Abstract
The growth in the number of completely sequenced microbial genomes (bacterial and archaeal) has generated a need for a procedure that provides UniProtKB/Swiss-Prot-quality annotation to as many protein sequences as possible. We have devised a semi-automated system, HAMAP (High-quality Automated and Manual Annotation of microbial Proteomes), that uses manually built annotation templates for protein families to propagate annotation to all members of manually defined protein families, using very strict criteria. The HAMAP system is composed of two databases, the proteome database and the family database, and of an automatic annotation pipeline. The proteome database comprises biological and sequence information for each completely sequenced microbial proteome, and it offers several tools for CDS searches, BLAST options and retrieval of specific sets of proteins. The family database currently comprises more than 1500 manually curated protein families and their annotation templates that are used to annotate proteins that belong to one of the HAMAP families. On the HAMAP website, individual sequences as well as whole genomes can be scanned against all HAMAP families. The system provides warnings for the absence of conserved amino acid residues, unusual sequence length, etc. Thanks to the implementation of HAMAP, more than 200 000 microbial proteins have been fully annotated in UniProtKB/Swiss-Prot (HAMAP website: http://www.expasy.org/sprot/hamap).
INTRODUCTION
The increasing number of completely sequenced microbial genomes represents an unparalleled opportunity to achieve a better understanding of prokaryotes, including their metabolic pathways, virulence factors, phylogeny, etc. However, the sequences themselves are not enough. It is of fundamental importance that these genomes be annotated with high quality and that the nomenclature be standardized.
Since the publication in 1995 of the complete Haemophilus influenzae genome (1), more than 700 bacterial and archaeal genomes have been entirely sequenced; the development of new sequencing techniques, such as parallel pyrosequencing of 454 Life Sciences (2) and Solexa/Illumina Genome Analyzer sequencing-by-synthesis technology (3), has greatly increased the amount of sequenced data that is generated, and they complement the classic Sanger DNA sequencing method (4). Public databases currently hold more than 100Gb of sequence and this amount will continue to increase exponentially as sequencing centres will soon have an annual throughput of several gigabases each.
Most of the proteins coming from these sequencing projects will probably never be characterized, and the annotation at the DNA level is succinct. Sequencing centres have developed automated pipelines from a combination of methods, such as sequence similarity, presence of domains and pathway prediction, among many other sequence analysis methods usually employed (5) to attempt to annotate the proteome of a certain microorganism. Though the prediction of coding sequences (CDSs) is usually very good, the quality of the functional annotation attached to them is very variable.
Many methods have been developed to improve genome functional annotation, including the use of genomic context information (6), mapping of pathways in orthologous groups (7), or defining protein function based on protein–protein interactions (8). Genome annotation by the scientific community using Wiki software has lately been the focus of several initiatives (9–11), but one of the major hurdles is the establishment of common standards for the annotation provided by each expert. Since sequencing centres and users in general rely on large protein databases, and especially on UniProtKB/Swiss-Prot (12), to annotate new genomes and identify new proteins, we consider it to be an important mission of UniProtKB to provide as many annotated proteins as possible, with the highest possible quality.
In order to address this need, we have implemented HAMAP (High-quality Automated and Manual Annotation of microbial Proteomes), a semi-automated pipeline system within UniProtKB/Swiss-Prot, dedicated to high-throughput, high-quality annotation of proteins from microbial complete proteomes, that also provides complete proteome sets that are consistent and non-redundant. Its aim is to maximize the complementarity between manual and automated annotation; the HAMAP system is composed of two databases and an automatic annotation pipeline. It targets proteins from bacteria, archaea and plastids, the latter being included due to their bacterial origin.
On the HAMAP website (http://www.expasy.org/sprot/hamap), two databases are available: one that provides curated information on all bacterial, archaeal and plastid proteomes—only fully sequenced and assembled genomes submitted to the public databases and whose CDSs have been annotated are taken into account—and a family database that contains all manually created protein families and annotation templates (also called ‘family rules’). There is also a tool for user-derived complete protein annotation (protein recommended name, gene name, function, subunit, membership to a protein family, sequence features, etc., as specified in the family annotation template) that is provided upon submission of either one protein sequence, if it belongs to one of the HAMAP families, or of a complete genome even before submission to the public DNA databases. Since the system provides not only annotation, but also warnings regarding atypical N-termini, lack of conserved residues and many other features, we believe that this tool can help the scientific community in the annotation of whole microbial genomes or any protein from bacteria, archaea and plastids.
THE PROTEOME DATABASE
The proteome database (http://www.expasy.org/sprot/hamap/proteomes.html) is developed jointly with the UniProt team at the European Bioinformatics Institute. Its aim is to provide, in a relational database, information on the biology, genome and taxonomy of each completely sequenced proteome that has been submitted to the public DNA databases. Whole-genome-shotgun genomes (WGSs) are not incorporated into the proteome database.
On the ‘HAMAP proteomes’ homepage, a list of all available proteomes is provided, plus a link to all sequenced microorganisms that are known to interact with other organisms (for example, a list of sequenced strains that are avirulent, animal intracellular parasites, plant symbionts, etc).
A page is provided for each complete proteome added; this page contains three sections: general information, genome(s) sequenced, and tools.
- The ‘General Information’ section contains:
- taxonomic information;
- information on the biology and genomics of the sequenced strain; and
- presence of some morphological characteristics.
The ‘Genome(s) sequenced’ section describes all DNA elements (chromosome and plasmids), with links to the DNA database and the reference to the paper, if the genome has been published, plus links to external databases that refer to the genome in question. This database is constantly updated: as papers are published, the references are added to the database and to the UniProtKB entries themselves.
- The ‘Tools’ section contains:
- the genome viewer, which allows the user to see the CDSs encoded on a particular region of the sequenced genome;
- BLAST searches against all proteins from the proteome;
- a link to download all UniProtKB entries for the proteome, either in UniProtKB format or in FASTA format;
- a link to retrieve all characterized or identified proteins from the proteome; this is based on the ‘Protein existence’ line present in each UniProtKB entry [for details see (12)]; and
- a link to retrieve all proteins from the proteome for which a 3D structure is available.
This database is extensively curated in several aspects: plasmids (which are not always submitted simultaneously with the chromosome sequences) are attached to the proteome sets to form complete genomes; extensive information on the sequenced strain is presented; cross-references to relevant sites are manually added and maintained, as is information on genome publications. The complete proteome sets presented contain both annotated entries from UniProtKB/Swiss-Prot and from its supplement, UniProtKB/TrEMBL (12).
At the time of writing, the proteome database contained pages for 622 bacterial proteomes, 53 archaeal proteomes and 133 plastid proteomes.
THE FAMILY DATABASE
The HAMAP annotation system was designed (13) to propagate manually generated annotation to all members of a given protein family in an automated, but controlled way. The system is based on protein families and their annotation templates, which are created manually by curators (see below) and which are used as the annotation template for the propagation of annotation to members of a protein family. Members of HAMAP families are identified using a profile collection (see below).
Three types of protein families are dealt with by the HAMAP annotation system:
Proteins that belong to well-characterized families, a family being a manually compiled collection of orthologs. Their function is known, i.e. has been described for at least one or several members, and has been well studied in one or more species;
UPFs, i.e. uncharacterized protein families, are conserved proteins found in several species but for which no function is known at present; and
proteins belonging to complex families, such as ABC transporters.
The main components of the HAMAP annotation system are the protein families and their annotation templates, the alignments and the profiles that are generated from them, and the annotation pipeline. Each component is explained in the following sections.
HAMAP protein families and annotation templates
The annotation templates are manually created and contain all the annotation that will be propagated to the members of a family. In order to create the annotation template, all characterized proteins that belong to this family are manually annotated according to UniProtKB/Swiss-Prot standards; this means that curators perform a thorough, detailed and in-depth review of the existing literature on a certain protein, including proteins from genomes that are not fully sequenced. The information available on these proteins provides the contents of each annotation template (family rule), and these proteins are listed in the field ‘Template’ in each family rule (see below). Most available papers are read and used to annotate the characterized proteins. This manual annotation and additional BLAST similarity searches (14) are used by curators to define what information can be safely propagated to other prokaryotes and to manually select the set of member sequences that will be used to build the seed alignment. In other words, curators determine the nature and extent of the annotation that can be propagated to orthologs.
The advantage of the manual intervention by curators who continuously revise the existing literature is that annotation templates and protein families are periodically revised to ensure that the annotation is as up-to-date as possible, and also to ensure that the organisms represented are as divergent as possible. This is important for the generation and maintenance of profiles. Also, if a curator comes across experimental evidence that contradicts the propagated annotation, the entire family is revised and the annotation template is updated taking into account the new available experimental evidence. Manual curation ensures that most available experimental knowledge is represented in the database, even though this is a slow, time-consuming process that usually lags behind the pace at which new evidence becomes available.
At present, more than 1500 protein families and their annotation templates are available on the HAMAP website (http://www.expasy.org/sprot/hamap/families.html).
Each annotation template for a HAMAP protein family (Figure 1) has a unique identifier of the format MF_xxxxx. They contain several fields, among which (for detailed information on all the fields present in HAMAP annotation templates see http://www.expasy.org/unirule/unirule_web_view.html#General):
general information, such as last revision date;
annotation that can be propagated to all members, such as protein name (which usually includes only the recommended name of a protein, but can also include some alternative, synonymous names if appropriate); gene name when available; general annotation lines such as function, catalytic activity, subunit, subcellular location, PTMs and the name of the family to which the protein belongs, among other information; keywords; relevant sequence features, such as active sites, metal-binding residues, domains, topology, etc.;
Gene Ontology (GO) terms (15), which are manually selected by the curators after thorough review of the existing literature and of the available terms;
cross-references to PROSITE (16), Pfam (17), TIGRFAMs (18), PRINTS (19) and/or PIRSF (20);
UniProtKB accession numbers of all entries (templates) that were manually annotated and for which there is experimental evidence or structural data that was used to build the family and its annotation template; and
sets of member sequences divided by taxonomic groups.
Figure 1.
Example of a HAMAP protein family annotation template (family rule), MF_00074 (http://www.expasy.org/unirule/MF_00074). Annotation templates contain three sections: ‘General rule information’, ‘Propagated annotation’ and ‘Additional information’. General information comprises: family identification number (MF_xxxxx), dates of creation and revision, ‘Data class’, i.e. that the whole protein is annotated by the family rule and not only a specific domain, and ‘Predictors’, which contain the distribution of matches and the alignment that was used to generate the family profile. The ‘Propagated annotation’ section contains the information that is propagated to all members of a protein family, or to some, if the field is preceded by ‘cases’ or ‘conditions’. For MF_00074, the function field will be different depending on the taxonomic origin, but all proteins will have ‘Cytoplasm’ as subcellular location and all belong to the family ‘RNA methyltransferase rsmG’. It also contains cross-references to other protein family databases, such as Pfam and TIGRFAMS, and manually selected GO terms. Additional information includes the size range of members of this family, if there are protein families related to this one, the list of characterized protein(s) that were used to compile information for the creation of the protein family and its annotation template (for MF_00074, literature is found for the proteins of E. coli, Bacillus subtilis, Microbacterium tuberculosis and Streptomyces coelicolor), the scope, i.e. the taxonomic groups covered by this family, if in at least one member this protein is fused to another protein either in the N-terminal or C-terminal region, and whether there are duplicates or whether in some species the protein is encoded on a plasmid. In the ‘UniProtKB rule member sequences’ section, complete sets of member proteins can be retrieved, taxonomic distribution can be browsed, and specific sets of proteins can be retrieved.
The use of conditional statements (‘cases’ and conditions) ensures that the annotation is only applied where appropriate, to guarantee the production of annotation of the same quality as that produced by manual curation (see Figure 2 for some examples).
Figure 2.
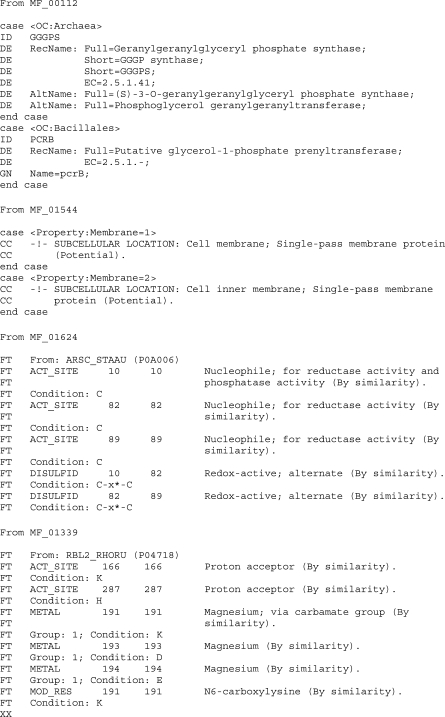
Examples of uses of the conditional statements ‘case’ and ‘conditions’ in family annotation templates (family rules). MF_00112 (http://www.expasy.org/unirule/MF_00112): an example of ID/protein name/gene name propagation depending on taxonomic distinction. In archaea, no gene name has been assigned but enzyme function has been proven in several different species, whereas the gene name pcrB is used only in Bacillales, with a function that has only been suggested for B. subtilis. Note that the reaction catalyzed by the archaeal protein has no biological significance in bacteria, since GGGP is a specific precursor of archaeal membrane lipids. MF_01544 (http://www.expasy.org/unirule/MF_01544): Subcellular location is predicted based on the number of membranes the bacterium possesses. MF_01624 (http://www.expasy.org/unirule/MF_01624): an example of conditions used for active site and disulfide bond feature propagation. If the indicated amino acid(s) are not present in the appropriate position(s) in the sequence, the feature is not propagated and a warning is generated, necessitating manual intervention. MF_01339 (http://www.expasy.org/unirule/MF_01339): an example of active site, metal and modified residue feature propagation. In the last two examples, the template entry used to derive the information is also indicated.
Cases and conditions are derived from relevant biological information collected from the literature; cases can restrict the propagation of annotation to a specific taxonomic group, for example, or be dependent (in this case a ‘condition’ statement exists in the annotation template) on the presence of a specific amino acid residue, or group of residues, for the annotation to be propagated. The annotation templates are designed to perform numerous checks on the sequences themselves as well, such as sequence length, aberrant N-termini, absence of expected sequence features, among others.
On the website, the protein families and their annotation templates can be browsed by protein name, gene name, pathway, scope (archaeal, bacterial and/or plastid families), etc.
Alignments and profiles
Once the seed members of a protein family are manually selected, the sequences are aligned using ClustalW (21), MUSCLE (22) or T-Coffee (23). The alignments are manually verified, and sometimes manually edited. The sequences themselves are also manually corrected if appropriate, for example if they result from a frameshift or are too long or too short at their N-terminus. The alignments are used both for the automated generation of identification profiles used to generate family matches [for details see (13)], and for the propagation of sequence features by similarity to the template sequence.
The whole collection of HAMAP profiles can be downloaded by ftp at ftp.expasy.org/databases/hamap/.
Detailed explanations about the database, the fields in the annotation templates and the annotation pipeline in general, plus a comprehensive user manual, can be found in the ‘Documents’ section (http://www.expasy.org/sprot/hamap/hamap_doc.html).
THE ANNOTATION PIPELINE
The annotation pipeline was set up to optimize the interaction between programs and curators and to ensure that ‘problematic’ sequences will always be re-directed to manual check and curation. The aim is to propagate annotation as carefully as possible; built-in checks and limitations will prevent a protein sequence from being annotated in case of doubt. The aim is always to achieve quality rather than maximal coverage.
In brief, the system works as follows (Figure 3): after a complete genome is deposited in DDBJ/EMBL/GenBank (24) entries are produced containing the original annotation that was provided by the submitter, plus, in some cases, automatically added additional annotation. These entries are stored in UniProtKB/TrEMBL, the unreviewed section of UniProtKB. All microbial and plastid protein sequences in UniProtKB/TrEMBL are run daily against the HAMAP profile collection and family members are identified. Matches with a score above the cutoff are annotated using the annotation templates and are integrated into UniProtKB/Swiss-Prot; problematic proteins (for example, sequences having unusual length, missing conserved amino acid residues or having aberrant N-termini) generate warnings and are channeled to manual review and annotation.
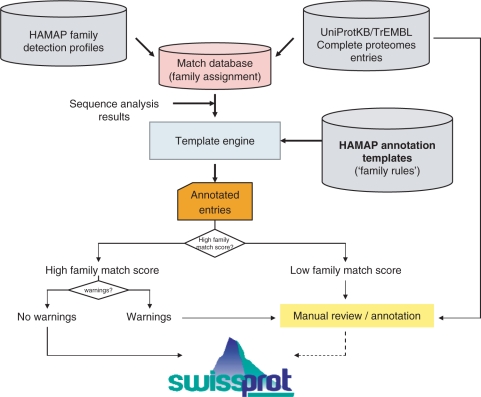
Figure 3.
The HAMAP annotation pipeline. UniProtKB/TrEMBL complete proteome entries matching a HAMAP family detection profile (derived from an alignment of manually selected family members; matches to those profiles are stored in a ‘Match database’, allowing assignment of family membership) are passed through a ‘template engine’ that applies the annotation found in the corresponding HAMAP annotation template (and resolving its conditional statements) to generate UniProtKB/Swiss-Prot annotation. If the system generates warnings, or if the matching score is low, the entry is channelled to manual annotation; entries without warnings are directly integrated into UniProtKB/Swiss-Prot. UniProtKB entries for which there is available literature are manually annotated.
UniProtKB/Swiss-Prot entries that belong to a HAMAP family, i.e. manually curated templates and entries that are the product of the automated annotation pipeline, can be identified by the cross-reference to HAMAP and the corresponding family number (in the ‘Cross references’ field, under ‘Family and domain databases’, MF_xxxxx).
TOOLS
On the website ‘Tools’ section, several analysis and retrieval tools are available: users can scan one protein sequence or a whole genome against the collection of HAMAP families; specific sets can be retrieved (characterized or identified proteins from specific proteomes, or sequences for which there are 3D structures available).
Submission of sequences or genomes for analysis
On the HAMAP tools page (http://www.expasy.org/sprot/hamap/index.html#tools), sequences can be submitted and checked whether they belong to any HAMAP family.
Two types of scan can be performed: ‘quick scan’, for one or a few sequences, and ‘advanced scan’, for whole microbial genomes.
After submission, results are displayed on the website. If a sequence hits one or more HAMAP families (a distinction is made between a ‘true’ membership, which is above the trusted cut-off, and a ‘weak’ match, below the trusted cut-off), the user is directed to the corresponding protein family and its annotation template containing the annotation that is applied to the respective family members.
If a whole genome is submitted, the results are password-protected and can be retrieved on the ‘HAMAP Scan results’ page, with full annotation and warnings regarding N-termini that are too long or too short, absence of conserved amino acid residues (which can be useful to check potential sequencing errors or frameshifts), absence of expected domains, etc.
Retrieval of sets of characterized/existent proteins or with 3D structures
With this tool, users can retrieve specific sets of proteins for which some characterization is available, i.e. the protein has been found to exist through mass spectrometry, in 2D gels, etc., or for which there is some literature, according to standards defined by the UniProtKB ‘Protein Existence’ line (12). A typical use would be to retrieve all ‘characterized’ proteins of a bacterium or archaeon (for example, retrieve all ‘characterized’ proteins of H. influenzae, or for a group of organisms, such as enterobacteria). The same can be done for retrieval of proteins for which there is at least one 3D structure available.
CONCLUSION
The HAMAP database makes available to the scientific community and genome sequencing centres a collection of manually curated microbial protein families and profiles that can be useful for the functional annotation of protein sequences or microbial genomes. The automated pipeline can be used to detect occasional sequence errors by making use of the warnings generated by the system.
The HAMAP system as a whole has greatly increased the speed at which microbial protein sequences are annotated in UniProtKB/Swiss-Prot and we believe that this has been achieved without lowering the standards for which UniProtKB/Swiss-Prot is renowned. The coverage of HAMAP families keeps increasing as new families are manually created—at the moment, about 25% of the Escherichia coli K-12 proteins belong to a HAMAP family.
We hope that the HAMAP resource can help the annotation of complete genomes, improving the quality of CDS prediction and functional annotation.
The development of the system and its website is an ongoing effort and future plans include the addition of phylogenetic analysis to help establish true orthology, checks of consistency within pathways and taking into account the conservation of gene neighborhoods, improvements in the generation of identification profiles and, especially, the coverage of all housekeeping genes.
FUNDING
The Swiss-Prot group is part of the Swiss Institute of Bioinformatics (SIB) and of the UniProt Consortium. Swiss-Prot group activities are supported by the Swiss Federal Government through the Federal Office of Education and Science, and by the National Institutes of Health (grant 2 U01 HG02712-04). Additional support comes from the European Commission contract FELICS (021902RII3) and from PATRIC BRC (NIH/NIAID contract HHSN 266200400035C). Funding for open access charges: Swiss Federal Government through the Federal Office of Education and Science.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Alexandre Gattiker for the design and implementation of the initial HAMAP pipeline, Sandrine Pilbout for all the taxonomy-related work, Nicole Redaschi, Thomas Kappler and Paul Kersey for database management, and Nicolas Hulo and Christian Sigrist for help with alignments and profiles.
REFERENCES
- 1.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb J.-F, Dougherty BA, Merrick JM, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 4.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stothard P, Wishart DS. Automated bacterial genome analysis and annotation. Curr. Opin. Microbiol. 2006;9:505–510. doi: 10.1016/j.mib.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 7.Mao F, Su Z, Olman V, Dam P, Liu Z, Xu Y. Mapping of orthologous genes in the context of biological pathways: an application of integer programming. Proc. Natl Acad. Sci. USA. 2006;103:129–134. doi: 10.1073/pnas.0509737102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 9.Salzberg SL. Genome re-annotation: a wiki solution? Genome Biol. 2007;8:102–102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsik CG, Worley KC, Zhang L, Milshina NV, Jiang H, Reese JT, Childs KL, Venkatraman A, Dickens CM, Weinstock GM, et al. Community annotation: procedures, protocols, and supporting tools. Genome Res. 2006;16:1329–1333. doi: 10.1101/gr.5580606. [DOI] [PubMed] [Google Scholar]
- 11.Mons B, Ashburner M, Chichester C, van Mulligen E, Weeber M, den Dunnen J, van Ommen GJ, Musen M, Cockerill M, Hermjakob H, et al. Calling on a million minds for community annotation in Wiki proteins. Genome Biol. 2008;9:R89–R89. doi: 10.1186/gb-2008-9-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gattiker A, Michoud K, Rivoire C, Auchincloss AH, Coudert E, Lima T, Kersey P, Pagni M, Sigrist CJ, Lachaize C, et al. Automated annotation of microbial proteomes in SWISS-PROT. Comput. Biol. Chem. 2003;27:49–58. doi: 10.1016/s1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gene Ontology Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief. Bioinform. 2008;9:210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- 18.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P, et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113–113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 24.Kulikova T, Akhtar R, Aldebert P, Althorpe N, Andersson M, Baldwin A, Bates K, Bhattacharyya S, Bower L, Browne P, et al. EMBL nucleotide sequence database in 2006. Nucleic Acids Res. 2007;35:D16–D20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]