Abstract
RTPrimerDB (http://www.rtprimerdb.org) is a freely accessible database and analysis tool for real-time quantitative PCR assays. RTPrimerDB includes records with user submitted assays that are linked to genome information from reference databases and quality controlled using an in silico assay evaluation system. The primer evaluation tools intended to assess the specificity and to detect features that could negatively affect the amplification efficiency are combined into a pipeline to test custom-designed primer and probe sequences. An improved user feedback system guides users and submitters to enter practical remarks and details about experimental evaluation analyses. The database is linked with reference databases to allow the submission of assays for all genes and organisms officially registered in Entrez Gene and RefSeq. Records in RTPrimerDB are assigned unique and stable identifiers. The content is provided via an interactive web-based search system and is available for download in the recently developed RDML format and as bulk export file. RTPrimerDB is a one-stop portal for high-quality and highly annotated real-time PCR assays.
INTRODUCTION
RTPrimerDB is an integrative publicly available database for the storage, retrieval and analysis of primer and probe information. RTPrimerDB provides unique identifiers (RTPrimerDB ID) for any submitted assay for a given application, detection chemistry, target and organism. All assay information is tracked and integrated into an interactive query system and is directly accessible by using a dedicated URL linking. The information maintained includes primer sequences, target gene and organism information, mapping data, in silico evaluation characteristics, user feedback, links to citations, assays for the same target and external databases. Data submissions are preferentially linked to citations in peer-reviewed journals to ensure the integrity of the information provided.
The information stored in RTPrimerDB results from assay submissions by users, the RTPrimerDB administrator and project collaborators, from automated analyses and text mining and from external genome reference databases maintained by the National Center for Biotechnology Information [Entrez Gene and RefSeq; (1,2)], The Wellcome Trust Sanger Institute [Ensembl; (3)] and the University of California, Santa Cruz (4).
DATABASE CONTENT AND FUNCTIONALITY
The RTPrimerDB project was initiated to address the issue of laborious primer design and assay evaluation for quantification or detection of the same nucleic acid target sequences by different individuals, which significantly impedes standardized and assay uniformity. As such, the initial goal of RTPrimerDB was to facilitate the dissemination of information related to experimentally validated primer and probe assays submitted by experts in the field of real-time PCR (5). The information available for a specific assay is required for understanding the purpose of an assay, interpreting its suitability and implementing it in a wet lab experiment. In a second phase, we introduced an in silico assay evaluation pipeline to streamline the quality control of custom designed primer and probe sequences prior to ordering and experimental evaluation (6). The following features are recently added to RTPrimerDB and constitute the 2009 upgrade: the database and in silico evaluation system are improved and remodeled to allow the (i) submission; (ii) analysis of assays targeting sequences in many more species than before; (iii) consequently, RTPrimerDB has grown more than 10-fold in the last year; (iv) we have enabled the submission of primer sets used for the detection and quantification of transcription factor binding to target regions using chromatin immunoprecipitation (ChIP); (v) a new and advanced user feedback system is introduced to facilitate the submission of experimental evaluation reports for a given assay; (vi) a user and assay community system is developed to group users employing the same set of assays or assemble assays based on specific criteria; (vii) the evaluation pipeline is extended with a link to BiSearch, a more powerful primer alignment algorithm for assay specificity assessment (7); (viii) finally, all real-time PCR assays can be exported into the newly developed Real-time PCR Data Markup Language (RDML) format, a structured and universal data standard for exchanging quantitative PCR (qPCR) data (8).
Real-time PCR assay retrieval and new applications
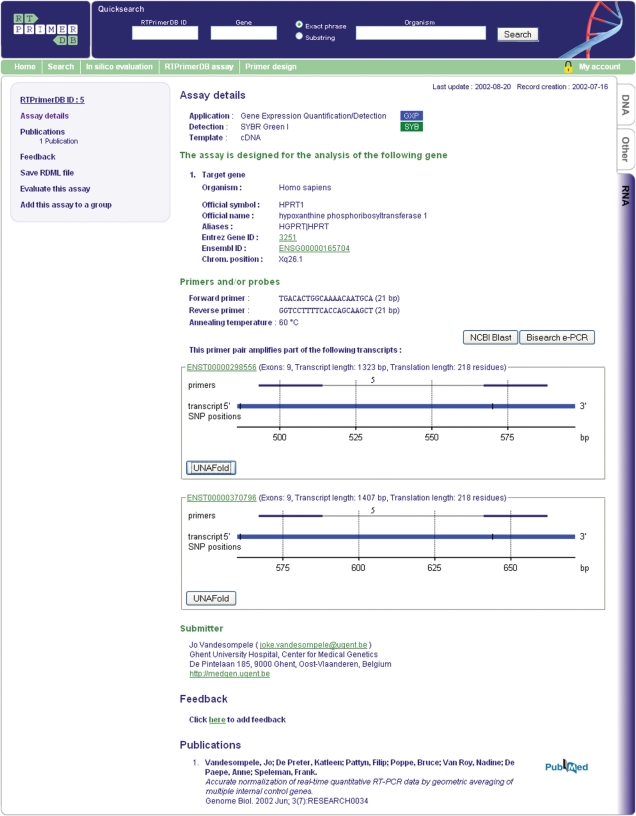
Figure 1 displays representative assay-specific information that can be retrieved through RTPrimerDB. The number of assays has significantly grown and now includes a multitude of target organisms. This greatly expands the potential use of RTPrimerDB to researchers in fields such as microbiology, plant genetics and veterinary sciences (Table 1). Although real-time quantitative PCR has been mostly used for gene expression profiling, the technology is now also being applied to increase the accuracy, ease of use and throughput of more advanced DNA quantification applications such as the detection of transcription factor target enrichment after ChIP. Users can submit and search for assays specifically designed for this application. These assays can be retrieved by searching on the target gene or on the transcription factor.
Figure 1.
Representative RTPrimerDB full-report page. The standard assay report starts with summary information on the target gene, the application and detection chemistry, a table listing the nucleotide sequences and experimental conditions, a mapping of the assay on the target gene, a list of citations, user remarks and finally the submitter's contact details. This page contains links to the various reference databases that enable a visitor to quickly find additional information about the target gene and the publications in which the assay is described. Additional links to BLAST and BiSearch allow a direct assessment of the assays specificity and an additional link to perform a secondary structure analysis is provided when the amplicon sequence is available.
Table 1.
Representative statistics of RTPrimerDB (September, 2008)
| Genes | Organisms | |
|---|---|---|
| Submitted assays | ||
| published in literature | 2970 | 10 |
| In silico assay evaluation | ||
| full featured | 323 147 | 11 |
| mapping and secondary structure analysis only | 329 789 | 16 |
| specificity search only | 475 204 | 20 |
| Assays published in literature | ||
| Detection chemistry use | SYBR Green I | 59% |
| TaqMan | 40% | |
| Others | 1% | |
| Target organism | Human | 70% |
| Mouse | 17% | |
| Rat | 10% | |
| Others | 3% | |
| Application | Gene expression | 97% |
| Others | 3% |
Real-time PCR assay submission
Data submission is possible after free registration to associate the assay information to a person. Additional links in the menu appear after successful login to guide the submitter through the submission process. Dependent on the application type, detection chemistry, target template (cDNA or DNA), the assay submission system requests all necessary information to build an assay report. In addition, when submitting primers for gene expression analysis of a gene for which the sequence and splicing information is available, the assays will be automatically mapped on the transcripts and screened for the presence of SNPs on the primer annealing sites. Thereafter, the amplicon sequences will be scrutinized using UNAFold for the presence of stable secondary structures putatively interfering with efficient primer annealing and amplification (9).
Real-time PCR assay feedback and community system
Users and submitters are invited to submit experimental feedback using a certain assay (melting curve analysis, agarose gel analysis, amplification efficiency assessment using a standard curve or single curve algorithms, etc.), or to provide the PubMed identifier of one or more publications employing an RTPrimerDB assay. This information is of great importance to score an assay on its usability and to confirm the results of the in silico assay evaluation analyses. This permits the introduction of an assay rank score based on the performance in different labs. We further introduced a community system for registered users to select a set of assays into a publicly visible or private group and to invite other users to join. A publicly visible group could be used to combine assays that are used as reference genes for a given condition. A private group is only visible for group members and allows combining private assays sets. We are convinced that this system will promote the selection of the most popular and best performing assays promoting their wide-scale use as a standard.
In silico assay evaluation
Real-time quantitative PCR became the gold standard for sequence detection and quantification due to its accuracy, speed and ease of use. It is important to realize that the technology relies on the efficiency of the primers used. Therefore, RTPrimerDB was extended with a primer and probe in silico evaluation system to detect potential assay features that could negatively influence the assay's efficiency or specificity (6). Here, we have introduced the use of the BiSearch alignment algorithm to evaluate assay specificity (7). This algorithm outperforms the classical BLAST alignment approach that is not efficient at analyzing and interpreting the alignment of two oligonucleotides on the same target sequence. BiSearch specifically screens for alignments of primers that are exponentially amplifiable.
Current statistics
As of September, 2008, there are more than 7000 records in RTPrimerDB, distributed among more than 10 different species (Table 1), a more than 2-fold increase compared to 2006. A detailed overview is available on the statistics page of the website. Assuming that RTPrimerDB's content of validated assays published in literature is representative for the general use of real-time PCR, it is clear that real-time PCR is generally used for gene expression analysis using SYBR Green I in human samples.
DATA ACCESS AND EXPORT
Access to RTPrimerDB data
The information in RTPrimerDB can be accessed in multiple ways. The most straightforward is to navigate to the database and to submit a query using the ‘search’ or ‘quick search’ page and display the results. The search engine can be queried by type of application, type of detection chemistry, organism, gene name, gene symbol (official or any synonym), Entrez Gene ID, SNP identifier or submitter's name. Each assay can be exported into an RDML file to allow the exchange of annotated qPCR primer and probe data between instrument software and third-party data analysis packages, between colleagues and collaborators, and between authors, peer reviewers, journals and readers (8,10). Direct links to an individual assay are available by using its unique RTPrimerDB ID in an URL (http://www.rtprimerdb.org/assay_report.php?assay_id=;<RTPrimerDB ID>). More complex direct query links can be constructed to select all assays targeting one specific gene, using a specific detection chemistry, used in a given application, submitted by a specific user, or cited in a given publication by adjusting the URL of the search results page. Finally, the database information is available from the download page as an export file that is updated on a weekly basis.
The database is freely accessible for the search and retrieval of assay information. The web interface is fully functional when using a recently updated internet browser. All figures displaying mapping information of the primers on their target sequence are constructed as scalable vector graphics (SVG), the web standard for zoomable and customizable high content vector graphics and supported by all recent internet browsers. Users are invited to submit their validated assays upon free registration to enable tracking of the submitted information and to provide advanced functionalities linked to a user account.
Data integrity through linking to external reference databases
The integrity of the information is maintained by linking assays to general reference databases storing nomenclature data (gene symbols and names), gene mapping information, SNP data, splicing information and publication data by using constant identifiers. Therefore, we are able to keep the information updated when new genome builds are released, or databases such as the SNP database, Entrez Gene or RefSeq are updated. We developed an extensive system to download, parse and integrate updated information from these external databases into RTPrimerDB. The frequency of the updates depends on the update policy utilized by the reference databases.
FEEDBACK
We welcome user feedback with respect to the RTPrimerDB interface, or any data contained therein. All comments can be posted using a form directly accessible from each page or by email to rtprimerdb@medgen.ugent.be.
FUNDING
Ghent University Special Research Fund (BOF) (doctoral grant to S.L.; postdoctoral grant to F.P.). Funding for open access charges: Ghent University Special Research Fund.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We are grateful to Joris S’heeren and Elke Van Vlierberghe from the Center for Medical Genetics, Ghent for their technical support.
REFERENCES
- 1.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pattyn F, Speleman F, De Paepe A, Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res. 2003;31:122–123. doi: 10.1093/nar/gkg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattyn F, Robbrecht P, De Paepe A, Speleman F, Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database, major update 2006. Nucleic Acids Res. 2006;34:D684–D688. doi: 10.1093/nar/gkj155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aranyi T, Tusnady GE. BiSearch: ePCR tool for native or bisulfite-treated genomic template. Methods Mol. Biol. 2007;402:385–402. doi: 10.1007/978-1-59745-528-2_20. [DOI] [PubMed] [Google Scholar]
- 8.Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz PA, Bogue M, Booth T, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat. Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 10.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]