- 12.Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in drosophila embryogenesis. J. Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- 13.Akam M. The molecular basis for metameric pattern in the drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- 14.Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to drosophila segmentation proteins. Dev. Genes Evol. 1998;208:290–294. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- 15.Janssens H, Kosman D, Vanario-Alonso CE, Jaeger J, Samsonova M, Reinitz J. A high-throughput method for quantifying gene expression data from early drosophila embryos. Dev. Genes Evol. 2005;215:374–381. doi: 10.1007/s00427-005-0484-y. [DOI] [PubMed] [Google Scholar]
- 16.Ingham PW. The molecular genetics of embryonic pattern formation in drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- 17.Surkova S, Myasnikova E, Janssens H, Kozlov K, Samsonova AA, Reinitz J, Samsonova M. Pipeline for acquisition of quantitative data on segmentation gene expression from confocal images. Fly. 2008;2:58–66. doi: 10.4161/fly.6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myasnikova E, Samsonova M, Kosman D, Reinitz J. Removal of background signal from in situ data on the expression of segmentation genes in drosophila. Dev. Genes Evol. 2005;215:320–326. doi: 10.1007/s00427-005-0472-2. [DOI] [PubMed] [Google Scholar]
- 19.Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of drosophila melanogaster. Development. 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- 20.Myasnikova E, Samsonova A, Samsonova M, Reinitz J. Support vector regression applied to the determination of the developmental age of a drosophila embryo from its segmentation gene expression patterns. Bioinformatics. 2002;18(Suppl.):S87–S95. doi: 10.1093/bioinformatics/18.suppl_1.s87. [DOI] [PubMed] [Google Scholar]
- 21.Kozlov K, Myasnikova E, Pisarev A, Samsonova M, Reinitz J. A method for two-dimensional registration and construction of the two-dimensional atlas of gene expression patterns in situ. In Silico Biol. 2002;2:125–141. [PubMed] [Google Scholar]
- 22.Samsonova M, Pisarev A, Blagov M. Processing of natural language queries to a relational database. Bioinformatics. 2003;19:241–249. doi: 10.1093/bioinformatics/btg1033. [DOI] [PubMed] [Google Scholar]