Abstract
The antimicrobial peptide database (APD, http://aps.unmc.edu/AP/main.php) has been updated and expanded. It now hosts 1228 entries with 65 anticancer, 76 antiviral (53 anti-HIV), 327 antifungal and 944 antibacterial peptides. The second version of our database (APD2) allows users to search peptide families (e.g. bacteriocins, cyclotides, or defensins), peptide sources (e.g. fish, frogs or chicken), post-translationally modified peptides (e.g. amidation, oxidation, lipidation, glycosylation or d-amino acids), and peptide binding targets (e.g. membranes, proteins, DNA/RNA, LPS or sugars). Statistical analyses reveal that the frequently used amino acid residues (>10%) are Ala and Gly in bacterial peptides, Cys and Gly in plant peptides, Ala, Gly and Lys in insect peptides, and Leu, Ala, Gly and Lys in amphibian peptides. Using frequently occurring residues, we demonstrate database-aided peptide design in different ways. Among the three peptides designed, GLK-19 showed a higher activity against Escherichia coli than human LL-37.
INTRODUCTION
The antimicrobial peptide field is growing rapidly in response to the demand for novel antimicrobial agents (1–5). To promote the research, education and information exchange in the field, we established the multifunctional antimicrobial peptide database (APD) in 2003 and published a brief description in the database issue of Nucleic Acid Research in 2004 (6). Although there are other databases (7–14), APD has been widely utilized by users around the world (∼15 000 web hits per year) (15–17). APD is a general database dedicated to antimicrobial peptides from all biological sources, ranging from bacteria, plants, to animals, including humans. The database collects only ‘mature and active’ peptides (<100 amino acid residues), rather than a mixture of mature and precursor proteins. Furthermore, the peptides in APD are classified based on their biological activities such as anticancer, antiviral, antifungal and antibacterial, allowing users to readily obtain a list of peptides of special interest to them. In addition, the database provides statistical information for all listed peptides, or a group of peptides with desired properties such as anticancer. Finally, the database has interfaces for both peptide prediction and peptide design. APD also provides ‘Links’ that allow users to access other databases dedicated to antimicrobial peptides.
Since the publication of the first version of APD in 2004, a significant number of antimicrobial peptides have been discovered at both the gene and protein levels. Therefore, it is proper to include the newly identified members into APD. In total, more than 700 additional peptides have been registered. To meet the new requirements of the field as well as based on users' feedback, we have constructed additional search capabilities for the database. These include peptide families, sources, posttranslational modified peptides, and peptide-binding targets. Finally, we illustrate that novel antimicrobial peptides can be designed with the aid of APD. Among the three peptides designed, one of them showed a higher activity against Escherichia coli than human LL-37. Here we briefly describe the new features and findings of the second version of APD (hereinafter referred to as APD2). For new users, please also read our original article (6) or read ‘About’ (a brief guide to APD2) online, as the previously described functions and findings are not reiterated here.
DATABASE UPDATE
To facilitate database update and maintenance, we purchased the NAVICAT 8 software (PremiumSoft CyberTech Limited, Hong Kong) and solved the compatibility problem with our database by writing additional programs. These programs also enable a recalculation of peptide parameters should the original peptide sequence have been corrected, updated or completely replaced. By September 2008, APD2 possesse 1228 peptide entries. Relative to the original 525 entries (6), the total number of antimicrobial peptides in APD2 has been more than doubled. Among them, the number of antibacterial peptides increased from 498 to 944 (i.e. by 90%), antifungal peptides from 155 to 327 (by 111%), antiviral peptides from 28 to 76 (by 171%) and anticancer peptides from 18 to 65 (by 261%). It appears that the interest in developing antiviral and anticancer peptides on the basis of naturally occurring antimicrobial peptides is growing (18–20).
NEW FEATURES ADDED
The ‘name’ field of APD2 has been expanded significantly through extensive information registration. Peptide synonyms such as LL-37 and LL37 are included. This feature should benefit the ongoing effort in standardizing the nomenclature of antimicrobial peptides from amphibians (21,22). Because both the old and new names of the same peptides are entered, APD2 will facilitate the transition from the old to the new names.
Not all antimicrobial peptides have complete amino acid sequences. A list of peptides with incomplete amino acid sequences can be viewed by entering the letter string ‘BWQ’ into the name field followed by clicking on the ‘search’ button. Currently, APD2 collects 18 such entries.
APD2 allows the search for peptides from a particular life form by using common names: ‘bacteria, plant, ants, fish, cow, insect, crabs, toad and frog’ (actual words for search), to list just a few. For example, when the word ‘frog’ was searched in the name field, 398 peptides (32% in APD2) appeared, consistent with the notion that amphibians are an important source for natural antimicrobial peptides (1,21,22). Similarly, we found 203 peptides from plants, 118 from insects and 112 from bacteria. Therefore, the majority of the naturally occurring antimicrobial peptides collected in APD2 are originated from animals. To advance our understanding of the structure-activity relationships of antimicrobial peptides as a basis for peptide design, a select set of synthetic peptide derivatives were also collected. After update, the database contains 38 synthetic peptides (3% in APD2). Most of the synthetic peptides have known 3D structures, determined primarily by solution NMR spectroscopy (23,24). In total, there are 160 peptides (152 by NMR and eight by X-ray diffraction) with known 3D structures (70 originally). As long as the structural coordinates are deposited with the Protein Data Bank (PDB) (25), the structure of a particular peptide can be viewed directly by clicking on the PDB link provided in APD2 for each peptide entry.
In addition, ‘peptide families’ can be searched, also via the name field. When the word ‘cyclotide’ (small circular proteins from plants) (19) was entered, 126 peptides appeared. Likewise, we found 131 defensins and 51 cathelicidins when such words were searched.
To allow for the search for ‘chemically modified antimicrobial peptides’, a coding system was created by starting with XX. Thus, phosphorylation, lipidation, glycosylation, C-terminal amidation, peptide cyclization, oxidation and d-amino acids are represented by a set of unique letter strings (Table 1). Such modifications are described in the ‘Additional Info’ field so that the unmodified peptide sequence can be entered into the database. In total, 259 chemically modified peptides were found when XX was used to search in the name field. Among them, 135 peptides are cyclic, 113 peptides are C-terminally amidated (XXA), seven peptides are oxidized (XXO), and nine peptides contain d-amino acids (XXD). The number of d-amino acid residues in a peptide is indicated by an Arabic number after the code XXD. This annotation is very important since some antimicrobial peptides such as gassericin A (APD2 entry 766) and reutericin 6 (APD2 entry 930) share the same amino acid sequence and differ only in the number of d-amino acids. Note that the actual numbers of chemically modified peptides in the database will vary due to continued updating of our database.
Table 1.
Letter keys for searching antimicrobial peptides with chemical modification and peptide binding targets in APD2
| Code | Chemical modification | Code | Binding target |
|---|---|---|---|
| XXA | C-end amidation | BBMm | Membranes |
| XXC | Cyclization | BBB | Peptide aggregation |
| XXO | Oxidation | BBPP | Proteins/enzymes |
| XXP | Phosphorylation | BBN | RNA/DNA |
| XXD | d-amino acids | BBS | Sugars |
| XXG | Glycosylation | BBL | Lipopolysaccharides (LPS) |
| XXL | Lipidation | BBII | Metal ions (e.g. Fe3+) |
It is now accepted that ‘molecular targets’ of antimicrobial peptides are not restricted to bacterial membranes and can be other molecules as well (1–5). To enable the search for peptide targets, we also defined a set of unique letter strings (starting BB) for different targets such as proteins, nucleic acids, sugars and metal ions (Table 1) and entered such information into the database. A typical example is human LL-37, which is capable of binding to a variety of molecules such as proteins (represented by BBPP), DNA (BBD) and lipopolysaccharides or LPS (BBL) (reviewed in reference 15). In addition, the peptide is able to form oligomers by itself (BBB) in aqueous solutions upon increase in peptide concentrations or salts. Likewise, the string BBBm is coined as an indicator for peptide oligomerization in membrane environments. A further description of the interactions between antimicrobial peptides and different targets is given in the field of ‘Additional Info’ (e.g. see entry 310 for LL-37). This change will enrich the key information on biochemical and physical property, activity, 3D structure and mechanism of action of each antimicrobial peptide in APD2.
It is also useful to search for peptides that target a particular microbe by using the letter ‘Z’. When ‘ZZH’ was used to search, 53 anti-HIV peptides appeared. A further classification of the peptide sources was also made: ZZHa, anti-HIV peptides from animals (14 entries); ZZHb, anti-HIV peptides from bacteria (three entries); ZZHp, anti-HIV peptides from plants (24 entries, mainly cyclotides); ZZHh, anti-HIV peptides from humans (six entries); ZZHs, anti-HIV peptides from chemical synthesis (six entries). Additional letters can be appended to indicate the binding targets of HIV-active peptides. Such a feature is useful for users to choose a peptide template for engineering anti-HIV peptides by mutagenesis (20). Likewise, future database annotations will allow users to search for peptides with toxicity against a particular drug-resistant bacterial strain (ZZB), parasites (ZZP), or certain type of cancer cells (ZZC).
FREQUENTLY USED RESIDUES, MOTIFS AND APPLICATIONS IN PEPTIDE DESIGN
Frequently used amino acid residues in antimicrobial peptides
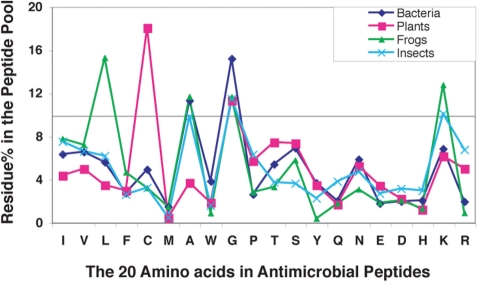
The search capability for peptide sources, families, and activities can be combined with the peptide analysis tools in APD2. A statistical analysis was conducted for a group of peptides from bacteria, plants, insects, and frogs. These life kingdoms were chosen because of a relatively large pool of peptides (>100 members) (Table 2). While the details are provided in the Supplementary table, the percentages for the 20 amino acids in each group of peptides from the four kingdoms are plotted in Figure 1. We define a residue in antimicrobial peptides as ‘frequently used’ if its percentage is ∼10% or greater. Thus, residues Ala and Gly are most abundant in bacterial peptides (dark blue line in Figure 1); residues Cys and Gly are more common in plant peptides (red line); residues Ala, Gly and Lys are frequently occurring in insect peptides (light blue line); and residues Leu, Ala, Gly and Lys have percentages higher than 10% in frog peptides (green line). A summary of these results is given in Table 2. It is notable that residue Gly is highly used in antimicrobial peptides from all the four kingdoms. In contrast, residue Met is rarely used (<1.5%). Interestingly, cationic Lys is preferred over Arg in all the kingdoms. In particular, residues Arg and Tyr are rarely used (<1%) in frog peptides. The reasons for these observations remain to be elucidated in the future. However, the lists of highly used residues derived from antimicrobial peptides from different kingdoms (Table 2) contain structural information. The abundance in residue Cys in plant peptides suggests that disulfide bonds (related to β-sheets) are common, rendering the peptide structure more stable (19). Indeed, the percentage of the Cys residue is also high in peptides with known β-sheet structures. Likewise, the frequently used residues from frogs (Leu, Ala, Gly and Lys) have the strong tendency to adopt an amphipathic helix. Interestingly, a search of APD2 for antimicrobial peptides with known helical structures followed by statistical analysis of the 170 peptides revealed the same set of frequently occurring residues.
Table 2.
Frequently used amino acid residues in naturally occurring antimicrobial peptides isolated from bacteria, plants, insects, or frogs
| Kingdom | Number of peptides in APD2 | Frequently used amino acids | Predicted structure |
|---|---|---|---|
| Bacteria | 112 | A, G | |
| Plants | 203 | C, G | β-Sheet |
| Insects | 118 | A, G, K | |
| Frogs | 398 | A, G, K, L | α-Helix |
The bold face is letter ‘G’ indicates amino acid is common in all life kingdom.
Figure 1.
The percentages of the 20 amino acid residues in antimicrobial peptides discovered from bacteria, plants, frogs and insects. The cutoff line at 10% defines frequently occurring residues (∼10% or greater). A summary of the frequently used residues is listed in Table 2.
Database-aided peptide design
The frequently used residues identified in each kingdom (Table 2) may be applied to peptide design in different ways. First, new antimicrobial peptides can be designed based on ‘templates derived from naturally occurring antimicrobial peptides’. Recently, we have identified the smallest antibacterial peptide, KR-12 (Table 3), from human LL-37 (26). Using KR-12 as the template, we designed KL-12 (Table 3) by converting all hydrophobic residues to leucines and all charged and hydrophilic residues to lysines. Unfortunately, antibacterial assays using the standard microdilution method (27) found that KL-12 was inactive against Gram-negative bacteria such as E. coli K12 or Gram-positive bacteria such as methicin-resistant Staphyloccocus aureus (MRSA) USA-300 until 160 μM. However, this method may be successful with other more active peptide templates. Second, new antimicrobial peptides can be designed by ‘combining database-derived peptide motifs’, consisting of frequently used residues (Table 2). Using the motifs consisting of only residues Gly, Leu and Lys, a second peptide was designed. GLK-19 (Table 3) was found to be active against E. coli and also weakly active against MRSA. Third, new peptides can be designed based on ‘amino acid percentages obtained from statistical analysis of antimicrobial peptides’ from a specific kingdom or peptide family (e.g. see Supplementary table). Thus, a third peptide was designed in three steps. (i) The amino acid composition for MULTI-18 was obtained by multiplying the percentage of each residue with the total number of residues (18 here) in the target peptide. Thus, we obtained 2 Gly, 2 Ala, 3 Leu, 3 Lys, 2 Ile, 1 Phe, 1 Asp, 1 Val, 1 Thr, 1 Ser and 1 Asn residues based on the statistical values obtained. (ii) We searched peptide motifs, consisting of these residues and found that sequence combinations such as GLFD, AAK, KIV, GKL, ITS and LN all exist in at least five peptides in APD2. (iii) Multiple peptides can be obtained by combining the above motifs. We only designed one peptide by following the amphipathic pattern for helical peptides. The resulting peptide MULTI-18 showed a low activity against E. coli. Although we only designed three peptides, one of the peptides (GLK-19) was found to have a higher antibacterial activity against E. coli than human LL-37 (Table 3). Therefore, our results prove the principle that APD2 is a useful tool for peptide design. In the previous version of our database (6), we already observed that antimicrobial peptides with toxic effects on mammalian cells are more hydrophobic. That finding, consistent with the results from experiments (28), should be useful in improving the therapeutic indexes of the designed peptides such as GLK-19 in the next step.
Table 3.
Minimal inhibitory concentrations (μm) of a few peptides designed based on frequently occurring residues and their combinations
| Name | Peptide sequence | E. coli K12 | MRSA USA-300 |
|---|---|---|---|
| LL-37 | LLGDLLRKSKEKIGKEF KRIVQRIKDFLRNLVPRTES | 40a | – |
| KR-12 | KRIVQRIKDFLR | 40a | >190 |
| KL-12 | KKLLKKLKKLLK | >160 | >160 |
| GLK-19 | GLKKLLGKLLKKL GKLLLK | 10 | 160 |
| MULTI-18 | GLFDAAKKIVGKLITSLN | 80 | >160 |
aData from reference 26. The region in the LL-37 sequence that corresponds to KR-12 is underlined.
SUMMARY AND DATABASE AVAILABILITY
Our database update increased the number of antimicrobial peptides from 525 to 1228. This increase should improve the reliability of the results from the statistical interface. New search capabilities such as peptide families, peptide sources and anti-HIV activity have been created via extensive information annotations and registration for peptide entries. In addition, both chemically modified peptides and molecular targets of antimicrobial peptides can be searched. In combination with the statistical interface, we found that frequently used amino acid residues in antimicrobial peptides differ in different kingdoms (Table 2). Using the frequently occurring residues from amphibian peptides, we demonstrate that the APD2 findings are useful in peptide design.
In conclusion, this update illustrates nicely the flexibility of our original database that allows expansion in both database functions and the scope of peptides to be collected. While the database update and developments continue, you are welcome to email us your comments, suggestions, or corrections for which we are grateful. APD2 can be accessed at the same website http://aps.unmc.edu/AP/main.php.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for Open Access publication charges has been waived by the Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Joe Ziskovsky and Atul Rayamajhi (UNMC) for their consistent support to make the database accessible to users via the Internet, Erik Martin (a summer student from the University of Southern Maine) and Edward Ezell (UNMC) for collecting the information for a few peptide entries, and Dr Kenneth Bayles (UNMC) and Dr Alan Peterkofsky (NIH) for providing us the MRSA and E. coli strains used for antibacterial assays. We also thank Sophie Wang (Wellesley College) for designing a new web page for APD2.
REFERENCES
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Boman HG. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 3.Yamasaki K, Gallo RL. Antimicrobial peptides in human skin disease. Eur. J. Dermatol. 2008;18:11–21. doi: 10.1684/ejd.2008.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 5.Daly NL, Chen YK, Rosengren KJ, Marx UC, Phillips ML, Waring AJ, Wang W, Lehrer RI, Craik DJ. Retrocyclin-2: structural analysis of a potent anti-HIV theta-defensin. Biochemistry. 2007;46:9920–9928. doi: 10.1021/bi700720e. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Wang G. APD: the Antimicrobial peptide database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wade D, Englund J. Synthetic antimicrobial peptide database. Protein Pept. Lett. 2002;9:53–57. doi: 10.2174/0929866023408986. [DOI] [PubMed] [Google Scholar]
- 8.Brahmachary M, Krishnan SP, Koh JL, Khan AM, Seah SH, Tan TW, Brusic V, Bajic VB. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004;32:D586–D589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitmore L, Wallace BA. The Peptaibol Database: a database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32:D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gueguen Y, Garnier J, Robert L, Lefranc MP, Mougenot I, de Lorgeril J, Janech M, Gross PS, Warr GW, Cuthbertson B, et al. PenBase, the shrimp antimicrobial peptide penaeidin database: sequence-based classification and recommended nomenclature. Dev. Comp. Immunol. 2006;30:283–288. doi: 10.1016/j.dci.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Seebah S, Suresh A, Zhou S, Choong YH, Chua H, Chuon D, Beuerman R, Verma C. Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res. 2007;35:D265–D268. doi: 10.1093/nar/gkl866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fjell CD, Hancock REW, Cherkasov A. AMPer: a database and an automated discovery tool for antimicrobial peptides. Bioinformatics. 2007;23:1148–1155. doi: 10.1093/bioinformatics/btm068. [DOI] [PubMed] [Google Scholar]
- 13.Hammami R, Zouhir A, Ben Hamida J, Fliss I. BACTIBASE: a new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007;7:89. doi: 10.1186/1471-2180-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang CK, Kaas Q, Chiche L, Craik DJ. Cybase: a database of cyclic protein sequences and structures, with applications in protein discovery and engineering. Nucleic Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G. Tool developments for structure-function studies of host defense peptides. Protein Pept. Lett. 2007;14:57–69. doi: 10.2174/092986607779117182. [DOI] [PubMed] [Google Scholar]
- 16.Loose C, Jensen K, Rigoutsos I, Stephanopoulos G. A linguistic model for the rational design of antimicrobial peptides. Nature. 2006;443:867–869. doi: 10.1038/nature05233. [DOI] [PubMed] [Google Scholar]
- 17.Lata S, Sharma BK, Raghava GP. Analysis and prediction of antibacterial peptides. BMC Bioinformatics. 2007;8:263. doi: 10.1186/1471-2105-8-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoskin DW, Ramarmoorthy A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole AM, Cole AL. Antimicrobial polypeptides are key anti-HIV-1 effector molecules of cervicovaginal host defense. Am. J. Reprod. Immunol. 2008;59:27–34. doi: 10.1111/j.1600-0897.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang G, Waston KM, Buckheit R.W., Jr. Anti-human immunodeficiency virus type 1 activities of antimicrobial peptides derived from human and bovine cathelicidins. Antimicrob. Agents Chemother. 2008;52:3438–3440. doi: 10.1128/AAC.00452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlon JM. A proposed nomenclature for antimicrobial peptides from frogs of the genus Leptodactylus. Peptides. 2008;29:1631–1632. doi: 10.1016/j.peptides.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 22.Amiche M, Ladram A, Nicolas P. A consistent nomenclature of antimicrobial peptides isolated from frogs of the subfamily Phyllomedusinae. Peptides. 2008 doi: 10.1016/j.peptides.2008.06.017. (in press) [DOI] [PubMed] [Google Scholar]
- 23.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 24.Wang G. Structural biology of antimicrobial peptides by NMR spectroscopy. Curr. Org. Chem. 2006;10:569–581. [Google Scholar]
- 25.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J. Biol. Chem. 2008;283:32637–32643. doi: 10.1074/jbc.M805533200. [DOI] [PubMed] [Google Scholar]
- 27.Wang G, Li Y, Li X. Correlation of three-dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J. Biol. Chem. 2005;280:5803–5811. doi: 10.1074/jbc.M410116200. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Li Y, Han H, Miller DW, Wang G. Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J. Am. Chem. Soc. 2006;128:5776–5785. doi: 10.1021/ja0584875. [DOI] [PubMed] [Google Scholar]