Abstract
It is 10 years since the IMGT/HLA database was released, providing the HLA community with a searchable repository of highly curated HLA sequences. The HLA complex is located within the 6p21.3 region of human chromosome 6 and contains more than 220 genes of diverse function. Many of the genes encode proteins of the immune system and are highly polymorphic. The naming of these HLA genes and alleles, and their quality control is the responsibility of the WHO Nomenclature Committee for Factors of the HLA System. Through the work of the HLA Informatics Group and in collaboration with the European Bioinformatics Institute, we are able to provide public access to this data through the website http://www.ebi.ac.uk/imgt/hla/. The first release contained 964 sequences, the most recent release 3300 sequences, with around 450 new sequences been added each year. The tools provided on the website have been updated to allow more complex alignments, which include genomic sequence data, as well as the development of tools for probe and primer design and the inclusion of data from the HLA Dictionary. Regular updates to the website ensure that new and confirmatory sequences are dispersed to the HLA community, and the wider research and clinical communities.
INTRODUCTION
The IMGT/HLA database was established to provide a locus-specific database (LSDB) for the allelic sequences of the genes in the HLA system, also known as the human major histocompatibility complex (MHC). This complex of over four megabases is located within the 6p21.3 region of the short arm of human chromosome 6 and contains in excess of 220 genes (1). The core genes of interest in the HLA system are 21 highly polymorphic HLA genes that mediate the host response to infectious disease and influence the outcome of cell and organ transplants. With a nomenclature spanning over 50 genes and 3000 alleles, there is an obvious need for a LSDB to curate these highly polymorphic variants. The sequencing of HLA alleles began in the late 1970s predominantly using protein-based techniques to determine the sequences of HLA class I allotypes. The first complete HLA class I allotype sequence, B7.2 now know as B*070201, was published in 1979 (2). The first HLA class II allele defined by DNA sequencing, DRA*0101, followed in 1982 (3). While the first HLA antigens were named during the 1960s (4), the first HLA DNA sequences or alleles were named by the WHO Nomenclature Committee for Factors of the HLA System (5) in 1987. At that time 12 class I alleles and 9 class II alleles were named. Two years later, in 1989, the Nomenclature Committee assigned official allele names to 56 novel class I alleles and 78 class II alleles (6). Advances and availability of sequencing technology meant that in 2007 the Nomenclature Committee was able to name over 400 new alleles, with the number of HLA-B alleles exceeding 1000 in 2008.
The dissemination of new allele names and sequences is of paramount importance in the clinical setting. The importance of a single recognized source for this data led to the first incarnation of the database, the HLA Sequence Databank (HLA-DB) (7), which allowed the periodic publication of HLA class I (8–11) and class II (12–17) sequence alignments in a variety of journals. By 1995, the numbers of new alleles being reported warranted the publication of monthly nomenclature updates (18), which continues to this day. That year also saw the first distribution of the HLA sequence alignments online through the web pages of the Tissue Antigen Laboratory at the Imperial Cancer Research Fund (ICRF), London, UK. This work transferred to the Anthony Nolan Research Institute (ANRI) in 1996 where it continues to this day. The latest incarnation, is the IMGT/HLA database (19–22), which began in 1997 as part of a collaboration involving the ICRF, ANRI and the European Bioinformatics Institute (EBI). The first public release of the IMGT/HLA database was made on the 16th December 1998 (23). Since then the database has been updated every 3 months, in a total of 40 releases, to include all the publicly available sequences officially named by the WHO Nomenclature Committee at the time of release. The previous releases are archived for reference.
IMGT/HLA CONTENT
The IMGT/HLA database contains entries for all HLA alleles, and alleles of some related genes, officially named by the Nomenclature Committee. These entries are derived from expertly annotated copies of the original EMBL-Bank/GenBank/DDBJ entries. This means that the IMGT/HLA database may contain multiple entries for any single allele. These component entries are submitted to the database either by the original author, or by our curators, when sequences of interest have been identified by data-mining but have yet to be submitted to the database. To distinguish each IMGT/HLA entry from the component EMBL entries, each new allele is assigned a unique accession number. The accession numbers follow the format HLA00000, where the ‘00000’ represents a numerical code.
It must be noted that all sequences within the IMGT/HLA database should also be available from the more general nucleotide sequence databases: EMBL-Bank (24,25), GenBank (26) and the DNA Database of Japan (DDBJ) (27,28). The main problem when accessing HLA sequences from these databases lies in the definition of the sequence. Despite the work of the members of the WHO Nomenclature Committee for Factors of the HLA System in monitoring HLA allele designations and maintaining the sequences, they have no control of how sequences are defined in these generalist databases. Readers should, therefore, be aware that entries in these generalist databases may be incorrectly named, contain unofficial designations or contain known, but uncorrected, sequencing errors.
Retrieving allele information and displaying polymorphisms
The main access point for the user is the World Wide Web (WWW), which allows users to employ a number of search tools and other facilities to retrieve, manipulate and analyse HLA data. The IMGT/HLA website can be split into three main areas. The first area comprises information and help pages that provide background on the database and provide in-depth help on the tools and data available and documentation of the IMGT/HLA file formats. The second area includes the tools designed specifically for the IMGT/HLA database. These core tools allow the users to perform sequence alignments, allele queries and sequence searches as well as queries more relevant to how the data are used and interpreted in a clinical setting. The third area comprises final pages that provide links to commonly used third-party applications such as the sequence-analysis tools at the EBI, including SRS, BLAST and FASTA.
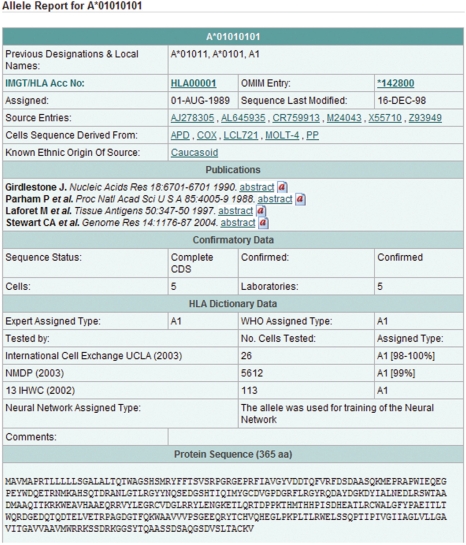
As the primary users of the database are members of the clinical HLA community involved in transplantation of tissues and organs, the most commonly accessed tools have been written to aid in their common queries. All tools are written in Perl as CGI scripts and access restricted views of the underlying Oracle relational database. The transplant and tissue typing community have two main queries; either to retrieve information on a particular allele or to view how a number of alleles differ in sequence. To answer these questions, the database provides a detailed report on any allele, as well as an interactive alignment tool to view how allelic sequences differ. The Allele Search tool provides a simple-to-use interface for retrieving allele information. The output, see Figure 1, for each allele includes the official allele designation, previously used designations and the unique IMGT/HLA accession number. Other information provided includes the date that the allele was named, current status (as some allele designations have been deleted) and information on the individual or cell line from which the sequence was derived. Links to all component EMBL-Bank/GenBank/DDBJ entries are also included. Recently, information from the HLA Dictionary (29) has also been added to some entries. The dictionary presents the serological equivalents of HLA-A, -B, -C, -DRB1, -DRB3, -DRB4, -DRB5 and -DQB1 allotypes. The data summarizes equivalents obtained by the WHO Nomenclature Committee for Factors of the HLA System, the International Cell Exchange (UCLA), the National Marrow Donor Program (NMDP), the 13th International Histocompatibility Workshop, recent publications and individual laboratories. Any citations are also included with, wherever possible, a link to the PubMed entry for that citation. The PubMed link provides an online version of the abstract as well as links to other citations by the author and to similar papers. This is also done for any other citations that appear on the website. The final section of the output details the official nucleotide and protein sequence as well as any genomic sequence for the allele that is available.
Figure 1.
Section of an HLA allele report. The report provides cross-references to a text flat-file in SRS (HLA0001), the OMIM entry for HLA-A, the source entries in EMBL-Bank (AJ278305-Z93949) and to the seminal citations in PubMed. Other information provided includes links to the source material the sequence was derived from. The full entry also contains the nucleotide sequence at both the CDS and genomic level.
HLA allele sequences can differ from each other by as little a single nucleotide substitution, within a genomic sequence of 3300 bases. Such nucleotide differences between the alleles of prospective transplant donors and recipients can make the difference between a successful transplant, graft failure and death. This means that the database must be able to quickly and easily display this information to the user. The HLA community is interested in seeing the polymorphisms in terms of the changes to the sequence rather than as a list of individual single nucleotide polymorphisms (SNPs). To this end, we have developed the alignment tool, rather than push the users into producing their own alignments for the sequences of interest or simply just reporting the polymorphic positions. These alignments allow a visual interpretation of sequence similarity so that polymorphic positions and motifs, found in multiple alleles, can easily be identified. The representation of HLA sequences in this manner can be useful when designing reagents for HLA typing, such as primers or oligonucleotide probes or comparing mismatches when looking at potential donors. The interface provided lets the user define a number of key variables for the alignments, these include the gene(s) to be aligned, the alleles of interest and the reference sequence they are aligned against, as well as the type of sequence: nucleotide coding region, nucleotide genomic and the amino-acid sequence of the protein, to be aligned. Further, specific regions like individual exons or signal peptides can be selected. The alignment tool uses standard formatting conventions for the display of sequence alignments and alignments adhere to standard conventions for displaying evolutionary events and numbering.
An example of alignments specially tailored to the HLA transplant community is in the presentation of alleles with an alternative splice site. For most alleles, the nucleotide sequence displayed as a coding sequence (CDS) represents the contiguous, correctly spliced exons. For alternatively spliced alleles, the sequence displayed will contain the spliced exons plus any alternatively spliced segment that lies within the traditional exon framework, when compared to a reference sequence. The otherwise missing sequence is also included and highlighted to emphasize the region of interest, rather than omit it, a feature important for the design of reagents that allow for typing of the alternatively spliced allele. Figure 2 illustrates how an alternatively spliced allele (A*0111N) is represented in the sequence alignments.
Figure 2.
CDS HLA Allele alignment indicating the alternatively spliced allele, A*0111N. The alternatively spliced A*0111N allele is shown aligned to the reference allele A*01010101. Identity to the A*01010101 allele is shown by hyphens (-) and the exon borders are indicated by a pipe (|). The single nucleotide mutation in the third base of codon 175, introduces a new splice site and as a result the region shaded in green is spliced out and is not part of the CDS. It is helpful to include the otherwise missing sequence and highlight it to emphasize the region of interest, rather than omit it, this feature is important for the design of reagents to allow for typing of this allele.
The previous text-only versions of the alignments are still requested and as a result, are available from the ANRI website and in a zipped file in the FTP directory. For users who prefer to use other existing software to produce their own alignments, then the FTP directory contains files in popular formats for them to download and import.
Recent developments and future applications
Recent developments to the website have seen the addition of a search tool for identifying primer and probe sequences. Many HLA typing laboratories who have designed their own reagents for HLA typing have spreadsheets detailing probe-hit patterns for different alleles. These are used when typing samples to identify known alleles based on the reaction patterns seen. Each time a new release of the database was made it was necessary to manually update these ever-expanding lists by cross-referencing the primer sequence with the sequence alignments, which with the rapidly increasing numbers of alleles was becoming a slow and laborious task. The new ‘Probe & Primer Search Tool’ allows users to enter a list of primer sequences and the tool will search the known alleles for the presence of these sequences and report any matches in a file format suitable for cutting-and-pasting into existing spreadsheets. The tool is currently limited to coding sequences but as the number of genomic sequences in the database expands it will be modified to search these regions as well.
The IMGT/HLA database is also involved in developing data format standards for HLA information exchange between the reference database, HLA typing laboratories and commercial typing-kit manufacturers. This work, which is being performed in collaboration with other immuno-informatics groups will provide both an XML output format for the IMGT/HLA database as well as XML reporting format for tissue typing laboratories. The XML output will contain similar information to that described for the flat files and allele output (30).
The rise of high-throughput genome typing has seen the expansion of genome browsers like ENSEMBL (31). These browsers have a different priority in how you view a gene, the alleles and any SNPs. The IMGT/HLA database is working with groups like ENSEMBL, EMBL-Bank and UniProt (32) to help define HLA references to suit all parties at the different levels through the development of Locus Reference Genomic Sequences (LRGS). A current project is to improve cross-referencing of the HLA data with that from other systems. The aim is to make sure that when users find an entry referring to an HLA allele in a third-party system they can also find a link back to the IMGT/HLA entry for that allele, which should be considered the primary reference for the sequence.
CONCLUSIONS
The IMGT/HLA database provides a centralized resource for everybody interested, clinically or scientifically, in the HLA system. The database and accompanying tools allow the study of all HLA alleles from a single site on the World Wide Web. It should aid in the management and continual expansion of HLA nomenclature, providing an ongoing resource for the WHO Nomenclature Committee. The earliest version of the IMGT/HLA database, December 1998, included only 964 alleles, covering 24 genes and was limited to much simpler tools and interfaces. The latest release, July 2008, contained over 3300 alleles for 34 genes, with this number set to grow as the database continues to receive and name over 450 new alleles a year. The expansion of the database content has been reflected in its use, in 1999 the website averaged just over 1500 visitors per month, in 2008 this had increased to over 7500 visitors viewing over 40 000 pages per month. The challenge for the database is to keep up with this increase in sequences, develop new tools for the visualization of the sequences whilst maintaining the high standards set in the presentation and quality of the HLA sequences and nomenclature to the research community.
FUNDING
This work was supported by Histogenetics; Abbott Laboratories Inc.; the American Society for Histocompatibility and Immunogenetics; the Anthony Nolan Trust; BAG Healthcare; Biotest; the European Federation for Immunogenetics; Innogenetics; Invitrogen; the Marrow Foundation; the National Marrow Donor Program; One Lambda Inc.; Qiagen and Tepnel Lifecodes. Initial support for the IMGT/HLA database project was from the Imperial Cancer Research Fund (now Cancer Research UK) and an EU Biotech grant (BIO4CT960037). Funding for open access charge: the Anthony Nolan Trust.
Conflict of interest statement. None declared.
APPENDIX: ACCESS AND CONTACT
IMGT/HLA Homepage: http://www.ebi.ac.uk/imgt/hla/
IMGT/HLA FTP Site: ftp://ftp.ebi.ac.uk/pub/databases/imgt/mhc/hla/
Contact: hladb@ebi.ac.uk
REFERENCES
- 1.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot C.C., Jr, Wright MW, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 2.Orr HT, Lopez de Castro JA, Lancet D, Strominger JL. Complete amino acid sequence of a papain-solubilized human histocompatibility antigen, HLA-B7. 2. Sequence determination and search for homologies. Biochemistry. 1979;18:5711–5720. doi: 10.1021/bi00592a030. [DOI] [PubMed] [Google Scholar]
- 3.Lee JS, Trowsdale J, Travers PJ, Carey J, Grosveld F, Jenkins J, Bodmer WF. Sequence of an HLA-DR alpha-chain cDNA clone and intron-exon organization of the corresponding gene. Nature. 1982;299:750–752. doi: 10.1038/299750a0. [DOI] [PubMed] [Google Scholar]
- 4.Payne R, Bodmer WF, Troup GM, Walford RL. Serologic activities and specificities of eleven human leukocyte antisera produced by planned immunization. Transplantation. 1967;5:597–605. doi: 10.1097/00007890-196707000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Bodmer WF, Albert E, Bodmer JG, Dupont B, Mach B, Mayr WR, Sasazuki T, Schreuder GMT, Svejgaard A, Terasaki PI. In: Immunobiology of HLA. Dupont B, editor. Vol. 1. New York: Springer; 1989. pp. 72–79. [Google Scholar]
- 6.Bodmer JG, Marsh SGE, Parham P, Erlich HA, Albert E, Bodmer WF, Dupont B, Mach B, Mayr WR, Sasasuki T, et al. Nomenclature for factors of the HLA system, 1989. Tissue Antigens. 1990;35:1–8. doi: 10.1111/j.1399-0039.1990.tb01749.x. [DOI] [PubMed] [Google Scholar]
- 7.Marsh SGE, Bodmer JG. HLA class II sequence databank. Hum. Immunol. 1993;36:44. [Google Scholar]
- 8.Zemmour J, Parham P. HLA class I nucleotide sequences, 1991. Tissue Antigens. 1991;37:174–180. doi: 10.1111/j.1399-0039.1991.tb01869.x. [DOI] [PubMed] [Google Scholar]
- 9.Zemmour J, Parham P. HLA class I nucleotide sequences, 1992. Tissue Antigens. 1992;40:221–228. doi: 10.1111/j.1399-0039.1992.tb02049.x. [DOI] [PubMed] [Google Scholar]
- 10.Arnett KL, Parham P. HLA class I nucleotide sequences, 1995. Tissue Antigens. 1995;46:217–257. doi: 10.1111/j.1399-0039.1995.tb03124.x. [DOI] [PubMed] [Google Scholar]
- 11.Mason PM, Parham P. HLA class I region sequences, 1998. Tissue Antigens. 1998;51:417–466. doi: 10.1111/j.1399-0039.1998.tb02983.x. [DOI] [PubMed] [Google Scholar]
- 12.Marsh SGE, Bodmer JG. HLA-DRB nucleotide sequences, 1990. Immunogenetics. 1990;31:141–144. doi: 10.1007/BF00211548. [DOI] [PubMed] [Google Scholar]
- 13.Marsh SGE, Bodmer JG. HLA class II nucleotide sequences, 1991. Tissue Antigens. 1991;37:181–189. doi: 10.1111/j.1399-0039.1991.tb01870.x. [DOI] [PubMed] [Google Scholar]
- 14.Marsh SGE, Bodmer JG. HLA class II nucleotide sequences, 1992. Tissue Antigens. 1992;40:229–243. doi: 10.1111/j.1399-0039.1992.tb02050.x. [DOI] [PubMed] [Google Scholar]
- 15.Marsh SGE, Bodmer JG. HLA class II region nucleotide sequences, 1994. Eur. J Immunogenet. 1994;21:519–551. doi: 10.1111/j.1744-313x.1994.tb00223.x. [DOI] [PubMed] [Google Scholar]
- 16.Marsh SGE, Bodmer JG. HLA class II region nucleotide sequences, 1995. Tissue Antigens. 1995;46:258–280. doi: 10.1111/j.1399-0039.1995.tb03125.x. [DOI] [PubMed] [Google Scholar]
- 17.Marsh SGE. HLA class II region sequences, 1998. Tissue Antigens. 1998;51:467–507. doi: 10.1111/j.1399-0039.1998.tb02984.x. [DOI] [PubMed] [Google Scholar]
- 18.Marsh SGE. Nomenclature for factors of the HLA system, update December 2007. Tissue Antigens. 2008;71:262–263. doi: 10.1111/j.1399-0039.2007.01004.x. [DOI] [PubMed] [Google Scholar]
- 19.Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 20.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–213. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SGE. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson J, Marsh SGE. The IMGT/HLA sequence database. Rev. Immunogenet. 2000;2:518–531. [PubMed] [Google Scholar]
- 23.Robinson J, Bodmer JG, Malik A, Marsh SGE. Development of the international immunogenetics HLA database. Hum. Immunol. 1998;59:17. [Google Scholar]
- 24.Kanz C, Aldebert P, Althorpe N, Baker W, Baldwin A, Bates K, Browne P, van den Broek A, Castro M, Cochrane G, et al. The EMBL nucleotide sequence database. Nucleic Acids Res. 2005;33:D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cochrane G, Akhtar R, Aldebert P, Althorpe N, Baldwin A, Bates K, Bhattacharyya S, Bonfield J, Bower L, Browne P, et al. Priorities for nucleotide trace, sequence and annotation data capture at the Ensembl Trace Archive and the EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2008;36:D5–D12. doi: 10.1093/nar/gkm1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugawara H, Ogasawara O, Okubo K, Gojobori T, Tateno Y. DDBJ with new system and face. Nucleic Acids Res. 2008;36:D22–D24. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tateno Y. International collaboration among DDBJ, EMBL Bank and GenBank. Tanpakushitsu Kakusan Koso. 2008;53:182–189. [PubMed] [Google Scholar]
- 29.Schreuder GM, Hurley CK, Marsh SGE, Lau M, Fernandez-Vina M, Noreen HJ, Setterholm M, Maiers M. The HLA Dictionary 2004: a summary of HLA-A, -B, -C, -DRB1/3/4/5 and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR and -DQ antigens. Int. J. Immunogenet. 2005;32:19–69. doi: 10.1111/j.1744-313X.2005.00497.x. [DOI] [PubMed] [Google Scholar]
- 30.Maiers M, Cano P, Shi J, Dinauer D, Culkin J, Goodridge D, De Clercq K, Rozemuller E, Robinson J, Liu A, et al. Bioinformatics infrastructure standards for immunogenetics: the HLA Information Exchange Data Standards (HIEDFS) working group. Tissue Antigens. 2008;71:384. [Google Scholar]
- 31.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]