Abstract
The ‘Human Immunodeficiency Virus Type 1 (HIV-1), Human Protein Interaction Database’, available through the National Library of Medicine at www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions, was created to catalog all interactions between HIV-1 and human proteins published in the peer-reviewed literature. The database serves the scientific community exploring the discovery of novel HIV vaccine candidates and therapeutic targets. To facilitate this discovery approach, the following information for each HIV-1 human protein interaction is provided and can be retrieved without restriction by web-based downloads and ftp protocols: Reference Sequence (RefSeq) protein accession numbers, Entrez Gene identification numbers, brief descriptions of the interactions, searchable keywords for interactions and PubMed identification numbers (PMIDs) of journal articles describing the interactions. Currently, 2589 unique HIV-1 to human protein interactions and 5135 brief descriptions of the interactions, with a total of 14 312 PMID references to the original articles reporting the interactions, are stored in this growing database. In addition, all protein–protein interactions documented in the database are integrated into Entrez Gene records and listed in the ‘HIV-1 protein interactions’ section of Entrez Gene reports. The database is also tightly linked to other databases through Entrez Gene, enabling users to search for an abundance of information related to HIV pathogenesis and replication.
INTRODUCTION
The year 2008 marks the 27th anniversary of the first case report of a new disease today known as acquired immunodeficiency syndrome (AIDS), whose etiological agent is human immunodeficiency virus type 1 (HIV-1) (1). An estimated 38.6 million people are now living with HIV or AIDS worldwide, and nearly 11 000 people are infected by HIV daily (Joint United Nations Programme on HIV/AIDS/World Health Organization). Since the documentation of the first AIDS case, numerous efforts have focused on vaccine and antiviral drug discovery and development, on identifying measures to prevent HIV transmission, on understanding HIV pathogenesis and the associated host immune responses, and on defining the interactions of HIV-1 proteins with human host cell proteins. The latter is crucial to understanding the individual steps of HIV-1 replication and pathogenesis, and provides an essential foundation for the development of safe and effective therapeutic and prevention strategies to combat AIDS. As a result of these efforts, thousands of published articles have addressed the interaction of HIV-1 proteins with human host proteins. However, each individual publication addresses only one or a few HIV protein–host protein interactions making it cumbersome to collect information on all interactions for one particular HIV or cellular protein.
The Division of Acquired Immunodeficiency Syndrome (DAIDS) of the National Institute of Allergy and Infectious Diseases (NIAID) recognized the need for a searchable platform to catalog the interactions of individual HIV proteins with host cell proteins. Therefore, the development of an HIV-1, Human Protein Interaction Database was initiated in collaboration with Southern Research Institute and the National Center for Biotechnology Information (NCBI).
DATABASE AND DATA DESCRIPTIONS
Development of the HIV-1, Human Protein Interaction Database from the peer-reviewed scientific literature available in PubMed was a 7-year effort starting in 2000. A short communication detailing the development of the database and including a visualization of the HIV-1, human protein interaction network has been published recently (2). Briefly, more than 100 000 journal abstracts and publications were identified and screened for original research describing interactions between HIV-1 and human host proteins. In addition, new literature is routinely reviewed to identify interactions described in current publications. Review of publications by scientific curator staff is organized by individual HIV-1 proteins and catalogued into an Access database by extracting the interaction information from the continuous text. As review of individual interactions is completed, data are provided to NCBI incrementally as a set of comprehensive tab-delimited text files and loaded to a MS SQL Server 2005 database. The loading process validates the RefSeq, PubMed and NCBI Entrez Gene identifiers. Validated interaction data are integrated into appropriate records in Entrez Gene and provided as custom reports and downloads per HIV-1 protein through the ‘Reports and Downloads’ tools at http://www.ncbi.nlm.nih.gov/projects/RefSeq/HIVInteractions/. The complete dataset is also available for ftp (ftp://ftp.ncbi.nih.gov/gene/GeneRIF/hiv_interactions.gz). An update to the database released on 13 November 2007, which included the interaction data set for the HIV-1 Env proteins, marked the milestone of completion of the comprehensive ‘HIV-1, Human Protein Interaction Database’ based on original research articles published since 1984. Updates to the database based on interactions described in new scientific reports will be released on a recurring basis.
The goal in developing this database was to provide scientists in the field of HIV/AIDS research a concise, yet detailed, summary of all known interactions between HIV-1 and host cell proteins and it has therefore been designed to track the following information for each protein–protein interaction identified in the literature:
NCBI Reference Sequence (RefSeq) protein accession numbers;
NCBI Entrez Gene ID numbers;
Brief description of the protein–protein interaction;
Keywords to support searching for interactions;
National Library of Medicine (NLM) PubMed identification numbers (PMIDs) for all journal articles describing the interaction.
The information compiled into the database is made publicly available through the NCBI website.
DATA DISSEMINATION AND EXPORT
The purpose of the database is to serve as a central interactive interface for viewing an ensemble of the known interactions between individual HIV-1 proteins and human proteins. The HIV-1, Human Protein Interaction Database home page (http://www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions/) enables users to simultaneously view and download a variety of reports detailing interactions for each HIV-1 protein. The database is structured by initial searches for the nine HIV proteins (e.g. Gag, Pol, Env, Tat, Rev, Nef, Vif, Vpr and Vpu), listed in the top right panel of the home page. An alphabetical report of all interacting human proteins is accessed by following the link for any of the HIV-1 proteins. The HIV-1 proteins can also be searched based on their components, for example HIV-1 Envelope can be searched either for the entire protein gp160, or separately for the gp120 surface glycoprotein or the gp41 transmembrane protein, which result from proteolytic cleavage of gp160. The HIV-to-human protein interactions are categorized by 43 interaction keywords (e.g. activates, associates with, binds, cleaves, complexes with, deglycosylates, inhibits, modulates, upregulates, etc.). A query interface allows for searching of the database to identify cellular proteins that have a specific type of interaction with a viral protein based on these keywords. The report can be customized to categories of interest by selecting a specific HIV protein and interaction keywords from the drop down menus. Reports can be viewed as a web page, or downloaded as a text file for later use. In addition, to help facilitate the retrieval of related data, links to other database resources, such as the Database of Interacting Proteins (DIP; 3), the Molecular INTeraction Database (MINT; 4), the Binding Database (5) and the Los Alamos National Laboratories (LANL) HIV Databases (6), are provided on the home page.
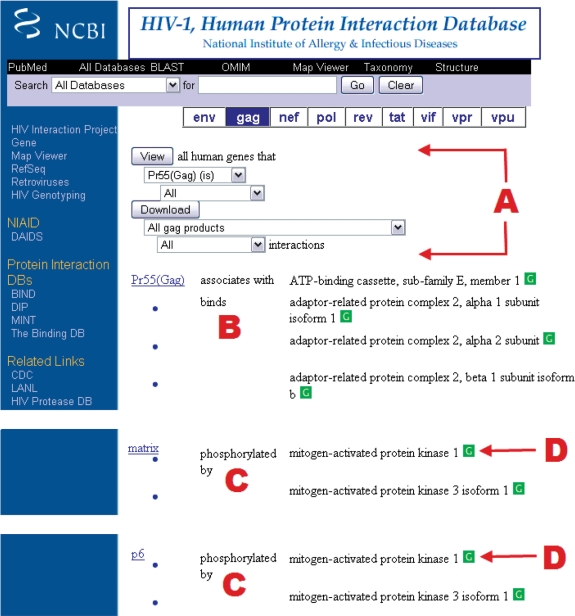
Figure 1 depicts the report and search interface page for the HIV-1 Gag polyprotein and its cleavage products. As mentioned earlier, the drop down menus (Figure 1A) allow for the selection of data related to the individual Gag cleavage products (e.g. matrix, capsid, nucleocapsid, p1 and p6) and also facilitate searching by specific keywords (e.g. associates with, binds and inhibits) that represent the relationship between the viral proteins and the interacting human proteins (Figure 1B). Reports can either be viewed online or downloaded in ASCII format and contain the HIV-1 Tax ID, HIV-1 Gene ID, HIV-1 protein accession number, HIV-1 protein name, the Interaction Keyword, the human Tax ID, human Gene ID, human protein accession number, human protein name, the PMID(s), the modification date and the interaction description.
Figure 1.
Partial report page of HIV-1 Gag interactions with human proteins. (A) All or part of the interaction data available for an HIV-1 protein can be accessed using the drop down menus. (B) The interacting relationship between HIV-1 and human proteins is reported below the menus. The figure illustrates a query section to display all interactions catalogued for the HIV-1 Pr55 (Gag) protein. The display is sorted alphabetically by the interaction term. For example, the first two interactions shown are: (i) Pr55 (Gag) protein associates with ATP-binding cassette, sub-family E, member 1; and (ii) Pr55 (Gag) protein binds to adaptor-related protein complex 2, alpha 1 subunit isoform 1. (C) Further down, the display shows the association of HIV-1 matrix and p6 with the mitogen-activated protein kinase 1 (MAPK1). (D) The arrow points to the link for the Entrez Gene reports (the green ‘G’ icon).
DATA SEARCH, ANALYSIS AND VISUALIZATION TOOLS
Currently, the database is composed of 1434 human genes encoding 1448 proteins that directly (e.g. bind, inhibit) or indirectly (e.g. upregulate, modify) interact with HIV-1 proteins. It was found that the majority of the interactions reported are indirect (68%), whereas the rest are direct (2). In addition, the database comprises 2589 unique HIV-1 to human protein interactions and 5135 brief descriptions of the interactions, with a total of 14 312 PMID references to the original articles that reported the interactions. A network of links to supporting literature and cross-references allows users to navigate concomitantly between this database and other resources at NCBI (7), such as Entrez Gene (8), RefSeq (9) and PubMed. Reports in Entrez Gene that contain HIV-1 interaction data can be retrieved with the query ‘hiv1interactions’[Properties] AND ‘Homo sapiens’[Organism]. Navigation to a target human protein interaction can be accomplished via one of two primary routes: an ‘HIV-1, Human Protein Interaction Database’ search or an Entrez Gene text query. For illustration purposes, two search scenarios for the signaling protein mitogen-activated protein kinase 1 (MAPK1), which displays a high magnitude of interactions with ten different HIV-1 proteins, are provided subsequently.
Search scenario 1 begins with an ‘HIV-1, Human Protein Interaction Database’ search. To view interactions between MAPK1 and Gag or its cleavage products, users may select ‘gag’ in the horizontal selection bar on the top right panel of the database home page (http://www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions/), which makes a direct link to the illustration as shown in Figure 1. Using the scroll down mouse menu, MAPK1 can be identified since interacting proteins reported in each interaction session (e.g. associates with and binds in Figure 1B) are alphabetic. As a searching result, MAPK1 is involved in the process of matrix and p6 phosphorylation (Figure 1C). Users may click on links to Entrez Gene (the green ‘G’ icon; Figure 1D) to view the MAPK1 full report.
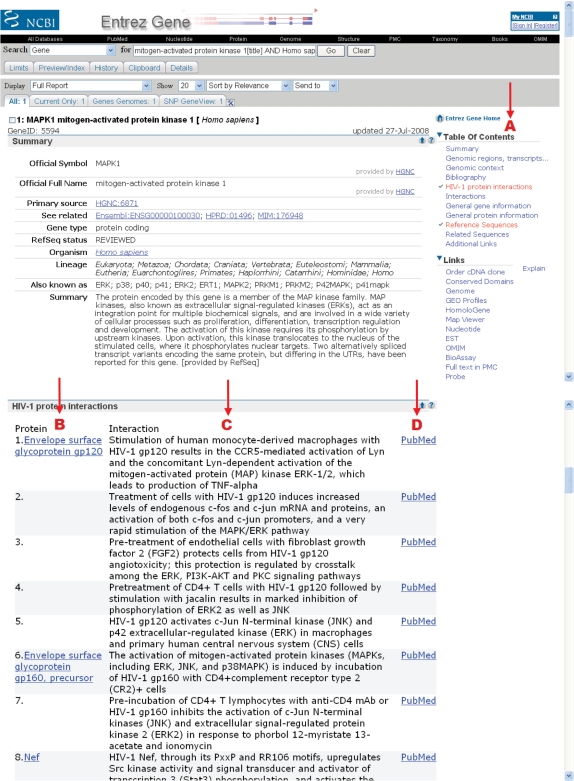
Search scenario 2 begins with a text-based search in Entrez Gene. Entrez Gene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene) is NCBI's database for gene-specific information. Users may begin with the following query: mitogen-activated protein kinase 1[title] AND Homo sapiens [organism]. The entries (e.g. MAPK1 and MAPK1IP1L) identified with the query are displayed on the Entrez Gene results page. Adding ‘AND hiv1interactions[prop]’ to the query restricts the results to only those entries that have HIV-1 interaction data, and in this example returns a single match to the MAPK1 gene report shown in Figure 2. The protein–protein interactions associated with MAPK1 are listed on the Entrez Gene report page in the ‘HIV-1 protein interactions’ section (Figure 2); a link to this section is included in the right column ‘Table of Contents’ provided on the full report display (Figure 2A). Individual HIV-1 proteins (e.g. Envelope surface glycoprotein gp120) that interact with MAPK1 are listed (Figure 2B) along with brief descriptions of the interactions (Figure 2C) and links to the supporting literature in PubMed (Figure 2D).
Figure 2.
Partial Entrez Gene report page for MAPK1. (A) The report page includes a link to the ‘HIV-1 protein interactions’ section in the Table of Contents. (B) The ‘HIV-1 protein interactions’ section shows the interaction of MAPK1 with different HIV-1 proteins. (C) Summary descriptions of the interactions are provided. (D) The interactions and descriptions are linked to the supporting literature in PubMed.
By integrating the HIV-1 interaction data into the Entrez Gene database, researchers benefit from the additional computation NCBI provides. For example, from the ‘HIV-1, Human Protein Interaction Database’ home page, there are automatic queries provided to PubMed and the NCBI sequence databases for recent records of interest. Via Entrez Gene, information can be easily obtained about genomic context, pathway membership and protein domain structure. The representative Entrez Gene search strategies summarized in the following table demonstrate the strength of the data integration and provide examples of how specific subsets of data can be retrieved:
| Query to Enter in Entrez Gene | Explanation |
|---|---|
| hiv1interactions[prop] AND human[organism] AND 5[chr] AND 1000000:12000000[Base Position] | Genes for which products interact with HIV-1 proteins, based on chromosome location. The value before [chr] gives the chromosome, and the range separated by : gives the location in base pairs on that chromosome. |
| hiv1interactions[prop] AND human[organism] AND cytoplasm*[go] | Genes for which products interact with HIV-1 proteins, and are coded by the GO Consortium with at least one term starting with ‘cytoplasm’. |
| hiv1interactions[prop] AND human[organism] AND immunoglobulin[Domain Name] | Genes for which products interact with HIV-1 proteins, and are calculated by NCBI's Conserved Domain Database group as having an immunoglobulin domain. |
| hiv1interactions[prop] AND human[organism] AND (kegg OR reactome) | Genes for which products interact with HIV-1 proteins and for which pathways data are available from the KEGG or Reactome groups. |
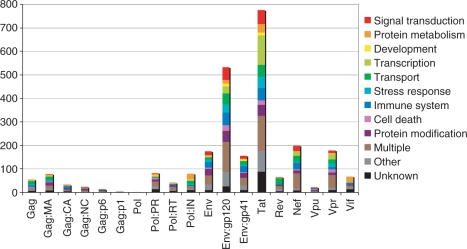
Data visualization can be accomplished in multiple ways utilizing the information stored in this database. Figure 3 shows an example of data visualization using biological process Gene Ontology (GO) terms (10, http://www.geneontology.org) and individual HIV-1 proteins. This bar chart also demonstrates that a large portion of interactions catalogued in the database are associated with the HIV envelope surface (gp120) and Tat proteins. The human cellular proteins interacting with HIV span a wide variety of functional categories, (e.g. signal transduction, protein metabolism, development, etc.) with an overrepresentation of interactions between Tat and cellular proteins involved in transcription. In addition, envelope and Tat proteins also have a high number of interactions with proteins representing multiple biological processes.
Figure 3.
Distribution of interactions based on biological process Gene Ontology (GO) terms and individual HIV-1 proteins. The x-axis shows the individual HIV-1 structural proteins Gag, Pol and Env and their cleavage products, and the regulatory and accessory HIV-1 proteins, Tat, Rev, Nef, Vpu, Vpr and Vif. The y-axis displays the number of interacting human proteins. The various colors represent the biological process categories according to GO terms.
VALUE OF THE DATABASE TO THE AIDS RESEARCH COMMUNITY
The HIV-1, Human Protein Interaction Database represents an important step towards a more detailed understanding of HIV-1 replication and pathogenesis. A recent example of the value of the database includes the work of Brass et al. (11,12), who used the database as a tool to help analyze and categorize human proteins required for HIV-1 replication. Similarly, in order to support their analysis of human–pathogen protein–protein interactions, Dyer et al. (13) were able to use a subset of the HIV-1 interaction data that has been incorporated into the Biomolecular Interaction Network Database (BIND; 14). Systematic mapping of human–pathogen protein–protein interactions has recently been studied in detail and such maps have revealed global and local networks that relate to known biological properties. Studies have indicated that both viral and bacterial proteins tend to interact with hubs (proteins with many interacting partners) and bottlenecks (proteins that are central to many pathways in the network) in human–pathogen protein–protein interaction networks (13,15–17). Development of such global and local pathway networks by utilizing the information provided in the HIV-1, Human Protein Interaction Database will provide additional insights into HIV-1 replication and disease mechanisms at a systems biology level. These networks may reconfirm and extend known pathways, as well as uncover previously unknown pathway components. In addition, these networks may serve as a starting point for a systems biology modeling of the development of effective therapeutic and prophylactic interventions.
FUTURE DEVELOPMENTS
The content, website display and bulk reporting from the ‘HIV-1, Human Protein Interaction Database’ will be continuously updated to keep the database populated with interactions newly reported in the literature. Current efforts are also focused on incorporating these data into Canada's Biomolecular Object Network Database (BOND) (http://bond.unleashedinformatics.com; successor to BIND; 14), a database cataloguing the interactions between all known cellular proteins. Feedback with respect to the ‘HIV-1, Human Protein Interaction Database’, or any data contained therein can be provided by using the ‘Write to the Help Desk’ link at the bottom of the database and Entrez Gene web pages.
FUNDING
National Institutes of Health, National Institute of Allergy and Infectious Diseases, Division of AIDS (N01-AI-05415 and N01-AI-70042 to W.F., B.E.S.-B. and R.G.P.); Intramural Research Program of the National Institutes of Health, National Library of Medicine (to K.S.K., D.R.M. and K.D.P.). Funding for open access charges: Southern Research Institute.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Roger Miller and Dr Carl Dieffenbach, NIH/NIAID/DAIDS, for discussions and intellectual input throughout this project; Dr Mikhail Rozanov, NCBI, for support in updating the HIV-1 RefSeq record; Joel Gillman, NCBI, for providing database support; and Dr David Robertson and Dr John Pinney, University of Manchester, UK, for help with Figure 3.
REFERENCES
- 1.Gayle HD. AIDS anniversaries in 2006 mark the time to deliver. Lancet. 2006;368:425–427. doi: 10.1016/S0140-6736(06)69127-7. [DOI] [PubMed] [Google Scholar]
- 2.Ptak RG, Fu W, Sanders-Beer BE, Dickerson JE, Pinney JW, Robertson DL, Rozanov MN, Katz KS, Maglott DR, Pruitt KD, Dieffenbach CW. AIDS Res. Hum. Retroviruses. Cataloguing the HIV-1 human protein interaction network. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuiken C, Korber B, Shafer RW. HIV sequence databases. AIDS Rev. 2003;5:52–61. [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Church DM, DiCuccio M, Edgar R, Federhen S, Helmberg W, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maglott DR, Ostell J, Pruitt KD, Tatusova TA. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruitt KD, Tatusova TA, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts, and proteins. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 12.Cohen J. HIV gets by with a lot of help from human host. Science. 2008;319:143–144. doi: 10.1126/science.319.5860.143. [DOI] [PubMed] [Google Scholar]
- 13.Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 16.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 17.Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, Holthaus AM, Ewence AE, Li N, Hirozane-Kishikawa T, et al. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl Acad. Sci. USA. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]