Abstract
Recently, body fluids have widely become an important target for proteomic research and proteomic study has produced more and more body fluid related protein data. A database is needed to collect and analyze these proteome data. Thus, we developed this web-based body fluid proteome database Sys-BodyFluid. It contains eleven kinds of body fluid proteomes, including plasma/serum, urine, cerebrospinal fluid, saliva, bronchoalveolar lavage fluid, synovial fluid, nipple aspirate fluid, tear fluid, seminal fluid, human milk and amniotic fluid. Over 10 000 proteins are presented in the Sys-BodyFluid. Sys-BodyFluid provides the detailed protein annotations, including protein description, Gene Ontology, domain information, protein sequence and involved pathways. These proteome data can be retrieved by using protein name, protein accession number and sequence similarity. In addition, users can query between these different body fluids to get the different proteins identification information. Sys-BodyFluid database can facilitate the body fluid proteomics and disease proteomics research as a reference database. It is available at http://www.biosino.org/bodyfluid/.
INTRODUCTION
In the post-genome era, proteomic technology has rapidly developed to be a powerful platform for the research of human physiology. It can be applied for identifying potential novel biomarkers for prognosis, diagnosis and therapeusis (1,2). And in recent years it is shown that body fluids have become one of the important targets for proteomics research (3). The body fluids include a wide variety of compositions like plasma/serum, urine, cerebrospinal fluid, saliva, bronchoalveolar lavage fluid, synovial fluid, nipple aspirate fluid, tear fluid, amniotic fluid and so on. Analysis of the protein composition in body fluids can help to understand human disease proteomics better. Hu et al.,(3) reviewed the body fluids research advances in proteome analysis and focused on its applications to human disease biomarker discovery. The importance of body fluids has also been appreciated by recent proteomics work (4). The database ‘MAPU: Max-Planck Unified database of organellar, cellular, tissue and body fluid’ (5) published in 2007 exhibit the close attention of the proteome researchers to the body fluids. The MAPU database stores the data from their own lab and contains several kinds of body fluids, such as urine and tear fluid. To collect more curated proteomics data in the related literatures of the body fluids and provide comprehensive protein annotation, as well as explore the relationships between the different body fluids, we constructed this database Sys-BodyFluid. Abundant proteomics data and in-depth protein annotation make Sys-BodyFluid to be a reference database for body fluid and clinical proteomics research.
DATABASE CONSTRUCTION
Sys-BodyFluid database was implemented through MySQL relational database (http://www.mysql.com). The web graphical user interface was constructed using JavaServer Pages technology (http://java.sun.com/products/jsp/). The manually curated body fluid protein data in the Sys-BodyFluid were imported to MySQL database by JAVA program. The protein annotation data were downloaded from International Protein Index (IPI) database, Gene Ontology (6), GOA database (7) and KEGG (8) pathway database. Open source JAVA library named as JFreeChart (http://www.jfree.org/jfreechart/) distributed under LGPL was adopted to plot the image of the statistics data in the web.
DATA SOURCE AND DATABASE CONTENTS
We searched PubMed and manually curated 50 related peer-review publications published online before May 2008. The primary sequences of the proteins were retrieved by the original ID from their corresponding databases in these publications. Due to the database updates, the protein sequences reported in the literatures may have changed or depleted in the current databases. Therefore, these protein sequences were manually validated before importing into the database. Each protein was mapping to the IPI database to uniform the protein ID in Sys-BodyFluid by blasting these protein sequences against the database (Human IPI Version 3.44) (the E-value cutoff was set to 10−8, the BLAST-HSP coverage was >0.9). Thus, each of the protein has a corresponding IPI ID in the Sys-BodyFluid database. The total unique proteins and paper numbers of the 11 kinds of body fluids in our database are summarized in Table 1. For example, there are 13 papers and 7748 proteins about the plasma/serum research in our database. Users can obtain this statistical information about the Sys-BodyFluid database in the ‘DATABASE’ web link in the website http://www.biosino.org/bodyfluid.
Table 1.
The data summary in Sys-BodyFluid database
| Body fluid name | Protein number | Paper number |
|---|---|---|
| Plasma/Serum (11–23) | 7748 | 13 |
| Saliva (24–31) | 2161 | 8 |
| Urine (32–40) | 1941 | 9 |
| Cerebrospinal fluid (41–46) | 1286 | 6 |
| Seminal fluid (47,48) | 916 | 2 |
| Amniotic fluid (49–51) | 899 | 3 |
| Tear (52,53) | 509 | 2 |
| Bronchoalveolar lavage fluid (54,55) | 411 | 2 |
| Milk (56,57) | 175 | 2 |
| Synovial fluid (58) | 114 | 1 |
| Nipple aspiration fluid (59,60) | 84 | 2 |
| Total | 10 138 | 50 |
DATA AVAILABILITY
The Sys-BodyFluid is accessed from graphical web interface (http://www.biosino.org/bodyfluid/) and the data are available for download through the ‘DOWNLOAD’ link in the website as a text file. Users could specify their interested body fluid data to download.
DATABASE UTILITY
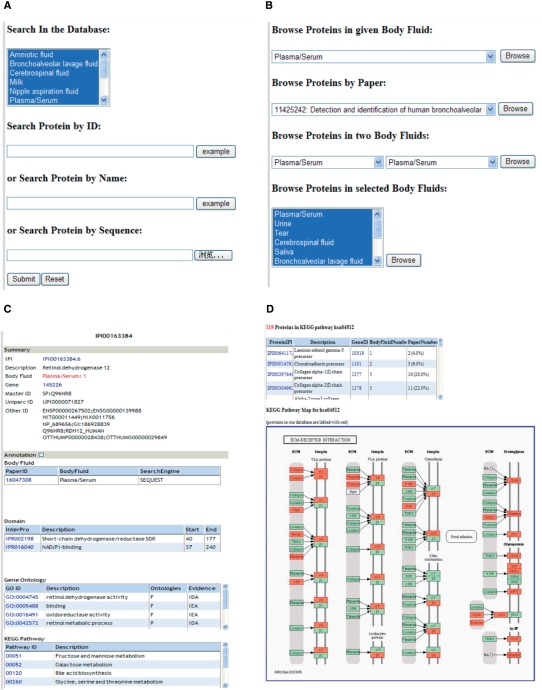
Sys-BodyFluid provides users the current database data statistics of different body fluids through the DATABASE link for the paper number and the unique protein number (DATABASE Link). As shown in Figure 1, Sys-BodyFluid offers users an optimal search function, including searching by protein ID, name and sequence similarity (SEARCH link, Figure 1A). The comprehensive browse option allows users to explore comparison analysis between two or more different body fluids data (Browse link, Figure 1B). For each protein in Sys-BodyFluid, we provide detailed annotation information, including protein description, involved body fluids, paper information, domain, Gene Ontology, pathway, sequence and so on (Figure 1C). Users can choose their interested body fluid to browse or download. Web page describing the body fluid provides users particular information. Furthermore, the availability of pathway analysis will assist users to investigate the difference between body fluids through involved metabolism and signal transduction pathway (Pathway link, Figure 1D). Proteins in our database are labeled with ‘red’ color. The body fluid number and paper number the proteins involved in are also showed in the web page.
Figure 1.
The web graphical user interface of Sys-BodyFluid database. (A) Search part and option. Users could search protein by protein ID, protein name and sequence similarity. (B) Browse part. Database allows user browse protein by their interested body fluid and interested paper. Protein existed in two body fluids could also be viewed and multi body fluids can be investigated. (C) Protein annotation part. There is detailed information in the database for each protein, including description, domain, Gene Ontology term, sequence and so on. (D) Pathway part. The proteins (colored by red) in different body fluids and their involved pathway are shown in pathway link. Proteins in our database are labeled with ‘red’ color. The body fluid number and paper number are also showed in the web page.
RESULTS AND DISCUSSION
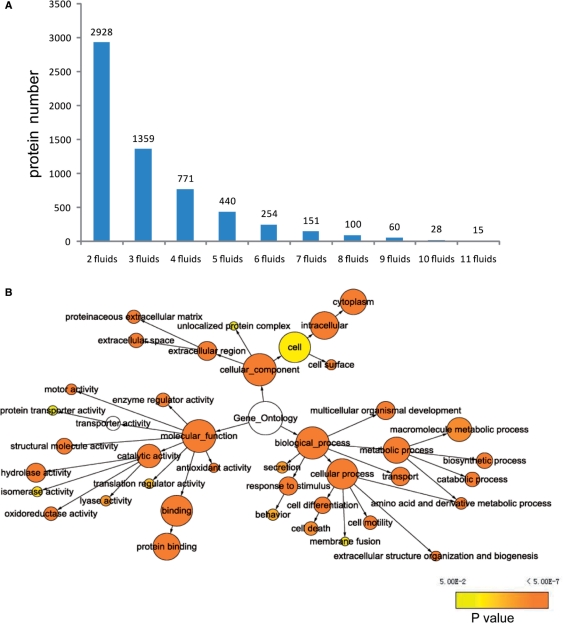
To get more comprehensive understanding of the relationship between body fluids, we compared the proteins composition in different body fluids. The result is shown in Figure 2A. There are 2928 proteins presented in at least two body fluids and 1359 proteins exist in at least three body fluids. Only 15 proteins exist in total 11 body fluids. For these 2928 proteins, GO annotation information were obtained and enrichment analysis was performed using BiNGO (9) and Cytoscape (10). Each node in Figure 2B represents a GO term. The node's size is scaled by protein number and node's color shows P-value of the enrichment analysis. The edge denotes the parent–children relationship between nodes. From this analysis, it is shown that some molecular functions like ‘protein binding’ and ‘enzyme regulator activity’ are over-presented in this dataset, as well as the biological process like ‘transport’ and ‘secretion’. Cellular component like ‘extracellular region’ is significantly enriched.
Figure 2.
(A) The data comparison in different body fluids. There are 2928 proteins presented in at least two body fluids and 1359 proteins existed in at least three body fluids. Only 15 proteins exist in total 11 body fluids. (B) Gene Ontology annotation statistical analysis for the 2928 proteins existing in at least two body fluids.
Human body fluids proteome analysis is still a challenge because dynamic range and the complexity of the body fluids protein composition. It is important to construct a body fluid reference database dedicated to biomarker discovery research. Previous work like MAPU is a great effort to integrate the data from their own lab and aim to provide a ‘gold standard’ reference proteome database. It is still necessary to refer to other proteomic literature data. For this reason, our database Sys-BodyFluid was build as a complementary database to the MAPU and aimed to provide users more information about the body fluids accompanied by protein abundant annotations. The relationship between different body fluids was also focused in our database. Users can access this database by http://www.biosino.org/bodyfluid.
PERSPECTIVES
As more and more body fluid proteome data have been produced recently, it is planned to update Sys-BodyFluid database every 6 months. New body fluid proteome data produced during the time will be added to our database. Furthermore, more annotation information like protein interaction data will also be included. In the future, we will collect more body fluid proteome data in the disease proteomics research, for example, cancer and diabetes proteome data. If possible, tissue proteomics data will be also included to look into the crosstalk between the tissue protein and the body fluid protein.
FUNDING
Basic Research Foundation (2006CB910700); CAS Project (KSCX2-YW-R-106, KSCX2-YW-R-112, KGCX1-YW-13); High-technology Project (2007AA02Z334). Funding for open access charge: CAS project KSCX2-YW-R-106.
Conflict of interest statement. None declared.
Footnotes
The authors wish it to be known that, in their opinion, the first three authors should be regarded as joint First Authors
REFERENCES
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Binz PA, Hochstrasser DF, Appel RD. Mass spectrometry-based proteomics: current status and potential use in clinical chemistry. Clin. Chem. Lab. Med. 2003;41:1540–1551. doi: 10.1515/CCLM.2003.237. [DOI] [PubMed] [Google Scholar]
- 3.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6:6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fusaro VA, Stone JH. Mass spectrometry-based proteomics and analyses of serum: a primer for the clinical investigator. Clin. Exp. Rheumatol. 2003;21:S3–S14. [PubMed] [Google Scholar]
- 5.Zhang Y, Zhang Y, Adachi J, Olsen JV, Shi R, de Souza G, Pasini E, Foster LJ, Macek B, Zougman A, et al. MAPU: Max-Planck Unified database of organellar, cellular, tissue and body fluid proteomes. Nucleic Acids Res. 2007;35:D771–D779. doi: 10.1093/nar/gkl784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maere S, Heymans K, Kuiper M. BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 10.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin WH, Dai J, Li SJ, Xia QC, Zou HF, Zeng R. Human plasma proteome analysis by multidimensional chromatography prefractionation and linear ion trap mass spectrometry identification. J. Proteome Res. 2005;4:613–619. doi: 10.1021/pr049761h. [DOI] [PubMed] [Google Scholar]
- 12.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, et al. The human plasma proteome: a nonredundant list developed by combination of four separate sources. Mol. Cell. Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Barnea E, Sorkin R, Ziv T, Beer I, Admon A. Evaluation of prefractionation methods as a preparatory step for multidimensional based chromatography of serum proteins. Proteomics. 2005;5:3367–3375. doi: 10.1002/pmic.200401221. [DOI] [PubMed] [Google Scholar]
- 14.Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, et al. Different immunoaffinity fractionation strategies to characterize the human plasma proteome. J. Proteome Res. 2006;5:1379–1387. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 15.He P, He HZ, Dai J, Wang Y, Sheng QH, Zhou LP, Zhang ZS, Sun YL, Liu F, Wang K, et al. The human plasma proteome: analysis of Chinese serum using shotgun strategy. Proteomics. 2005;5:3442–3453. doi: 10.1002/pmic.200401301. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Valentine SJ, Plasencia MD, Trimpin S, Naylor S, Clemmer DE. Mapping the human plasma proteome by SCX-LC-IMS-MS. J. Am. Soc. Mass Spectrom. 2007;18:1249–1264. doi: 10.1016/j.jasms.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, et al. Overview of the HUPO plasma proteome project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 18.Sennels L, Salek M, Lomas L, Boschetti E, Righetti PG, Rappsilber J. Proteomic analysis of human blood serum using peptide library beads. J. Proteome Res. 2007;6:4055–4062. doi: 10.1021/pr070339l. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka Y, Akiyama H, Kuroda T, Jung G, Tanahashi K, Sugaya H, Utsumi J, Kawasaki H, Hirano H. A novel approach and protocol for discovering extremely low-abundance proteins in serum. Proteomics. 2006;6:4845–4855. doi: 10.1002/pmic.200500774. [DOI] [PubMed] [Google Scholar]
- 20.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Tu CJ, Dai J, Li SJ, Sheng QH, Deng WJ, Xia QC, Zeng R. High-sensitivity analysis of human plasma proteome by immobilized isoelectric focusing fractionation coupled to mass spectrometry identification. J. Proteome Res. 2005;4:1265–1273. doi: 10.1021/pr0497529. [DOI] [PubMed] [Google Scholar]
- 22.Valentine SJ, Plasencia MD, Liu X, Krishnan M, Naylor S, Udseth HR, Smith RD, Clemmer DE. Toward plasma proteome profiling with ion mobility-mass spectrometry. J. Proteome Res. 2006;5:2977–2984. doi: 10.1021/pr060232i. [DOI] [PubMed] [Google Scholar]
- 23.Zhou M, Prieto DA, Lucas DA, Chan KC, Issaq HJ, Veenstra TD, Conrads TP. Identification of the SELDI ProteinChip human serum retentate by microcapillary liquid chromatography-tandem mass spectrometry. J. Proteome Res. 2006;5:2207–2216. doi: 10.1021/pr060061h. [DOI] [PubMed] [Google Scholar]
- 24.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang X, Yang L, Wang W, Song T, Lee CS, DeVoe DL, Balgley BM. Comparison of electrokinetics-based multidimensional separations coupled with electrospray ionization-tandem mass spectrometry for characterization of human salivary proteins. Anal. Chem. 2007;79:5785–5792. doi: 10.1021/ac070611a. [DOI] [PubMed] [Google Scholar]
- 26.Guo T, Rudnick PA, Wang W, Lee CS, Devoe DL, Balgley BM. Characterization of the human salivary proteome by capillary isoelectric focusing/nanoreversed-phase liquid chromatography coupled with ESI-tandem MS. J. Proteome Res. 2006;5:1469–1478. doi: 10.1021/pr060065m. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 28.Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, Amado FM. Identification of human whole saliva protein components using proteomics. Proteomics. 2004;4:1109–1115. doi: 10.1002/pmic.200300638. [DOI] [PubMed] [Google Scholar]
- 29.Walz A, Stuhler K, Wattenberg A, Hawranke E, Meyer HE, Schmalz G, Bluggel M, Ruhl S. Proteome analysis of glandular parotid and submandibular-sublingual saliva in comparison to whole human saliva by two-dimensional gel electrophoresis. Proteomics. 2006;6:1631–1639. doi: 10.1002/pmic.200500125. [DOI] [PubMed] [Google Scholar]
- 30.Wilmarth PA, Riviere MA, Rustvold DL, Lauten JD, Madden TE, David LL. Two-dimensional liquid chromatography study of the human whole saliva proteome. J. Proteome Res. 2004;3:1017–1023. doi: 10.1021/pr049911o. [DOI] [PubMed] [Google Scholar]
- 31.Xie H, Rhodus NL, Griffin RJ, Carlis JV, Griffin TJ. A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol. Cell. Proteomics. 2005;4:1826–1830. doi: 10.1074/mcp.D500008-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castagna A, Cecconi D, Sennels L, Rappsilber J, Guerrier L, Fortis F, Boschetti E, Lomas L, Righetti PG. Exploring the hidden human urinary proteome via ligand library beads. J. Proteome Res. 2005;4:1917–1930. doi: 10.1021/pr050153r. [DOI] [PubMed] [Google Scholar]
- 34.Khan A, Packer NH. Simple urinary sample preparation for proteomic analysis. J. Proteome Res. 2006;5:2824–2838. doi: 10.1021/pr060305y. [DOI] [PubMed] [Google Scholar]
- 35.Oh J, Pyo JH, Jo EH, Hwang SI, Kang SC, Jung JH, Park EK, Kim SY, Choi JY, Lim J. Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics. 2004;4:3485–3497. doi: 10.1002/pmic.200401018. [DOI] [PubMed] [Google Scholar]
- 36.Pieper R, Gatlin CL, McGrath AM, Makusky AJ, Mondal M, Seonarain M, Field E, Schatz CR, Estock MA, Ahmed N, et al. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4:1159–1174. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 37.Ru QC, Katenhusen RA, Zhu LA, Silberman J, Yang S, Orchard TJ, Brzeski H, Liebman M, Ellsworth DL. Proteomic profiling of human urine using multi-dimensional protein identification technology. J. Chromatogr. A. 2006;1111:166–174. doi: 10.1016/j.chroma.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 38.Spahr CS, Davis MT, McGinley MD, Robinson JH, Bures EJ, Beierle J, Mort J, Courchesne PL, Chen K, Wahl RC, et al. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics. 2001;1:93–107. doi: 10.1002/1615-9861(200101)1:1<93::AID-PROT93>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Sun W, Li F, Wu S, Wang X, Zheng D, Wang J, Gao Y. Human urine proteome analysis by three separation approaches. Proteomics. 2005;5:4994–5001. doi: 10.1002/pmic.200401334. [DOI] [PubMed] [Google Scholar]
- 40.Zerefos PG, Vougas K, Dimitraki P, Kossida S, Petrolekas A, Stravodimos K, Giannopoulos A, Fountoulakis M, Vlahou A. Characterization of the human urine proteome by preparative electrophoresis in combination with 2-DE. Proteomics. 2006;6:4346–4355. doi: 10.1002/pmic.200500671. [DOI] [PubMed] [Google Scholar]
- 41.Ogata Y, Charlesworth MC, Higgins L, Keegan BM, Vernino S, Muddiman DC. Differential protein expression in male and female human lumbar cerebrospinal fluid using iTRAQ reagents after abundant protein depletion. Proteomics. 2007;7:3726–3734. doi: 10.1002/pmic.200700455. [DOI] [PubMed] [Google Scholar]
- 42.Ogata Y, Charlesworth MC, Muddiman DC. Evaluation of protein depletion methods for the analysis of total-, phospho- and glycoproteins in lumbar cerebrospinal fluid. J. Proteome Res. 2005;4:837–845. doi: 10.1021/pr049750o. [DOI] [PubMed] [Google Scholar]
- 43.Pan S, Wang Y, Quinn JF, Peskind ER, Waichunas D, Wimberger JT, Jin J, Li JG, Zhu D, Pan C, et al. Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J. Proteome Res. 2006;5:2769–2779. doi: 10.1021/pr060251s. [DOI] [PubMed] [Google Scholar]
- 44.Wenner BR, Lovell MA, Lynn BC. Proteomic analysis of human ventricular cerebrospinal fluid from neurologically normal, elderly subjects using two-dimensional LC-MS/MS. J. Proteome Res. 2004;3:97–103. doi: 10.1021/pr034070r. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Goodlett DR, Peskind ER, Quinn JF, Zhou Y, Wang Q, Pan C, Yi E, Eng J, Aebersold RH, et al. Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiol. Aging. 2005;26:207–227. doi: 10.1016/j.neurobiolaging.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, Kumar C, Mann M. Integrated analysis of the cerebrospinal fluid peptidome and proteome. J. Proteome Res. 2008;7:386–399. doi: 10.1021/pr070501k. [DOI] [PubMed] [Google Scholar]
- 47.Fung KY, Glode LM, Green S, Duncan MW. A comprehensive characterization of the peptide and protein constituents of human seminal fluid. Prostate. 2004;61:171–181. doi: 10.1002/pros.20089. [DOI] [PubMed] [Google Scholar]
- 48.Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho CK, Shan SJ, Winsor EJ, Diamandis EP. Proteomics analysis of human amniotic fluid. Mol. Cell. Proteomics. 2007;6:1406–1415. doi: 10.1074/mcp.M700090-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Michaels JE, Dasari S, Pereira L, Reddy AP, Lapidus JA, Lu X, Jacob T, Thomas A, Rodland M, Roberts C.T., Jr., et al. Comprehensive proteomic analysis of the human amniotic fluid proteome: gestational age-dependent changes. J. Proteome Res. 2007;6:1277–1285. doi: 10.1021/pr060543t. [DOI] [PubMed] [Google Scholar]
- 51.Nilsson S, Ramstrom M, Palmblad M, Axelsson O, Bergquist J. Explorative study of the protein composition of amniotic fluid by liquid chromatography electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. J. Proteome Res. 2004;3:884–889. doi: 10.1021/pr0499545. [DOI] [PubMed] [Google Scholar]
- 52.de Souza GA, Godoy LM, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006;7:R72. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li N, Wang N, Zheng J, Liu XM, Lever OW, Erickson PM, Li L. Characterization of human tear proteome using multiple proteomic analysis techniques. J. Proteome Res. 2005;4:2052–2061. doi: 10.1021/pr0501970. [DOI] [PubMed] [Google Scholar]
- 54.Plymoth A, Yang Z, Lofdahl CG, Ekberg-Jansson A, Dahlback M, Fehniger TE, Marko-Varga G, Hancock WS. Rapid proteome analysis of bronchoalveolar lavage samples of lifelong smokers and never-smokers by micro-scale liquid chromatography and mass spectrometry. Clin. Chem. 2006;52:671–679. doi: 10.1373/clinchem.2005.060715. [DOI] [PubMed] [Google Scholar]
- 55.Sabounchi-Schutt F, Astrom J, Eklund A, Grunewald J, Bjellqvist B. Detection and identification of human bronchoalveolar lavage proteins using narrow-range immobilized pH gradient DryStrip and the paper bridge sample application method. Electrophoresis. 2001;22:1851–1860. doi: 10.1002/1522-2683(200105)22:9<1851::AID-ELPS1851>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 56.Fortunato D, Giuffrida MG, Cavaletto M, Garoffo LP, Dellavalle G, Napolitano L, Giunta C, Fabris C, Bertino E, Coscia A, et al. Structural proteome of human colostral fat globule membrane proteins. Proteomics. 2003;3:897–905. doi: 10.1002/pmic.200300367. [DOI] [PubMed] [Google Scholar]
- 57.Palmer DJ, Kelly VC, Smit AM, Kuy S, Knight CG, Cooper GJ. Human colostrum: identification of minor proteins in the aqueous phase by proteomics. Proteomics. 2006;6:2208–2216. doi: 10.1002/pmic.200500558. [DOI] [PubMed] [Google Scholar]
- 58.Gobezie R, Kho A, Krastins B, Sarracino DA, Thornhill TS, Chase M, Millett PJ, Lee DM. High abundance synovial fluid proteome: distinct profiles in health and osteoarthritis. Arthritis Res. Ther. 2007;9:R36. doi: 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander H, Stegner AL, Wagner-Mann C, Du Bois GC, Alexander S, Sauter ER. Proteomic analysis to identify breast cancer biomarkers in nipple aspirate fluid. Clin. Cancer Res. 2004;10:7500–7510. doi: 10.1158/1078-0432.CCR-04-1002. [DOI] [PubMed] [Google Scholar]
- 60.Varnum SM, Covington CC, Woodbury RL, Petritis K, Kangas LJ, Abdullah MS, Pounds JG, Smith RD, Zangar RC. Proteomic characterization of nipple aspirate fluid: identification of potential biomarkers of breast cancer. Breast Cancer Res. Treat. 2003;80:87–97. doi: 10.1023/A:1024479106887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Sys-BodyFluid is accessed from graphical web interface (http://www.biosino.org/bodyfluid/) and the data are available for download through the ‘DOWNLOAD’ link in the website as a text file. Users could specify their interested body fluid data to download.