Abstract
The composition of isolated floras has long been thought to be the result of relatively rare long-distance dispersal events. However, it has recently become apparent that the recruitment of lineages may be relatively easy and that many dispersal events from distant but suitable habitats have occurred, even at an infraspecific level. The evolution of the flora on the high mountains of Africa has been attributed to the recruitment of taxa not only from the African lowland flora or the Cape Floristic Region, but also to a large extent from other areas with temperate climates. We used the species rich, pan-temperate genera Carex, Ranunculus and Alchemilla to explore patterns in the number of recruitment events and region of origin. Molecular phylogenetic analyses, parametric bootstrapping and ancestral area optimizations under parsimony indicate that there has been a high number of colonization events of Carex and Ranunculus into Africa, but only two introductions of Alchemilla. Most of the colonization events have been derived from Holarctic ancestors. Backward dispersal out of Africa seems to be extremely rare. Thus, repeated colonization from the Northern Hemisphere in combination with in situ radiation has played an important role in the composition of the flora of African high mountains.
Keywords: biogeography, test of monophyly, dispersal, colonization, Afroalpine, island floras
1. Introduction
Isolated floras such as on Hawaii, the Galápagos and the Macaronesian Islands have long been thought to have evolved following a minimum of rare long-distance dispersal events (Cowie & Holland 2006; Lomolino et al. 2006). However, recent evidence indicates that long distance dispersal may be more frequent than previously recognized (de Queiroz 2005; Levin 2006; Heaney 2007; Harbaugh et al. 2009; Schaefer et al. 2009). Alsos et al. (2007) even suggested that plant recruitment to the isolated Arctic island Svalbard is limited by establishment and not by dispersal. They showed additionally that Svalbard was colonized from various source areas. This importance of geographically widespread recruitment areas in the assembly of floras was also demonstrated for the Cape flora, where lineages were traced from South America, North America, Eurasia, Africa and Australasia (Galley & Linder 2006). Similarly, far flung source areas have been documented for the flora of Hawaii (Wagner et al. 1990) and New Zealand (McGlone et al. 2001; Winkworth et al. 2005; McDowall 2008).
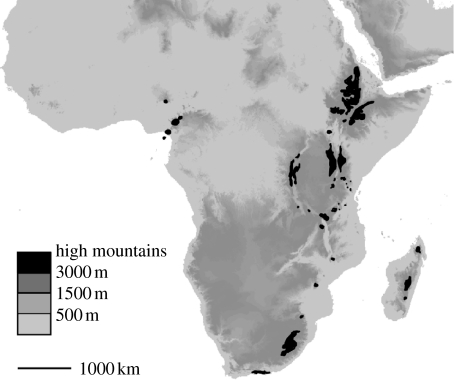
The cool-adapted flora of the high mountains in tropical and southern Africa (figure 1) is sharply distinct from the lowland tropical flora. Whereas the tropical flora is mainly composed of pan-African genera belonging to Takhtajan's African subkingdom (Takhtajan 1986), the mountain flora contains elements derived from the Cape Floristic Region, the Mediterranean, the Northern Hemispheric and the Southern Hemispheric temperate regions, in addition to some lineages derived from the lowland tropical flora (Engler 1892, 1904; Hedberg 1961, 1965, 1970). These scattered high mountains and mountain ranges can, by analogy to oceanic islands, be referred to as ‘sky islands’. Their cool-adapted floras have always been well separated from each other, and also from extra-African areas with similar climatic conditions and vegetation types, by large expanses of much drier surrounding lowland habitats (Hedberg 1970; Brühl 1997; Cohen et al. 1997; Menocal de 2004). There have only been few biogeographical analyses of taxa from the African mountain flora. A number of groups from the Cape Floristic element, for example, were shown to be derived from several unidirectional colonization events, which entered the tropical mountains from the south (Galley et al. 2007). Most of these colonizations from the Cape Floristic Region resulted in small radiations in the tropical mountains. Other species (e.g. Arabis alpina, Lychnis and Cardamine) have been shown to have been derived from colonizations from the north (Koch et al. 2006; Carlsen et al. 2007; Popp et al. 2008). However, patterns of recruitment of the pan-temperate element, which constitutes the largest component of this flora (Hedberg 1961), are not known.
Figure 1.
Distribution of the high mountain areas in Tropical and Southern Africa (black).
Here, we use three typical pan-temperate genera (Carex, Ranunculus and Alchemilla) to investigate the patterns in the recruitment of this floral element. Considering the enormous distances between the African mountains to other locations of the pan-temperate flora, we ask whether the pan-temperate recruitment was by a minimal number of dispersals (comparable with the situation in Hawaii) or repeated dispersal by the same species (comparable with the Svalbard model). Secondly, we investigate whether the pan-temperate taxa are recruited from all areas where they occur (as seems to be the normal situation), or from a limited subset of regions. Finally, we investigate whether reverse colonization, that is, out of Africa, occurs.
2. Material and methods
(a) Taxon sampling
We focused on the more or less globally distributed, mostly cool-climate (i.e. pan-temperate) genera Carex, Ranunculus and Alchemilla, which are species rich, geographically widespread and have a wide altitudinal range in the African high mountain flora. We refer to the African high mountain flora as species found above 2300 m in Tropical Africa, 1500 m in Southern Africa outside the Cape Floristic Region as well as on Madagascar above 2000 m. We included as many taxa as possible from outside the African high mountains to improve the reliability of the monophyly test (appendices 1 and 2 in the electronic supplementary material). Where possible, multiple accessions for each species from geographically distant locations in Africa were included. This was done to test the monophyly of the species and to detect problems arising from introgression and paralogues that are commonly found in highly polyploid groups such as Carex, Ranunculus and Alchemilla.
The genus Carex (or the tribe Cariceae, if the endemic African genus Schoenoxiphium is included) is, with approximately 2000 species, the largest clade in the Cyperaceae. The section Vigneastra, also referred to as subgenus Indocarex, comprises mainly tropical or subtropical Carex species, the African distribution of which is largely outside the African high mountain regions. Taxa of Vigneastra were therefore only included to test the affiliation of individual species to the section, and were excluded from the biogeographical inferences made. There is no recent revision or monograph of Carex; however, Gehrke's (2008) synopsis of sub-Saharan and Madagascan Carex lists 81 species for the area. Out of these 81 species, 43 occur on the high mountains, the remaining species either belong to section Vigneastra (30 species), occur only in lowland areas of Eritrea and Madagascar (2 species), the Cape Floristic Region (4 species) or have been introduced very recently (2 species). Within the tribe Cariceae, an additional 20 of the approximate 25 species of Schoenoxiphium are present in the African high mountain areas. Owing to the availability of data from a number of recent molecular phylogenetic studies of Carex (Yen & Olmstead 2000; Roalson et al. 2001; Hendrichs et al. 2004a,b; Roalson & Friar 2004; Starr et al. 2004), we were able to include over 600 accessions representing approximately 550 species in our analyses. Cariceae from the high African mountains are represented by 82 accessions of 43 taxa, which accounts for 68 per cent of the 63 taxa from the high mountains in Africa in the subtribe. An additional 18 accessions represent 14 taxa of Indocarex All other sequences were downloaded from NCBI GenBank (appendix 1 in the electronic supplemetary material). The total matrix of 602 accessions included 554 sequences of nuclear ITS and 365 of the chloroplast marker trnL–trnF.
With approximately 600 species, Ranunculus (the buttercups) is the largest genus within Ranunculaceae (Tamura 1995). Buttercups are cosmopolitan. Most species occur in temperate to Arctic/subantarctic regions (Paun et al. 2005), in terrestrial or aquatic habitats and in coastal to alpine vegetation. In the tropics, they are restricted to tropic-alpine or high mountain habitats. Two hundred and thirty Ranunculus species were included in the analyses, representing material from all continents and including representatives of all subgenera and sections (Hörandl et al. 2005; Paun et al. 2005). Out of the 232 accessions of Ranunculus analysed, 27 represent 16 out of the 20 African taxa (80%), missing only four species endemic to Ethiopia and the Rwenzori Mountains. New sequences for African material were generated for nuclear ITS and a chloroplast marker (matK; appendix 2 in the electronic supplemetary material), and all other accessions were downloaded from NCBI GenBank.
The genus Alchemilla has a global distribution (Fröhner 1995). According to Gehrke et al. (2008), the African members form two clades, referred to informally as ‘the Afromilla clade’ and the ‘Aphanes clade’. The Afromilla clade is endemic to tropical Africa (approx. 20–25 species), southern Africa (4–6 species) and Madagascar (6 endemic species). Most herbaceous Alchemilla species in Africa are widespread, whereas many woody taxa are found only on single mountains (i.e. Rwenzori) or montane regions (Ethiopia or Tropical East Africa). In addition to the Afromilla clade, a single species of Alchemilla subgenus Aphanes occurs in Ethiopia (A. bachiti). However, it is rare and we were not able to sample it for this study. Sequence alignment of nuclear ITS and chloroplast trnL–trnF, as well as the molecular phylogenetic reconstruction, was as described in Gehrke et al. (2008).
(b) DNA extraction, PCR amplification and sequencing
Leaf material was pulverized using a Regget Machine and DNA was extracted using DNeasy extraction kit (Qiagen). Polymerase chain reactions (PCR) were performed in 25 μl reactions (1×PCR buffer, with 2.5 mM MgCl2, 0.25 mM dNTPs, 1.6 μM primers and 1 unit of Taq polymerase (Sigma-Aldrich, USA) in a Biometra Thermocycler TGradient (Biometra, Göttingen, Germany). Either 2 μl BSA or 1 μl BSA and 1 μl EDTA was added to each reaction. The ITS1–5.8S–ITS2 region and the trnL-F intergenic spacer together with the trnL intron were amplified and sequenced using primers ITS-I and ITS4 (Urbatsch et al. 2000) and trnLF-c and trnLF-f (Taberlet et al. 1991) for Carex. For Ranunculus, the protocol of Hörandl et al. (2005) for the ITS and matK-region was used. PCR products were purified using DNA band purification kit (Amersham Biosciences, Otelfingen, Switzerland). Cycle sequencing was carried out on an ABI Prism, LA, USA 3100 Genetic Analyzer (Applied Biosciences, Foster City, CA, USA) using BigDye Terminator v. 3.1. Forward and reverse strand sequences were edited using Sequencer v. 4.2. (Genecode) and aligned by hand using Sequence Alignment Editor v. 2.0 (Rambaut 2002).
(c) Phylogenetic analyses
Simple indel coding (Simmons & Ochoterena 2000) was performed by hand for Cariceae and using SeqState v. 1.32 for Ranunculus (Müller 2005), excluding ambiguous areas in the alignment. An initial analysis of Cariceae including all taxa for the ITS and trnLF separately using TNT with standard ‘new technology’ settings (Goloboff et al. 2003) identified three clades of taxa corresponding to subgenera Carex (including Vigneastra), Vignea and Psyllophora, with the latter including Kobresia, Schoenoxiphium and Uncinia sensu (Kükenthal 1909; appendix 4 in the electronic supplemetary material). Subsequent parsimony analyses were performed on these three clades separately as well as on Ranunculus, using PAUP* v. 4.0.b10 (Swofford 2001), with heuristic search of 1000 replicates, random sequence addition, tree bisection-reconnection branch swapping, MULTREE on (keeping multiple, equally parsimonious trees), saving a maximum of 50 trees per replicate. Support was assessed using 1000 replicates of non-parametric bootstrap analysis using the same settings as in the heuristic search. The nuclear and plastid loci of Cariceae and Ranunculus were analysed separately, and since no conflicting nodes received bootstrap support 70 per cent or more, the loci were combined. The phylogenetic trees of Alchemilla, based on ITS and trnL–trnF data, were taken from Gehrke et al. (2008) and were constructed using the same methods as those for Cariceae and Ranunculus.
Bayesian analysis was performed as implemented in MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001). We identified the general time-reversible model with gamma-distributed rates (G) and a proportion of invariable characters (I) as best fitting the sequence data of the individual loci in the Cariceae and Ranunculus matrices, using Modeltest v. 3.7 (Posada & Crandall 1998) applying the Akaike information criterion. The prior values for the number of parameters in the DNA substitution models were therefore set to NST=6+G+I. The dataset was partitioned into ITS and trnLF loci, and parameter values of each varied independently. These settings were duplicated for two or three independent Markov chain Monte Carlo runs of each matrix (as below), with three heated and one cold chain each, sampling every 1000 generations from the cold chain.
In Cariceae, Bayesian analyses were carried out separately for each of the subgenus taxon sets with the combined data, and with the following number of generations and runs: Psyllophora taxon set, 2×2 000 000 generations; Carex subgenus Vignea, 3×5 000 000 generations; for subgenus Carex, 2×10 000 000 generations. Ranunculus was analysed using two separate, independent runs of 5 000 000 generations. Convergence and sampling of model parameters were estimated using the program Tracer v. 1.3 (Rambaut & Drummond 2005). Topologies sampled prior to convergence to the optimal mean log likelihood plateau were discarded as burn-in (2×3 000 000 generations for Carex subgenus Carex, 3×1 000 000 generations for Carex subgenus Vignea, 2×1 000 000 generations for Carex section Psyllophora and 2×1 000 000 generations for Ranunculus), and clade posterior probabilities were computed from the remaining trees as a measure of node support.
(d) Parametric bootstrapping as a test of monophyly
We determined the minimum number of colonization events needed to constitute the African high mountain floras of the study groups, by locating the largest possible clades with an ancestral area in the high mountains of Tropical and Southern African. We used parametric bootstrapping (Huelsenbeck et al. 1996) to evaluate whether the monophyly of even larger, more inclusive clades is rejected by the data (which would reduce the number of colonization events needed). We investigated whether the increase in tree length caused by constraining the monophyly of such groups of African taxa could be due to stochastic processes of sequence evolution. For details on parametric bootstrapping and the different constraints used, see appendix 4 in the electronic supplemetary material.
(e) Ancestral area reconstruction
In order to locate the geographical origin of each African high mountain clade, the distribution ranges of their ancestral nodes were reconstructed using unambiguous parsimony optimization as implemented in Mesquite v. 2.0 (Maddison & Maddison 2006). We incorporated phylogenetic uncertainty by using all most parsimonious trees, summarizing the resulting optimizations on the Adams consensus tree, one of the most parsimonious trees and the majority rule consensus. The Adams consensus was used because it is particularly useful for identifying common tree structure when one or more taxa have very different positions in the trees of the profile. The areas used were (i) African high mountains, (ii) western temperate and boreal Eurasia, (iii) eastern temperate and boreal Eurasia (iv) the Mediterranean region including North Africa and Macaronesia, (v) North America, (vi) South America and (vii) Australasia. The Cape Floristic Region was not coded separately since only one of the taxa included the analyses is restricted to this area.
3. Results
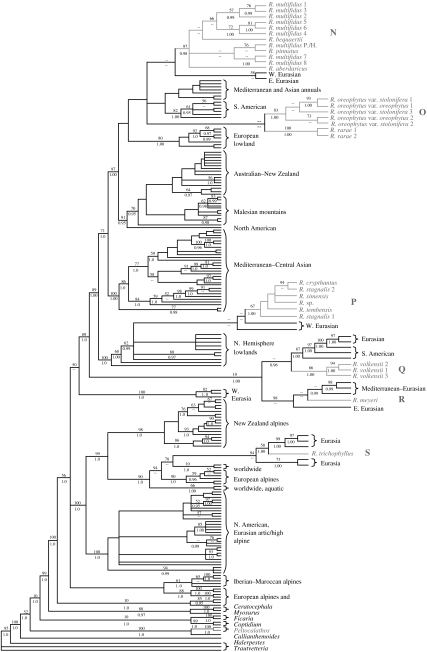
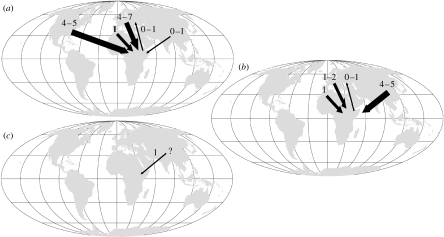
A summary of each clade of African high mountain taxa used in our analyses can be found in table 1, and the consensus tree of Ranunculus from the parsimony analysis is presented in figure 2 as an example of the phylogenetic trees. For a more detailed description of these clades and the phylogenetic trees, see the electronic supplementary material (appendices 3 and 5 in the electronic supplemetary material). There were at least 13 colonization events into the African high mountains by Carex and 4–6 colonization events by Ranunculus (figure 3). Parametric bootstrapping confirmed the non-monophyly of the African high mountain clades, indicating that they have been derived from different non-African high mountain ancestors (appendix 4 in the electronic supplemetary material). In some cases, the closest related species could not be identified due to weak support values or incomplete taxon sampling (appendix 5 in the electronic supplemetary material). However, all African high mountain clades are nested within Northern Hemisphere clades, and the inferred colonizations were mainly from temperate Eurasia (appendix 6 in the electronic supplemetary material). In two cases, dispersals out of the African high mountains could not be rejected; however, no such event is supported (appendix 3 in the electronic supplemetary material). Parsimony optimizations revealed four to eight colonizations by Carex from temperate Eurasia, one to three from the Mediterranean and four to five from North America. For Ranunculus, parsimony optimizations suggest five to six colonizations from temperate and boreal Eurasia and one from the Mediterranean, none from North America and none from South America (figure 3). Alchemilla (i.e. the Afromilla clade) was reconstructed as originating from eastern Eurasia (appendix 6 in the electronic supplemetary material).
Table 1.
African high mountain clades in molecular phylogenetic analysis of Carex, Ranunculus and Alchemilla. (Bootstrap support values and Baysian clade credibilities of the monophyly of the clades in the combined analyses of ITS and trnLF are given. Bootstrap support values are indicated at the left and Bayesian clade credibilities in brackets. Clades with higher support for one marker region are marked with an asterisk and the value given after the slash. E. Eura., eastern Eurasia; W. Eura., western Eurasia; Med., Mediterranean and Northern Africa; N-Am., North America.)
| clade names | clade support | reconstructed ancestral area |
|---|---|---|
| Carex subgenus Carex | ||
| A | −/67* (0.93) | Med. |
| B | one accession | W.Eura. |
| C | 99 (1.00) | W.Eura. |
| D | 70 (−) | N-Am. |
| E | 66/89*(−) | N-Am./E.Eura. |
| F | 58 (−) | N-Am. |
| G | 99 (1.00) | W.Eura |
| Carex subgenus Primocarex and other Cariceae | ||
| H | 100 (1.00) | N-Am. |
| I (Schoenoxiphium) | − (0.99) | W.Eura. |
| J | 100 (1.00) | W.Eura. |
| Carex subgenus Vignea | ||
| K | 81/91*(0.99) | N-Am. |
| L | 100 (−) | W.Eura./Med. |
| M | 97 (1.00) | W.Eura/Med. |
| Ranunculus | ||
| N | 97/0.98 | E.Eura. |
| O | − (−) | E.Eura. (0–2) |
| P | 67/1.00 | Med. |
| Q | 86 (1.00) | E.Eura. |
| R | one accession | E.Eura. |
| S | one accession | W.Eura. |
| Alchemilla | ||
| Afromilla clade | 100 (1.00) | E. Eur.? |
Figure 2.
Strict consensus tree of Ranunculus based on the combined nuclear ITS and matK dataset reconstructed using parsimony; bootstrap support values above 50 are given above branches and Bayesian clade credibility values above 0.95 are given below branches. Accessions from Africa are indicated by grey type. Non-African taxa are represented by their distribution according to Hörandl et al. (2005). A full tree description is found in the electronic supplementary material appendix 5.
Figure 3.
Results of ancestral area reconstructions. Thickness of arrows represents the number of inferred colonization events into the African high mountain areas. (a) Carex/Cariceae; (b) Ranunculus; (c) Alchemilla (Afromilla clade).
4. Discussion
(a) Multiple recruitments into the high mountains in Tropical and Southern African
We show that Alchemilla, Carex and Ranunculus colonized the high mountains of Africa several times. This pattern has also been reported for other temperate groups present in the African mountains such as Myosotis, Swertia, Cardamine and Trifolium (Chassot et al. 2001; Bleeker et al. 2002; Winkworth et al. 2002a; Ellison et al. 2006; Carlsen et al. 2007), as well as for clades centred in the Cape Floristic Region but which reach into the high African mountains, e.g. Pentaschistis, Disa, Iridaceae (Galley & Linder 2006). Hedberg (1965) postulated that two-thirds of all species of the high African mountains are derived from other areas, a proportion greatly in excess of that estimated here for Alchemilla, Carex and Ranunculus. However, the numbers of dispersal events to ‘islands’ are usually underestimated, especially in the absence of phylogeographic studies. Where there is no evidence to the contrary, members of a taxon are assumed to have resulted from single-colonization event followed by in situ radiation (Grant 1996; Alsos et al. 2007), as has been reported for most taxa of the Hawaiian flora (Price & Wagner 2004). This assumption may be incorrect. For example, A. alpina colonized the African high mountains from the Holarctic at least twice (Koch et al. 2006). Furthermore, the number of recruitments can be underestimated when taxon delimitation itself has been influenced by geographical distribution, as is often the case in large groups with extensive distribution areas (such as Carex and Ranunculus). Consequently, even the high numbers of colonization events reported here (13 for Carex and 4–6 for Ranunculus) are likely to be underestimates of the real number of colonizations, and therefore of dispersal events, into the high mountain flora of Africa.
These results illustrate the frequency of multiple recruitments into the African mountains. Nevertheless, single colonizations followed by in situ differentiation, which are often associated with the assembly of island floras, have been reported for some plant groups, e.g. Lychnis (Popp et al. 2008) and the Giant Senecio, Dendrosenecio (Knox & Palmer 1995; Pelser et al. 2007). Many African genera or supra-generic taxa on the high mountains have been established from single colonizations, e.g. the alpine Chironomidae (Insecta: Diptera; Eggermont & Verschuren 2007), several flightless insect genera (Brühl 1997), the Ethiopian wolf (Canis simensis; Gottelli et al. 2004) and several groups of birds (Fjeldså & Lovett 1997; Roy 1997; Johansson et al. 2007).
(b) Lack of support for dispersal out of the African high mountains
We have little evidence for dispersal out of Africa in general and out of the African high mountains in particular. Increased taxon sampling and better phylogenetic resolution might yet reveal such events, in particular in Carex acutiformis and in the clades N and O of Ranunculus (figure 2). The former was thought to occur naturally only in Europe and hence introduced to South Africa and North America by European settlers. However, C. acutiformis may be nested in a clade with the southern African Carex aethiopica and Carex drakensbergensis. Consequently, the most parsimonious interpretation is dispersal from Africa to Europe. Phylogenetic uncertainty also limits our conclusions with regard to Ranunculus praemorsus and Ranunculus vaginatus. The ancestral area reconstructions for some of the most parsimonious trees (though not for the Adams consensus) lead us to infer dispersal out of the African high mountains (for the Adams consensus see appendix 6 in the electronic supplemetary material, others not shown). Reverse dispersal appears to be similarly rare towards the south, and no instances of dispersal from the African high mountains into the Cape flora have been substantiated (Galley et al. 2007).
Contrary to these results, dispersal from islands to the mainland has been reported for both animals (Heaney 2007) and plants (Canary Islands: Allan et al. 2004; New Zealand: Winkworth et al. 2002b). There are several possible explanations for this unusual pattern in the African high mountain flora. First, this might be the result of the small total area occupied by the flora of the high mountains in African, and consequently of the low number of species and relatively small populations they support compared with other temperate areas. Second, the relative youth of the system (which was probably formed during the Plio–Pleistocene) might have reduced the opportunity for dispersal from the high mountains of tropical and southern Africa to other areas.
(c) The Holarctic as a source of recruitment of the high mountains in Africa
All clades from the high mountains in Africa in the three pan-temperate genera investigated here were recruited from temperate areas in the Northern Hemisphere. The source areas of Carex clades include not only Eurasia but also North America, which is surprising given the distance between North America and the African mountains (approx. 10 000 km, table 2). This suggests that either distance alone is not the limiting factor for dispersal in Carex or that our ancestral area reconstructions might have been influenced by limited taxon sampling. For example, the high number of inferred colonization events of Carex from North America could be due to the North American bias in taxon sampling, since eastern Eurasian Carex is relatively undersampled. Dispersal from North American to Africa is, however, not without precedent, for example in Pomaria (Simpson et al. 2006). It is interesting that there does not seem to be any recruitment from the mountain areas in the tropics (such as the Andean páramo or Malesian mountain regions) and extra-African southern temperate and mountainous areas (e.g. the Australia or New Zealand Alps, or the southern Andes), even though all three genera are well represented in these areas. The same pattern is shared by other temperate elements in the African sky-islands, such as A. alpina, Trifolium, Cardamine and Lychnis (Ellison et al. 2006; Koch et al. 2006; Carlsen et al. 2007; Popp et al. 2008). The predominance of a northern origin for high-montane or alpine floras has also been demonstrated for the Andes (Von Hagen & Kadereit 2001; Hughes & Eastwood 2006; Sklenar & Balslev 2007) and New Zealand (Winkworth et al. 2005). However, clades once established on these mountains often colonize successfully across the southern Pacific Ocean, and across the Tasman Sea (Von Hagen & Kadereit 2001; Crisp et al. 2004; Glenny 2004; Winkworth et al. 2005; Eggens et al. 2007), forming part of a worldwide alpine-temperate element (Smith & Cleef 1988).
Table 2.
Distance between different putative source regions and the African high mountains.
| area | distance (km) |
|---|---|
| Western Eurasia | 3500–5000 |
| Mediterranean | 2000–3500 |
| Eastern Eurasia | 4000–5000 |
| temperate North America | 11 000–13 000 (Cameroon 10 000) |
| temperate Hawaii | 16 000–18 000 |
| temperate South America | 8000–9000 |
| Páramo | 10 000–12 000 |
| temperate Malesia | 7000–10 000 (Madagascar 6000) |
| aseasonal-wet Australia | 10 000–12 000 (Madagascar 9000) |
| New Zealand | 10 500–13 000 |
Our results show that the African pan-temperate element as identified by Engler (1904) and Hedberg (1961) is probably entirely Northern Hemisphere derived and therefore Holarctic, making the Holarctic the most important source of lineage recruitment of the African high mountain flora of Tropical and Southern Africa. We predict that this also applies to other important temperate elements worldwide, such as Poa, Festuca, Agrostis, Cardamine, Cerastium and Luzula.
Acknowledgements
We thank Muthama Muasya, Eric Knox, Adane Assefa, Mulugeta Kebede, Sebsebe Demissaw, Desalegn Chala, Cyril Guibert, Timo van der Niet, Chloé Galley, Jaqueline Razanatsoa, Hassam I. Patal, Frank Mbago, Michael Pirie and Jonathan Kissling for their help in the field. We would also like to thank Cyril Guibert for sequence data of a number of Carex species. The manuscript was improved following helpful and detailed comments from Michael Pirie, Anthony Rees, Susanne Renner and an anonymous reviewer. The research was funded by the University of Zurich, and the fieldwork supported by the Georges- and Antoine-Claraz-Schenkung and the Swiss National Science Council (SNCAT). Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University, which is partially funded by Microsoft Corporation. Species identification was carried out with the help of SYNTHESYS (made available by the European Community—Research Infrastructure Action) grants to Berit Gehrke BE-TAF-1531, FR-TAF-836 and SE-TAF-837.
Supplementary Material
Accession details of taxa used in molecular phylogenetic analyses including authors, subgeneric classification of Carex, voucher details, references to first publication and NCBI Genebank numbers
Overview of Carex, Ranunculus and Alchemilla, their distribution and species numbers. Additionaly, the total number of taxa per group and the number of accessions are reported
All clades retrieved from the molecular phylogenetic analyses which include accessions from African are described and discussed in detail
A detailed account of the methodology of parametric bootstrap analyses is given together with all details of the constraints performed
Detailed tree description of all molecular phylogenetic trees are given as well as the relevant parts of the trees from the ancestral area reconstructions
References
- Allan G.J., Francisco-Ortega J., Santos-Guerra A., Boerner E., Zimmer E.A. Molecular phylogenetic evidence for the geographic origin and classification of Canary Island Lotus (Fabaceae: Loteae) Mol. Phylogenet. Evol. 2004;32:123–138. doi: 10.1016/j.ympev.2003.11.018. doi:10.1016/j.ympev.2003.11.018 [DOI] [PubMed] [Google Scholar]
- Alsos I.G., Eidesen P.B., Ehrich D., Skrede I., Westergaard K., Jacobsen G.H., Landvik J.Y., Taberlet P., Brochmann C. Frequent long-distance plant colonization in the changing Arctic. Science. 2007;316:1606–1608. doi: 10.1126/science.1139178. doi:10.1126/science.1139178 [DOI] [PubMed] [Google Scholar]
- Bleeker W., Franzke A., Pollmann K., Brown A.H.D., Hurka H. Phylogeny and biogeography of Southern Hemisphere high-mountain Cardamine species (Brassicaceae) Aust. Syst. Bot. 2002;15:575–581. doi:10.1071/SB01026 [Google Scholar]
- Brühl C. Flightless insects: a test case for historical relationships of African mountains. J. Biogeogr. 1997;24:233–250. doi:10.1046/j.1365-2699.1997.00073.x [Google Scholar]
- Carlsen, T., Bleeker, W., Hurka, H., Elven, R. & Brochmann, C. 2007 Biogeography and phylogeny of Cardamine (Brassicaceae) In Are there any trees in the Arctic? Reconstruction of evolutionary histories in a young biome (ed. T. P. t. Carlsen ), paper III. Oslo, Norway: Faculty of Mathematics and Natural Science, University of Oslo.
- Chassot P., Nemomissa S., Kupfer P. High paraphyly of Swertia L. (Gentianaceae) in the Gentianella-lineage as revealed by nuclear and chloroplast DNA sequence variation. Plant Syst. Evol. 2001;229:1–21. doi:10.1007/s006060170015 [Google Scholar]
- Cohen A.S., Lezzar K.-E., Tiercelin J.-J., Soreghan M. New palaeogeographic and lake-level reconstructions of Lake Tanganyika: implications for tectonic, climatic and biological evolution in a rift lake. Basin Res. 1997;9:107–132. doi:10.1046/j.1365-2117.1997.00038.x [Google Scholar]
- Cowie R.H., Holland B.S. Dispersal is fundamental to biogeography and the evolution of biodiversity on oceanic islands. J. Biogeogr. 2006;33:193–198. doi:10.1111/j.1365-2699.2005.01383.x [Google Scholar]
- Crisp M., Cook L., Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Phil. Trans. R. Soc. Lond. B. 2004;359:1551–1571. doi: 10.1098/rstb.2004.1528. doi:10.1098/rstb.2004.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz A. The resurrection of oceanic dispersal in historical biogeography. Trends Ecol. Evol. 2005;20:68–73. doi: 10.1016/j.tree.2004.11.006. doi:10.1016/j.tree.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Eggens F., Popp M., Nepokroeff M., Wagner W.L., Oxelman B. The origin and number of introductions of the Hawaiian endemic Silene species (Caryophyllaceae) Am. J. Bot. 2007;94:210–218. doi: 10.3732/ajb.94.2.210. doi:10.3732/ajb.94.2.210 [DOI] [PubMed] [Google Scholar]
- Eggermont H., Verschuren D. Taxonomy and diversity of Afroalpine Chironomidae (Insecta: Diptera) on Mount Kenya and the Rwenzori mountains, East Africa. J. Biogeogr. 2007;34:69–89. doi: 10.1111/j.1365-2699.2006.01590.x. doi:10.1111/j.1365-2699.2006.01590.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison N.W., Liston A., Steiner J.J., Williams W.M., Taylor N.L. Molecular phylogenetics of the clover genus (Trifolium—Leguminosae) Mol. Phylogenet. Evol. 2006;39:688–705. doi: 10.1016/j.ympev.2006.01.004. doi:10.1016/j.ympev.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Engler A. Verlag der Königl. Akademie der Wissenschaften; Berlin, Germany: 1892. Über die Hochgebirgsflora des tropischen Afrika. [Google Scholar]
- Engler A. Plants of the northern temperate zone in their transition to the high mountains of Tropical Africa. Ann. Bot. 1904;18:523–540. [Google Scholar]
- Fjeldså J., Lovett J.C. Geographical patterns of old and young species in African forest biota: the significance of specific montane areas as evolutionary centres. Biodivers. Conserv. 1997;6:325–346. doi:10.1023/A:1018356506390 [Google Scholar]
- Fröhner, S. E. 1995 Alchemilla In Illustrierte Flora von Mitteleuropa, vol. 4 (eds H. Scholz, H. J. Conert, E. J. Jäger, J. W. Kadereit, W. Schultze-Motel, G. Wagenitz & H. E. Weber), 13–242. Berlin, Germany: Paul Parey.
- Galley C., Linder H.P. Geographical affinities of the Cape flora, South Africa. J. Biogeogr. 2006;33:236–250. doi:10.1111/j.1365-2699.2005.01376.x [Google Scholar]
- Galley C., Bytebier B., Bellstedt D.U., Linder H.P. The cape element in the Afrotemperate flora: from Cape to Cairo? Proc. R. Soc. B. 2007;274:535–543. doi: 10.1098/rspb.2006.0046. doi:10.1098/rspb.2006.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke, B. 2008 Pan-temperate plant elements on the African high mountains. PhD thesis, at the Institute of Systematic Botany, University of Zurich, Zurich, Switzerland.
- Gehrke B., Bräuchler C., Romoleroux K., Lundberg M., Heubl G., Eriksson T. Molecular phylogenetics of Alchemilla, Aphanes and Lachemilla (Rosaceae) inferred from plastid and nuclear intron and spacer DNA sequences, with comments on generic classification. Mol. Phylogenet. Evol. 2008;47:1030–1044. doi: 10.1016/j.ympev.2008.03.004. doi:10.1016/j.ympev.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Glenny D. A revision of the genus Gentianella in New Zealand. NZ J. Bot. 2004;42:361–530. [Google Scholar]
- Goloboff, P., Farris, S. & Nixon, K. 2003 T.N.T. Tree analysis using new technology. Program and documentation. See http://www.zmuc.dk/public/phylogeny/tnt/
- Gottelli D., Marino J., Sillero-Zubiri C., Funk S.M. The effect of the last glacial age on speciation and population genetic structure of the endangered Ethiopian wolf (Canis simensis) Mol. Ecol. 2004;13:2275–2286. doi: 10.1111/j.1365-294X.2004.02226.x. doi:10.1111/j.1365-294X.2004.02226.x [DOI] [PubMed] [Google Scholar]
- Grant P.R. Oxford University Press; Oxford, UK: 1996. Evolution on islands. [Google Scholar]
- Harbaugh D.T., Wagner W.L., Allan G.J., Zimmer E.A. The Hawaiian Archipelago is a stepping stone for dispersal in the Pacific: an example from the plant genus Melicope (Rutaceae) J. Biogeogr. 2009;36:230–241. doi:10.1111/j.1365-2699.2008.02008.x [Google Scholar]
- Heaney L.R. Is a new paradigm emerging for oceanic island biogeography? J. Biogeogr. 2007;34:753–757. doi:10.1111/j.1365-2699.2007.01692.x [Google Scholar]
- Hedberg O. The phytogeographical position of the afroalpine flora. Rec. Adv. Bot. 1961;1:914–919. [Google Scholar]
- Hedberg O. Afroalpine flora elements. Webbia. 1965;19:519–529. [Google Scholar]
- Hedberg O. Evolution of the afroalpine flora. Biotropica. 1970;2:16–23. doi:10.2307/2989783 [Google Scholar]
- Hendrichs M., Michalski S., Begerow D., Oberwinkler F., Hellwig F.H. Phylogenetic relationships in Carex, subgenus Vignea (Cyperaceae), based on ITS sequences. Plant Syst. Evol. 2004a;246:109–125. doi:10.1007/s00606-004-0127-1 [Google Scholar]
- Hendrichs M., Oberwinkler F., Begerow D., Bauer R. Carex, subgenus Carex (Cyperaceae)—a phylogenetic approach using ITS sequences. Plant Syst. Evol. 2004b;246:89–107. doi:10.1007/s00606-004-0128-0 [Google Scholar]
- Hörandl E., Paun O., Johansson J.T., Lehnebach C., Armstrong T., Chen L., Lockhart P. Phylogenetic relationships and evolutionary traits in Ranunculus s.l. (Ranunculaceae) inferred from ITS sequence analysis. Mol. Phylogenet. Evol. 2005;36:305–327. doi: 10.1016/j.ympev.2005.02.009. doi:10.1016/j.ympev.2005.02.009 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Hillis D.M., Nielsen R. A likelihood-ratio test of monophyly. Syst. Biol. 1996;45:546–558. doi:10.2307/2413530 [Google Scholar]
- Hughes C., Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl Acad. Sci. USA. 2006;103:10 334–10 339. doi: 10.1073/pnas.0601928103. doi:10.1073/pnas.0601928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson U.S., Fjeldså J., Lokugalappatti L.G.S., Bowie R.C.K. A nuclear DNA phylogeny and proposed taxonomic revision of African greenbuls (Aves, Passeriformes, Pycnonotidae) Zool. Scr. 2007;36:417–427. doi:10.1111/j.1463-6409.2007.00290.x [Google Scholar]
- Knox E.B., Palmer J.D. The origin of Dendrosenecio within the Senecioneae (Asteraceae) based on chloroplast DNA evidence. Am. J. Bot. 1995;82:1567–1573. doi:10.2307/2446185 [Google Scholar]
- Koch M.A., Kiefer C., Ehrlich D., Vogel J., Brochmann C., Mummenhoff K. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae) Mol. Ecol. 2006;15:825–839. doi: 10.1111/j.1365-294X.2005.02848.x. doi:10.1111/j.1365-294X.2005.02848.x [DOI] [PubMed] [Google Scholar]
- Kükenthal G. Das Pflanzenreich IV. Wilhelm Engelmann; Leipzig, Germany: 1909. Cyperaceae-Cariocoideae. [Google Scholar]
- Levin D.A. Ancient dispersals, propagule pressure, and species selection in flowering plants. Syst. Bot. 2006;31:443–448. [Google Scholar]
- Lomolino M.V., Riddle B.R., Brown J.H. Sinauer Associates; Sunderland, MA: 2006. Biogeography. [Google Scholar]
- Maddison, W. P. & Maddison, D. R. 2006 Mesquite: a modular system for evolutionary analysis. 2. See http://mesquiteproject.org
- McDowall R. Process and pattern in the biogeography of New Zealand—a global microcosm? J. Biogeogr. 2008;35:197–212. doi:10.1111/j.1365-2699.2007.01830.x [Google Scholar]
- McGlone M.S., Duncan R.P., Heenan P.B. Endemism, species selection and the origin and distribution of the vascular plant flora of New Zealand. J. Biogeogr. 2001;28:199–216. doi:10.1046/j.1365-2699.2001.00525.x [Google Scholar]
- Menocal de P.B. African climate change and faunal evolution during the Pliocene–Pleistocene. Earth Planet. Sci. Lett. 2004;220:3–24. doi:10.1016/S0012-821X(04)00003-2 [Google Scholar]
- Müller K. Incorporating information from length-mutational events into phylogenetic analysis. Mol. Phylogenet. Evol. 2005;38:667–676. doi: 10.1016/j.ympev.2005.07.011. doi:10.1016/j.ympev.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Paun O., Lehnebach C., Johansson J.T., Lockhart P., Hörandl E. Phylogenetic relationships and biogeography of Ranunculus and allied genera (Ranunculaceae) in the Mediterranean region and in the European Alpine System. Taxon. 2005;54:911–930. [Google Scholar]
- Pelser P.B., Nordenstam B., Kadereit J.W., Watson L.E. An ITS phylogeny of tribe Senecioneae (Asteraceae) and a new delimitation of Senecio L. Taxon. 2007;56:1077–1104. (E1–E14) [Google Scholar]
- Popp M., Gizaw A., Nemomissa S., Suda J., Brochmann C. Colonization and diversification in the African ‘sky islands’ by Eurasian Lychnis L. (Caryophyllaceae) J. Biogeogr. 2008;35:1016–1029. doi:10.1111/j.1365-2699.2008.01902.x [Google Scholar]
- Posada D., Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Price J.P., Wagner W.L. Speciation in Hawaiian angiosperm lineages: cause, consequence, and mode. Evolution. 2004;58:2185–2200. doi: 10.1111/j.0014-3820.2004.tb01597.x. doi:10.1554/03-498 [DOI] [PubMed] [Google Scholar]
- Rambaut, A. 2002 Sequence Alignment Editor 2.0. See andrew.rambaut@zoo.ox.ac.uk
- Rambaut, A. & Drummond, A. 2005 MCMC Trace File Analyser 1.3. See http://evolve.zoo.ox.ac.uk/beast/
- Roalson E.H., Friar E.A. Phylogenetic relationships and biogeographic patterns in North American members of Carex section Acrocystis (Cyperaceae) using nrDNA ITS and ETS sequence data. Plant Syst. Evol. 2004;243:175–187. doi:10.1007/s00606-003-0089-8 [Google Scholar]
- Roalson E.H., Columbus T.J., Friar E.A. Phylogenetic relationships in Cariceae (Cyperaceae) based on ITS (nrDNA) and trnT-L-F (cpDNA) region sequences: assessment of subgeneric and sectional relationships in Carex with emphasis on section Acrocystis. Syst. Bot. 2001;26:318–341. [Google Scholar]
- Roy M.S. Recent diversification in African greenbuls (Pycnonotidae: Andropadus) supports a montane speciation model. Proc. R. Soc. Lond. B. 1997;264:1337–1344. doi:10.1098/rspb.1997.0185 [Google Scholar]
- Schaefer H., Heibl C., Renner S.S. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc. R. Soc. B. 2009;276:843–851. doi: 10.1098/rspb.2008.1447. doi:10.1098/rspb.2008.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M.P., Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000;49:369–381. doi:10.1016/j.ympev.2007.07.017 [PubMed] [Google Scholar]
- Simpson B., Larkin L., Weeks A., McDill J. Phylogeny and biogeography of Pomaria (Caesalpinioideae: Leguminosae) Syst. Bot. 2006;31:792–804. doi:10.1600/036364406779695915 [Google Scholar]
- Sklenar P., Balslev H. Geographic flora elements in the Ecuadorian superparamo. Flora (Jena) 2007;202:50–61. doi:10.1016/j.flora.2006.03.002 [Google Scholar]
- Smith J.M.B., Cleef A.M. Composition and origins of the world's tropicalpine floras. J. Biogeogr. 1988;15:631–645. doi:10.2307/2845441 [Google Scholar]
- Starr J.R., Harris S.A., Simpson D.A. Phylogeny of the unispicate taxa in Cyperaceae tribe Cariceae I: generic relationships and evolutionary scenarios. Syst. Bot. 2004;29:528–544. doi:10.1600/0363644041744455 [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2001. PAUP*: phylogenetic analysis using parsimony. [Google Scholar]
- Taberlet P., Gielly L., Pautou G., Bouvet J. A set of universal primers for amplifications of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Plant Mol. Biol. 1991;17:1105–1119. doi: 10.1007/BF00037152. doi:10.1007/BF00037152 [DOI] [PubMed] [Google Scholar]
- Takhtajan A. University of California Press; Los Angeles, CA: 1986. Floristic regions of the world. [Google Scholar]
- Tamura M. Angiospermae. Ordnung Ranunculales. Fam. Ranunculaceae. 2. Systematic part. In: Hiepko P., editor. Natürliche Pflanzenfamilien. Duncker & Humblot; Berlin, Germany: 1995. pp. 223–519. [Google Scholar]
- Urbatsch L.E., Baldwin B.G., Donoghue M.J. Phylogeny of coneflowers and relatives (Heliantheae: Asteraceae) based on nuclear rDNA internal transcribed spacer (ITS) sequences and chloroplast DNA restriction site data. Syst. Bot. 2000;25:539–565. doi:10.2307/2666695 [Google Scholar]
- Von Hagen K.B., Kadereit J.W. The phylogeny of Gentianella (Gentianaceae) and its colonization of the southern hemisphere as revealed by nuclear and chloroplast DNA sequence variation. Org. Div. Evol. 2001;1:61–79. doi:10.1078/1439-6092-00005 [Google Scholar]
- Wagner W.L., Herbst D.R., Sohmer S.H. University of Hawaii Press and Bishop Museum Press; Honolulu, HI: 1990. Manual of the flowering plants of Hawai'i. [Google Scholar]
- Winkworth R.C., Grau J., Robertson A.W., Lockhart P.J. The origins and evolution of the genus Myosotis L. (Boraginaceae) Mol. Phylogenet. Evol. 2002a;24:180–193. doi: 10.1016/s1055-7903(02)00210-5. doi:10.1016/S1055-7903(02)00210-5 [DOI] [PubMed] [Google Scholar]
- Winkworth R.C., Wagstaff S.J., Glenny D., Lockhart P.C. Plant dispersal N.E.W.S from New Zealand. Trends Ecol. Evol. 2002b;17:514–520. doi:10.1016/S0169-5347(02)02590-9 [Google Scholar]
- Winkworth R.C., Wagstaff S.J., Glenny D., Lockhart P.J. Evolution of the New Zealand mountain flora: origins, diversification and dispersal. Org. Div. Evol. 2005;5:237–247. doi:10.1016/j.ode.2004.12.001 [Google Scholar]
- Yen A.C., Olmstead R.G. Molecular systematics of Cyperaceae tribe Cariceae based on two chloroplast DNA regions: ndhF and trnL intron-intergenic spacer. Syst. Bot. 2000;25:479–494. doi:10.2307/2666691 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession details of taxa used in molecular phylogenetic analyses including authors, subgeneric classification of Carex, voucher details, references to first publication and NCBI Genebank numbers
Overview of Carex, Ranunculus and Alchemilla, their distribution and species numbers. Additionaly, the total number of taxa per group and the number of accessions are reported
All clades retrieved from the molecular phylogenetic analyses which include accessions from African are described and discussed in detail
A detailed account of the methodology of parametric bootstrap analyses is given together with all details of the constraints performed
Detailed tree description of all molecular phylogenetic trees are given as well as the relevant parts of the trees from the ancestral area reconstructions