Abstract
The Human Metabolome Database (HMDB, http://www.hmdb.ca) is a richly annotated resource that is designed to address the broad needs of biochemists, clinical chemists, physicians, medical geneticists, nutritionists and members of the metabolomics community. Since its first release in 2007, the HMDB has been used to facilitate the research for nearly 100 published studies in metabolomics, clinical biochemistry and systems biology. The most recent release of HMDB (version 2.0) has been significantly expanded and enhanced over the previous release (version 1.0). In particular, the number of fully annotated metabolite entries has grown from 2180 to more than 6800 (a 300% increase), while the number of metabolites with biofluid or tissue concentration data has grown by a factor of five (from 883 to 4413). Similarly, the number of purified compounds with reference to NMR, LC-MS and GC-MS spectra has more than doubled (from 380 to more than 790 compounds). In addition to this significant expansion in database size, many new database searching tools and new data content has been added or enhanced. These include better algorithms for spectral searching and matching, more powerful chemical substructure searches, faster text searching software, as well as dedicated pathway searching tools and customized, clickable metabolic maps. Changes to the user-interface have also been implemented to accommodate future expansion and to make database navigation much easier. These improvements should make the HMDB much more useful to a much wider community of users.
INTRODUCTION
Over the past 3 years, metabolomics has evolved from a little-known branch of analytical chemistry to a main-stream enterprise being practiced by hundreds of laboratories around the world. Thanks to technical advances in NMR spectroscopy, mass spectrometry and compound separation, it is now possible to identify and quantify hundreds of metabolites (i.e. the metabolome) from many different types of biological samples in relatively short order. This information can be used in a variety of applications including biomarker identification, drug discovery or development, clinical toxicology, nutritional studies and quantitative phenotyping of plants or microbes (1, 2). When combined with genomic, transcriptomic and/or proteomic studies, metabolomics can also help in the interpretation and understanding of many complex biological processes. Indeed, metabolomics is now widely recognized as being a cornerstone to all of systems biology (3).
As with any ‘omics’ discipline, metabolomics is highly dependent on the availability and quality of electronic databases. Furthermore, because metabolomics combines molecular biology with chemistry and physiology, there is a need for not just one type of database, but a wide variety of electronic resources. Currently, there are at least five types of databases used in metabolomics research. These include: (i) metabolic pathway databases; (ii) compound-specific databases; (iii) spectral databases; (iv) disease/physiology databases; and (v) comprehensive, organism-specific metabolomic databases. KEGG database (4), the ‘Cyc’ databases (5) and the Reactome database (6) are examples of some of the more popular metabolic pathway databases. These resources contain carefully illustrated, hyperlinked metabolic pathways with synoptic metabolite information for a wide range of organisms. On the other hand, compound-specific databases such as Lipid Maps (7), KEGG Glycan (4), DrugBank (8), ChEBI (9) and PubChem (10) contain essentially no pathway information. Rather, they focus on providing detailed nomenclature, structural or physicochemical data on restricted classes of compounds, such as lipids, carbohydrates, drugs, toxins or other chemicals of biological interest. These somewhat specialized databases often contain metabolites or xenobiotics not found in most metabolic pathway databases. Spectral databases for metabolomics include the BMRB (11), MMCD (12), MassBank (13), the Golm Metabolome database (14) and Metlin (15). These very valuable resources contain reference NMR, GC-MS and/or LC-MS spectra for a wide variety of small molecules along with software to identify these compounds via spectral matching. Disease and physiology databases (or encyclopedias) commonly used in metabolomics include OMIM (16), METAGENE (17) and Scriver's OMMBID (18). These contain descriptions of the causes, clinical symptoms, diagnostic indicators or genetic mutations associated with many metabolic disorders. Finally, organism-specific, comprehensive metabolomic databases—or knowledgebases—attempt to combine all of the information from most of the four kinds of databases into a single resource. Examples of these include BiGG (19), SYSTONOMAS (20) and the Human Metabolome Database or HMDB (21).
First described in 2007, the HMDB is currently the largest and most comprehensive, organism-specific metabolomics database assembled to date. It contains spectroscopic, quantitative, analytic and molecular-scale information about human metabolites, their associated enzymes or transporters, their abundance and disease-related properties. Since its initial release, the HMDB has been used in a wide range of metabolomics applications including the characterization and rationalization of biomarkers for multiple sclerosis (22), the identification of metabolites with anticancer properties (23) and the network modeling of liver cancer (24). Feedback from users has led to many excellent suggestions on how to expand and enhance HMDB's offerings. Likewise, continued advances in the field of metabolomics along with ongoing data collection and curation by the Human Metabolome Project (HMP) team has led to a substantial expansion of the HMDB's content. Here, we wish to report on these developments as well as many additions and improvements appearing in the latest version of the HMDB (release 2.0).
DATABASE ENHANCEMENTS
Details regarding the HMDB's overall design, data presentation format, data sources, curation protocols, data management system, quality assurance and metabolite selection criteria have been described previously (21). These have largely remained the same between releases 1.0 and 2.0. Here, we shall focus primarily on describing the changes and improvements made to the HMDB. More specifically, we will describe the: (i) enhancements to the HMDB's content, completeness and coverage; (ii) improvements to the HMDB's interface; (iii) enhancements to its spectral databases and searching; and (iv) improvements to the HMDB's data querying and data viewing.
Expanded database content, completeness and coverage
A detailed content comparison between the HMDB (release 1.0) versus the HMDB (release 2.0) is provided in Table 1. As seen here, the latest release of the HMDB now has detailed information on 6826 experimentally confirmed metabolites, representing an expansion of nearly 300% over the previous database. This increase is primarily due to the addition of more than 3800 lipids that have recently been experimentally detected and/or quantified in human tissues and biofluids. The addition of so many lipids reflects the fact that lipid detection and identification technologies are rapidly improving, leading to a greater number of lipid species being reported in the literature or being accessible via commercial lipidomic assays (25). While these technological improvements are impressive, it is still important to remember that upwards of 20 000 lipids could theoretically exist in the human body. Therefore it appears that only ∼20% of all possible lipids are detectable with today's technology.
Table 1.
Content comparison of HMDB 1.0 with HMDB 2.0
| Database feature or content status | HMDB (v 1.0) | HMDB (v 2.0) |
|---|---|---|
| Number of metabolites | 2180 | 6826 |
| Number of unique metabolite synonyms | 27 700 | 43 882 |
| Number of compounds with disease links | 862 | 1002 |
| Number of compounds with biofluid or tissue concentration data | 883 | 4413 |
| Number of compounds with chemical synthesis references | 220 | 1647 |
| Number of compounds with urine concentration data | 231 | 472 |
| Number of compounds with serum concentration data | 174 | 3976 |
| Number of compounds with cerebrospinal fluid concentration data | 47 | 360 |
| Number of compounds with experimental reference 13C NMR spectra | 380 | 784 |
| Number of compounds with experimental reference 1H NMR spectra | 385 | 792 |
| Number of compounds with predicted NMR spectra | 1900 | 3044 |
| Number of compounds with reference MS-MS spectra | 390 | 799 |
| Number of compounds with GC-MS reference data | 0 | 279 |
| Number of human-specific pathway maps | 26 | 58 |
| Number of compounds in Human Metabolome Library (HML) | 607 | 920 |
| Number of HMDB data fields | 91 | 102 |
| Pathway search/browse | No | Yes |
| Disease search/browse | No | Yes |
| Chemical class search/browse | No | Yes |
| Chemical substructure search | No | Yes |
| Biofluid search/sort tools | No | Yes |
| Advanced (multipeak or multicompound) NMR search | No | Yes |
| Advanced (multipeak or multicompound) MS-MS search | No | Yes |
| Advanced (retention index or MS peak) GC-MS search | No | Yes |
Other classes of compounds that have seen substantial increases in numbers over the past 2 years include glucuronides, carnitines, bile acids and coenzyme A derivatives. In many cases, these additions do not represent the discovery of new compounds, but simply reflect improvements in the HMDB curation team's ability to identify (with the assistance of text mining tools) and archive metabolites previously reported in the literature. Currently ∼60% of the metabolites in the HMDB have been identified or confirmed by the HMDB's team of analytical chemists using NMR, LC-MS or GC-MS methods applied to a variety of human biofluids. Likewise, ∼45% (2900/6475) of the metabolites in the HMDB have been identified and archived through literature surveys or electronic data mining. It is also worth noting that many of the most commonly used metabolite databases (KEGG, HumanCyc, BiGG or Lipid Maps) only list about one-fifth the number of metabolites found in the HMDB. We believe this statistic underscores the uniqueness and comprehensiveness of the HMDB in describing human metabolism.
In addition to substantially increasing the number of metabolite entries, we have also increased the completeness of the HMDB's annotations for hundreds of metabolites by adding many more detailed compound descriptions, including more synonyms (60% increase), doubling the number of compounds with NMR and MS spectra, increasing the number of compounds with biofluid concentration data by a factor of five and increasing the number of compounds with synthesis records by a factor of eight. Beyond these changes, a substantial effort was also made to manually classify all compounds in the HMDB into chemicals ‘kingdoms’, ‘classes’ and ‘families’. The chemical class information is particularly useful for metabolite comparison and classification. Table 2 provides a list of the 52 metabolite classes used by the HMDB and the number of compounds found in each class. In choosing these chemical class names, the HMDB curation team assessed a number of previously published chemical classification schemes (used in plant and microbial metabolomics) and attempted to select those class names that were most commonly used or most chemically informative. Of course, no classification scheme is perfect and the current ontology simply represents a compromise of many competing needs, ideas and preferences. Nevertheless, we believe this kind of chemical ontology should help to provide a common language for large-scale mammalian metabolome comparisons.
Table 2.
Chemical classes in the HMDB (v 2.0)
| Compound class | Number | Compound class | Number |
|---|---|---|---|
| Minerals and elements | 58 | Polyphenols | 54 |
| Fatty acids | 126 | Dicarboxylic acids | 70 |
| Alcohols and polyols | 103 | Alkanes and alkenes | 26 |
| Keto acids | 31 | Glycolipids | 138 |
| Carbohydrates | 195 | Hydroxy acids | 97 |
| Purines and purine derivatives | 32 | Prostanoids | 54 |
| Catecholamines and derivatives | 34 | Peptides | 69 |
| Acyl phosphates | 37 | Nucleotides | 106 |
| Phospholipids | 2630 | Cyclic amines | 55 |
| Amino ketones | 45 | Nucleosides | 52 |
| Glycerolipids | 1163 | Aromatic acids | 71 |
| Retinoids | 26 | Amino alcohols | 27 |
| Pterins | 47 | Steroids and steroid derivatives | 323 |
| Carnitines | 48 | Leukotrienes | 79 |
| Amino acids | 234 | Indoles and indole derivatives | 32 |
| Porphyrins | 54 | Sugar phosphates | 66 |
| Coenzyme A derivatives | 117 | Glucuronides | 74 |
| Ketones | 23 | Sugar phosphates | 66 |
| Inorganic ions and gases | 34 | Miscellaneous | 118 |
| Sphingolipids | 19 | Bile acids | 84 |
| Alcohol phosphates | 22 | Amino acid phosphates | 10 |
| Aldehydes | 21 | Quinones and derivatives | 16 |
| Pyrimidines and pyrimidine derivatives | 13 | Pyridoxals and derivatives | 9 |
| Tricarboxylic acids | 9 | Acyl glycines | 37 |
| Cobalamin derivatives | 9 | Lipoamides and derivatives | 10 |
| Biotin and derivatives | 6 | Polyamines | 5 |
Thanks to the feedback provided by HMDB's user community, a number of new data fields have been added to each MetaboCard in order to facilitate certain types of queries or comparisons. These include chemical source information (endogenous versus exogenous), physiological charge, experimental and predicted logP, HMDB pathway images, general metabolite references and macromolecular interacting partners (such as transporters or proteins that use the metabolites as co-factors). New data fields have also been added for the BiGG database, Wikipedia and METLIN (for metabolites) while extra data fields for GeneCard IDs, GeneAtlas IDs and HGNC IDs have been added for each of the corresponding enzymes. In addition to these changes, new data fields for NMR assignment files (both 1H and 13C) in the BMRB NMR* exchange format (11) have been inserted as well as data fields for experimental 1H-13C HSQC spectra, simplified TOCSY spectra and BMRB TOCSY spectra. Over and above these changes, the normal and abnormal biofluid concentration data fields have also been consolidated (from 10 to 2) and reformatted for improved viewing.
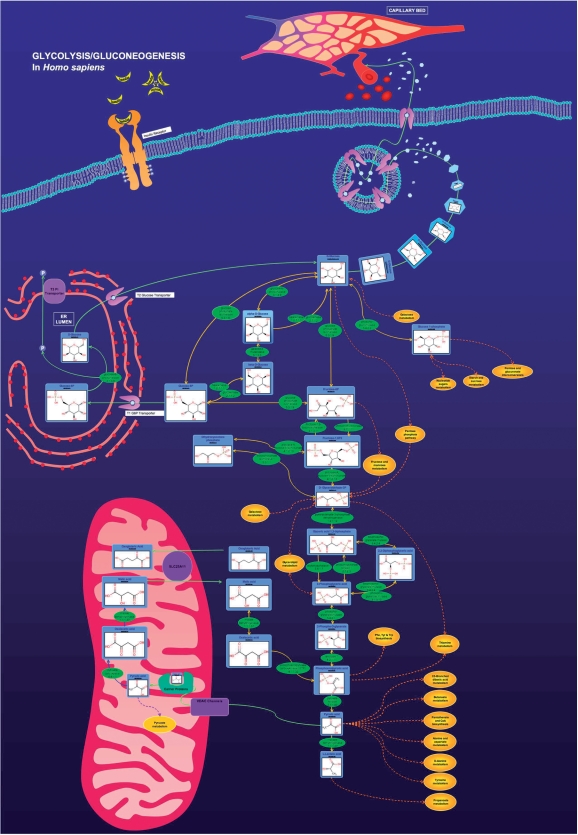
We believe that one of the more important improvements to the HMDB concerns the addition of nearly 60 hand-drawn, zoomable and fully hyperlinked human metabolic pathway maps (Fig. 1). While the HMDB still maintains full linkage to nearly 100 KEGG pathways, the addition of these ‘custom’ maps to the HMDB arose from requests by users who were dissatisfied with being unable to visualize the chemical structures on metabolic maps or unable to get detailed information about human metabolic enzymes. Unlike, most online metabolic maps, these HMDB pathway maps are quite specific to human metabolism and explicitly show the subcellular compartments where specific reactions are known to take place. All chemical structures in these pathway maps are hyperlinked to HMDB MetaboCards and all enzymes are hyperlinked to UniProt data cards for human enzymes. They are also searchable (via PathSearch) in a manner that is more conducive to typical metabolomics queries (see below).
Figure 1.
A screenshot of the HMDB pathway image for glycolysis/gluconeogenesis as found in humans. All metabolite structures and enzyme IDs are hyperlinked to the HMDB and UniProt, respectively.
In addition to these changes, a substantial effort has also been put into identifying and correcting a number of structural, image format, naming, annotation and spectral assignment errors in the HMDB. While a number of internal checking and editing procedures are used by the HMDB curation team [see (21) for details], we are particularly grateful to external users who identified more subtle errors or offered suggestions to improve the data quality. Interestingly, a number of errors were found to be ‘propagation’ errors arising from the transfer of erroneous data from one well-regarded database to another. In addition to these error corrections, a substantial update to the HMDB's metabolite–enzyme associations has also been completed. Indeed, all enzyme–metabolite associations that were automatically ‘text-mined’ have now been manually verified by multiple HMDB annotators. While it is difficult to formally quantify these changes or corrections, we can say that the quality of the data in release 2.0 is generally much better than the previous release.
User interface improvements
Both the front-end and selected components of the back-end of the HMDB have been substantially redesigned to accelerate searches, improve data visualization and allow greater flexibility in the number of query tools and links that can be provided by the database. The HMDB's navigation bar (located at the top of each page) has been simplified to just six pull-down menu tabs (‘Home’, ‘Browse’, ‘Search’, ‘About’, ‘Download’ and ‘Contact Us’). The ‘Browse’ tab allows users to select from six browsing options (HMDB Browse, Biofluid Browse, HML Browse, ClassBrowse, PathBrowse and Disease Browse) of which the last four are new. The HML Browse allows users to browse or search through the HML. The HML is a library of ∼1000 reference metabolites stored in −80°C freezers. Small amounts of these compounds are freely available to designated HMDB collaborators. They are also available on a cost-recovery basis to other laboratories on an as-needed basis. The second of the new browsing tools, ClassBrowse, allows users to view compounds according to their chemical class designation. Each displayed compound name is hyperlinked to the HMDB MetaboCard. Users may search for compounds (via a text box) or select to view certain compound classes using a pull-down menu located that the top of the ClassBrowse page. The third browsing tool, PathBrowse, allows users to browse through the custom-drawn HMDB pathway images. Each pathway is named and each image is zoomable and extensively hyperlinked. Users may also search PathBrowse using lists of compounds (obtained from a metabolomic experiment) and view hyperlinked tables that display all of the pathways that are potentially affected. The last browsing tool, Disease Browse, allows users to scroll and search through tables of diseases, which are co-listed with hyperlinked metabolite and enzyme/protein names. As with PathBrowse users may submit multiple lists of compounds and then view hyperlinked tables of diseases or conditions that may be associated with the observed metabolic changes.
The HMDB's ‘Search’ menu offers eight different querying tools including ChemQuery, TextQuery, SequenceSearch, DataExtractor, MS search, MS-MS search, GC-MS search and NMR search. While only the GC-MS and MS search features are new, significant improvements in terms of speed, accuracy and robustness have been made to many of the other query tools. These enhancements are described in detail in later sections of this article. Adjacent to the ‘Search’ menu, the ‘About’ pull-down menu contains information on the HMDB database, release notes, recent news or updates, database statistics, data source tables, data field explanations and links to other useful metabolomic databases. Finally, the ‘Download’ menu contains downloadable data for all HMDB compounds (in SDF format), all NMR spectra (in BMRB* format and as PNG images), all GC-MS spectra (in NIST format), all MS-MS spectra (as PNG images), all enzyme/protein sequences as well as complete flat file data sets of current and past HMDB releases.
Over and above these enhancements to the menu structure and database navigation scheme, improvements have also been made to the formatting and display of all of HMDB's MetaboCards. For instance, certain data fields have been reordered to bring logically similar data sets (such as structure files or pathway diagrams) closer together in each MetaboCard. Other data fields (such as the NMR and MS spectral data fields) have had extra information added to the data cell, such as collection conditions and FID data. In other cases, data fields have reformatted to provide more information in a more structured manner. For example, the information in normal and abnormal biofluid concentrations, data cell has been reformatted to display much more data in a more readable tabular format. A similar change has been made to the associated disorders field. Likewise all PubMed IDs and abbreviated chemical synthesis references have been replaced with full reference information (authors, title, journal, volume, page, year). In a similar manner, the SNP (single nucleotide polymorphism) data field (found in HMDB's Enzyme section) has also been modified so that SNPs are displayed in hyperlinked summary tables containing information on their type (synonymous, nonsynonymous), location, validation status and population distributions. This change to the SNP data field has also made the browsing of MetaboCards much faster and less taxing on our servers.
Enhancements to spectral databases and spectral searching
In genomics and proteomics, most genes and proteins are identified via sequence comparisons against libraries on known sequences. In metabolomics, most compounds are identified via spectral comparisons against libraries of known compound spectra. Consequently, there is a critical need by many metabolomics researchers for comprehensive, publicly accessible libraries of reference compound spectra. There is also an equally strong need for robust search algorithms to perform spectral matching and compound identification. Over the past 18 months, the HMDB's analytical chemistry team has been actively collecting, assigning and verifying reference NMR, GC-MS and MS-MS spectra for all compounds in the HML. As seen in Table 1, the number of compounds with experimentally acquired NMR and MS-MS spectra has more than doubled. Likewise, a completely new set of 279 experimentally acquired GC-MS spectra (with retention index data) has just been added. In another 6 months, the number of compounds with GC-MS spectra should nearly equal the number of compounds with NMR or MS-MS data.
In keeping with our open access mandate, all experimentally acquired NMR spectra in the HMDB are available in BMRB* format and as fully labeled PNG images. Likewise, all GC-MS spectra are available in NIST-AMDIS format, while all MS-MS spectra available as PNG images. What is particularly unique about the HMDB's NMR data is that all compounds are fully assigned (both 1H and 13C shifts) under standardized aqueous conditions. While reference spectral collection and deposition is continuing, it is expected that data for fewer than 100 compounds will be added over the coming year. This slowdown simply reflects the fact that pure standards of many metabolites are neither commercially available nor are they easily synthesized.
Thanks to suggestions from the user community, a number of enhancements to the MS-MS, MS and NMR search routines have been made. The HMDB's MS-MS search now allows users to search for compounds (with experimental MS-MS data) by name, synonym, molecular formula or parent ion mass. The complete, scrollable list of compounds with experimental MS-MS data is also viewable. The MS-MS peak search has also been improved by the addition of more search options and more detailed descriptions on how to use the query engine. The results from the MS-MS peak search query now return data on the spectral fit quality along with hyperlinks to the MetaboCards of the matching compounds. Also included is the corresponding MS-MS spectrum, the data collection protocol and the MS-MS peak list.
For the MS search, users can search for compounds by parent ion mass in three different modes (positive ion, negative ion and neutral) against four different databases including the HMDB, DrugBank, FooDB (a food additive and phytochemical database containing ∼2000 compounds) or all four databases together. Adducts (Na+, K+, NH4+, etc.) for all entries in each of the databases have been precalculated allowing users to identify potential adduct matches to the observed parent ion masses.
As with the MS-MS search, the NMR search supports queries for compounds (with experimental or predicted shifts) by name, synonym, molecular formula or molecular weight. Users may search against different types of NMR data including 1D 1H, 1D 13C, 2D TOCSY and 2D 1H-13C HSQC spectra. The input peak list may be for a pure compound or for a mixture of several dozen compounds (from a biofluid or tissue extract). Users may also select what kind of biofluid/extract they are analyzing (urine, CSF, plasma, cell extracts or undefined). The results from an NMR peak list query will return the name of the compound(s), the spectral matching score along with hyperlinks to each matching compound's spectral peak list and the category of spectrum matched (predicted or experimental). The algorithm used in the HMDB's NMR search combines peak matching with peak uniqueness and pairwise peak distance measures along with specific knowledge of specific biofluid compositions to identify compounds. The performance of the algorithm, when assessed with real and synthetic biofluid mixtures of up to 30 compounds (corresponding to several hundred peaks), was found to achieve >80% identification success using either TOCSY or 1H-13C HSQC data. This was 2-3X better than other NMR spectral matching algorithms. Additional details about the algorithm, the comparative performance and its limitations are given elsewhere (26).
Improvements in data querying and viewing
As mentioned earlier, improvements to the performance and speed for a number of HMDB query functions have been implemented with release 2.0. For both the general text search and the more specialized TextQuery functions, the HMDB now uses KinoSearch (27). This particular text query system is approximately five times faster than the previous system and supports text match rankings, misspellings (offering suggestions for incorrectly spelled words) and highlights text where the word is found. Consequently, general text queries now rapidly produce a table of hits that provides the HMDB ID, a MetaboCard link, the common name, the formula, the molecular weight and the text or sentence(s) where the query word is most frequently found. HMDB's TextQuery function not only uses the same KinoSearch engine, but also supports more sophisticated text querying functions (Boolean logic, multiword matching and parenthetical groupings) as well as data-field-specific queries (such as finding the query word only in the ‘Compound Source’ field). Additional details and examples are provided on the HMDB's TextQuery page. The Data Extractor has also been completely rewritten and the algorithm has been substantially sped up. This tool supports much more specialized queries and now provides users with the ability to output their data in HTML, HTML-printable and comma separated value (Excel compatible) formats.
The ChemQuery function has also been revamped, replacing the old, multistep conversion and query process with ChemAxon's single-step structure query tool. With this new and improved structure query system, users may draw a structure (using a chemical drawing applet) or paste a SMILES string directly into the structure drawing palette to query the HMDB structure database. Users can also select the type of search (exact or Tanimoto score) to be performed. We have found that the new structure querying tool is able to provide much more consistent structure matches than our ‘home-built’ structure matching tool used in release 1.0. The same ChemAxon structure querying applet is also used with the ‘Find Similar Structures’ button located at the top of every MetaboCard. Overall, we believe the improvements to many of the text and structure querying tools in this release of the HMDB should make data searching and data extraction much easier, more robust and significantly faster.
CONCLUSION
The HMDB is designed to be a comprehensive, web-accessible metabolomics database that brings together quantitative chemical, physical, clinical and biological data about all experimentally ‘proven’ or experimentally detected human metabolites. Over the past 2 years, a significant expansion to the content as well as a significant enhancement to the database's capabilities has taken place. Many of these content additions and content corrections are the result of continued experimental and literature mining efforts by the HMDB curatorial and analytical chemistry staff. Likewise, many of the graphical interface and query function improvements, which arose primarily from external user suggestions, are the result of significant programing efforts by the HMDB software development team. Overall, we believe these improvements to the query functions and enhancements to the database content should make the HMDB much more useful to a much wider collection of metabolomics researchers.
Unlike the human genome, the human metabolome is not a finite or easily defined entity (2). Certainly, as technology improves and detection limits decrease, it is likely that many more metabolites will be identified (by ourselves and others) or reported in the literature. What this particular release of the HMDB provides is a relatively complete picture of what is detectable in the human metabolome as of 1 January 2009. No doubt the size of the human metabolome will continue to grow (although, not as quickly as the past 2 years), as will the collection of reference compound spectra and our knowledge of metabolite concentrations, pathways, enzyme and disease associations. In an effort to keep the HMDB as current as possible, we intend to release database updates every 6 months (1 July and 1 January) for at least the next 2 years.
FUNDING
Alberta Advanced Education and Technology (AAET); Canadian Institutes of Health Research (CIHR); Alberta Ingenuity Centre for Machine Learning (AICML); Alberta Ingenuity Fund (AIF); Genome Alberta, a division of Genome Canada.
REFERENCES
- 1.German JB, Hammock BD, Watkins SM. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics. 2005;1:3–9. doi: 10.1007/s11306-005-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wishart DS. Current progress in computational metabolomics. Brief. Bioinform. 2007;8:279–293. doi: 10.1093/bib/bbm030. [DOI] [PubMed] [Google Scholar]
- 3.Quackenbush J. Extracting biology from high-dimensional biological data. J. Exp. Biol. 2007;210:1507–1517. doi: 10.1242/jeb.004432. [DOI] [PubMed] [Google Scholar]
- 4.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krummenacker M, Paley S, Mueller L, Yan T, Karp PD. Querying and computing with BioCyc databases. Bioinformatics. 2005;21:3454–3455. doi: 10.1093/bioinformatics/bti546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcántara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 13.Taguchi R, Nishijima M, Shimizu T. Basic analytical systems for lipidomics by mass spectrometry in Japan. Methods Enzymol. 2007;432:185–211. doi: 10.1016/S0076-6879(07)32008-9. [DOI] [PubMed] [Google Scholar]
- 14.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 15.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 16.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frauendienst-Egger G, Trefz FK. Metagene – knowledge base for inborn errors of metabolism (3.0) Indian J. Pharmacol. 1999;31:321. [Google Scholar]
- 18.The Online Metabolic and Molecular Basis of Inherited Disease (OMMBID) [last accessed date October 20, 2008]. http://genetics.accessmedicine.com/
- 19.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson B.Ø. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl Acad. Sci. USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi C, Münch R, Leupold S, Klein J, Siegel I, Thielen B, Benkert B, Kucklick M, Schobert M, Barthelmes J, et al. SYSTOMONAS–an integrated database for systems biology analysis of Pseudomonas. Nucleic Acids Res. 2007;35:D533–D537. doi: 10.1093/nar/gkl823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quintana FJ, Farez MF, Weiner HL. Systems biology approaches for the study of multiple sclerosis. J. Cell Mol. Med. 2008;12:1087–1093. doi: 10.1111/j.1582-4934.2008.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivekanandan P, Singh OV. High-dimensional biology to comprehend hepatocellular carcinoma. Expert Rev. Proteomics. 2008;5:45–60. doi: 10.1586/14789450.5.1.45. [DOI] [PubMed] [Google Scholar]
- 24.Arakaki AK, Mezencev R, Bowen NJ, Huang Y, McDonald JF, Skolnick J. Identification of metabolites with anticancer properties by computational metabolomics. Mol. Cancer. 2008;7:57. doi: 10.1186/1476-4598-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.German JB, Gillies LA, Smilowitz JT, Zivkovic AM, Watkins SM. Lipidomics and lipid profiling in metabolomics. Curr. Opin. Lipidol. 2007;18:66–71. doi: 10.1097/MOL.0b013e328012d911. [DOI] [PubMed] [Google Scholar]
- 26.Xia J, Bjorndahl TC, Tang P, Wishart DS. MetaboMiner – semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinformatics. doi: 10.1186/1471-2105-9-507. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.KinoSearch: A perl search engine library. [last accessed date October 20, 2008]. http://www.rectangular.com/kinosearch/