Abstract
Background
The base excision repair pathway is responsible for repairing small DNA base lesions caused by endogenous and exogenous damaging agents. Repair is initiated by DNA glycosylases that recognize and remove the lesions. NEIL3 is one of 11 mammalian DNA glycosylases identified to date and it was discovered on the basis of sequence homology to the E. coli Fpg and Nei glycosylases. Difficulties in purifying the protein have limited its biochemical characterization and in contrast to the other glycosylases, its function remains unclear.
Results
In this study we describe the expression pattern of Neil3 during mouse embryonic development with special focus on brain development. We have also looked at the expression of NEIL3 in several normal and tumor tissues. Quantitative real-time PCR and in situ hybridization revealed that Neil3 was highly expressed at embryonic days 12–13, when neurogenesis starts. The expression decreased during development and in the adult brain,Neil3 could not be detected in any of the brain areas examined by quantitative real-time PCR. During embryogenesis and in newborn mice specific expression was observed in areas known to harbour neural stem and progenitor cells such as the subventricular zone and the dentate gyrus. Finally, NEIL3 expression was higher in tumors compared to normal tissues, except for testis and pancreas.
Conclusion
Our findings indicate that mammalian NEIL3 is specifically expressed in brain areas where neurogenesis takes place during development and that its expression is tightly regulated both temporally and spatially. In addition, NEIL3 seems to be upregulated in tumor tissues compared to normal tissues. Altogether, mammalian NEIL3 seems to be highly expressed in cells with high proliferative potential.
Background
The base excision repair (BER) pathway, one of the major DNA repair pathways, is dedicated to the repair of damaged DNA bases arising from endogenous and exogenous insults [1-4]. The DNA bases may be subjected to oxidation, alkylation and deamination and oxidative DNA damage, in particular, has been implicated in the pathogenesis of many diseases including cancer, atherosclerosis, neurodegenerative diseases such as Parkinson's and Alzheimer's, and even aging [5-8]. DNA glycosylases are the key enzymes of BER; they initiate repair by catalysing the hydrolysis of the N-glycosylic bond between modified bases and the sugar-phosphate backbone [1,3,4,9]. To date, four mammalian DNA glycosylases that recognize and excise oxidized bases have been thoroughly characterized: OGG1, NTH1, NEIL1 and NEIL2 [10-12]. NEIL3 was identified together with NEIL1 and NEIL2 as a gene product with significant structural similarities to the E. coli Fpg and Nei DNA glycosylases [13-15]. However, in contrast to NEIL1 and NEIL2 no substantial DNA glycosylase activity has been detected [13,14,16,17]. Expression of human NEIL3 has been reported in thymus and testis, while mouse Neil3 has been demonstrated to be highly expressed in spleen, bone marrow, thymus, blood cells and brain regions that harbour progenitor cells [13,18,19]. NEIL3 has also been shown to be highly expressed in primary malignant melanomas associated with metastasis [20]. Most studies have been performed on adult organs and a recent paper described Neil3 expression in the developing mouse [17]. Here, we describe the expression pattern of Neil3 during embryonic development in mice with focus on the expression of Neil3 during brain development. The expression of NEIL3 in multiple human cancers was also examined.
Results
Neil3 is expressed during embryogenesis
The temporal expression pattern of Neil3 during embryonic development was examined by in silico electronic Northern blot (eNorthern) and Northern blot analysis. We used the UniGene collection http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene to generate in silico expression profiles for Neil3 at different stages during embryonic development. As seen in Figure 1A, there is a very high expression in the oocyte, unfertilised oocyte and zygote and this expression dramatically falls after the zygote stadium. During embryogenesis, Neil3 transcription is not detected until E8.5–11.5. Northern blot analysis was performed by using a northern blot containing poly(A)+ RNA isolated from whole mouse embryos at days 7, 11, 15 and 17 of gestation. Expression of Neil3 was not observed until E11 (Figure 1B).
Figure 1.
Expression analysis of Neil3 during embryonic development. (A) Expression profiles of Neil3 based on the EST profile viewer at the NCBI homepage. TPM, transcripts per million. (B) A multiple tissue northern blot containing poly(A)+ RNA from mouse embryo tissues at four different developmental stages was hybridized with 32P-labeled cDNA probes for Neil3 and β-actin.
Neil3 is highly expressed in developing brain
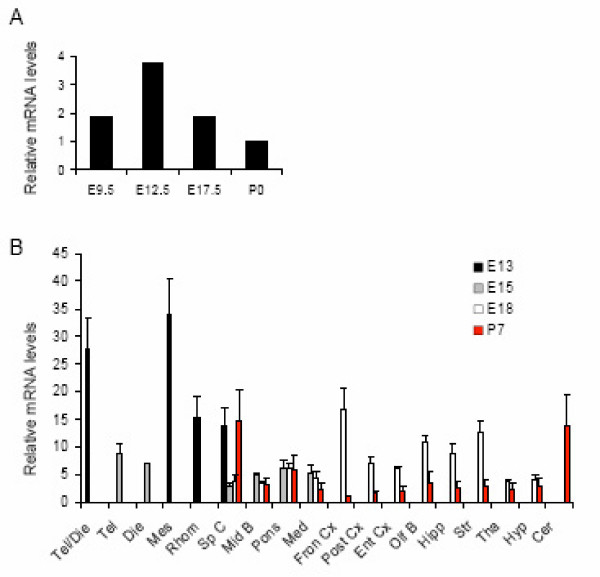
Next, we examined the expression of Neil3 in developing mouse brain. Messenger RNA was isolated from brains at days 9.5, 12.5 and 17.5 of gestation and at P0 (birth) and subjected to quantitative real-time PCR. Figure 2A shows the expression levels of Neil3 relative to Gapdh. The highest expression level of Neil3 was observed at E12.5. To dissect the sites of Neil3 expression, we evaluated the distribution of Neil3 by transcript profiling in various brain regions at five different stages of development: E13, E15 and E18 (embryo), P7 (neonatal) and 5 weeks (adult). As seen in Figure 2B, Neil3 expression was highest at E13 in the fore- and midbrain and the expression decreased during development. In neonatal mice the expression was relatively low in all areas examined, except for in the cerebellum and the spinal cord. In adult mice, Neil3 expression was below the detection limit (data not shown).
Figure 2.
Expression analysis of Neil3 in embryonic brain development. (A) Real-time quantitative PCR was performed on mRNA isolated from mouse brains at four different developmental stages. The data were normalized to Gapdh and expressed relative to the level of Neil3 at P0. (B) A detailed expression profile of Neil3 during brain development was obtained by using a mouse developmental tissue qPCR array. The array contains one concentration of cDNAs, normalized to Gapdh. Neil3 mRNA levels were calculated relative to the sample expressing the lowest level of Neil3 (Fron Cx, P7). The experiment was conducted twice and the standard deviations are indicated. Abbreviations: Tel, telencephalon; Die, diencephalon; Mes, mesencephalon (midbrain); Rhom, Rhombencephalon; Sp C, spinal cord; Mid B, midbrain; Med, medulla; Fron Cx, frontal cortex; Post Cx, posterior cortex; Ent Cx, entorhinal cortex; Olf B, olfactory bulb; Hipp, hippocampus; Str, Striatum; Tha, thalamus; Hyp, hypothalamus; Cer, cerebellum.
Neil 3 is highly expressed in regions of active neurogenesis in the developing brain
In order to characterize in more detail the expression of Neil3 mRNA in developing brain, in situ hybridization was performed on foetal mouse forebrains at E12, E17.5 and P0 (Figure 3). At E12 abundant Neil3 mRNA was detected in the subventricular zone (SVZ) of the lateral ventricles (LV). The expression of Neil3 decreased during development and at E17.5 and P0, expression was limited to distinct cells in the cortical SVZ, in cells of the secondary matrix (SM), the dentate gyrus migratory route (DGM) and the dentate gyrus (DG). Thus, Neil3 was expressed in regions that are rich in progenitor cells. No signals were detected for the sense DIG-RNA probe used as a control (data no shown).
Figure 3.

Expression of Neil3 in developing mouse forebrain. In situ hybridisation was performed on coronal sections of mouse brains at different embryonic stages. Magnifications of Neil3-positive cells are shown to the right. Abbreviations: CA, hippocampal differentiation fields; Ctx, cortex; DG, dentate gyrus; Hy, hypothalamus; LV, lateral ventricle; SM, secondary matrix; SVZ, subventricle zone.
NEIL3 is expressed in various tumor samples
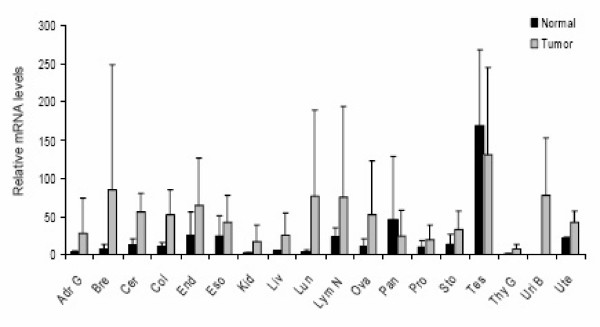
NEIL3 has recently been shown to be highly expressed in melanomas [20]. We therefore investigated the expression of NEIL3 in several tumor types and their normal counterparts by using qPCR arrays containing cDNA from diseased and normal tissues. Data shown in Figure 4 is from one of the two experiments performed and represents relative NEIL3 mRNA levels in normal and tumor tissues. Raw data and information about the samples examined are shown in Additional file 1. Results revealed that although we observed high variation within the different cancer samples from the same tissue, tumor samples in general displayed a higher expression of NEIL3 than the corresponding normal tissues, except for testis and pancreas. Furthermore, NEIL3 was highly abundant in normal testis tissue, as has been described before [13].
Figure 4.
Expression of NEIL3 in human cancers. Transcript profiling was performed by using a disease tissue qPCR array. cDNA concentrations are normalized to β-actin. Neil3 mRNA levels are calculated relative to the sample expressing the lowest level of Neil3 (one of the stomach cancer samples). Tumor and normal tissue samples were grouped according to their origin and the average and standard deviations were calculated for each group. The experiment was conducted twice and the results from one of the experiments are shown. Abbreviations: Adr G, adrenal gland; Bre, breast; Cer, cervix; Col, colon; End, endometrium; Eso, esophagus, Kid, kidney; Liv, liver; Lun, lung; Lym N, lymph node; Ova, ovary; Pan, pancreas; Pro, prostate; Sto, stomach; Tes, testis; Thy G, thyroid gland; Uri B, urinary bladder; Ute, uterus.
Discussion
In this study the expression of Neil3 during embryonic brain development was investigated. Our results showed that Neil3 was not uniformly expressed in the brain, but limited to specific cells in certain regions. The expression appeared to be tightly regulated both spatially and temporally during brain development.
Results from Northern blot analysis of total embryos revealed the presence of Neil3 after E7, while RT-PCR experiments showed the presence of Neil3 in the developing brain from E9. Thus, the expression of Neil3 appears to coincide with organogenesis. This is also supported by the in silico analysis, where Neil3 transcripts were not detected until E8.5–E11.5. Intriguingly, the in silico analysis revealed high expression of Neil3 during preimplantation. Mammalian embryos undergo major changes in their gene expression patterns throughout most stages of preimplantation development and massive maternal degradation of mRNA characterizes the transition from oocyte to early embryo before the onset of new transcription [21,22]. Genes that follow this expression pattern are suggested to have specific functions either in oogenesis, oocyte maturation, fertilization, and/or early phases of preimplantation development [23-25]. Our results suggest that Neil3 can be classified with this group of genes since Neil3 mRNA is highly expressed in unfertilised eggs and is abundant up to the zygote stadium before it is apparently degraded.
In mice, embryonic neurogenesis occurs from E12 to E17 [26,27]. There are two major proliferative populations in the developing brain [28]. The first to appear is a region called the ventricular zone (VZ) in the lateral ventricle. The VZ is lined by a population of cells that contains multipotent neural stem cells that give rise to most neurons and glial cells of the cortex. These cortical precursor cells constitute a heterogeneous population and there is increasing evidence that different precursor cells generate distinct differentiated cell types [29-33]. Another proliferative population is found in the subventricular zone (SVZ), located adjacent to the VZ along the lateral ventricle. Notably, neurogenesis continues in this region throughout adulthood, and in the adult mammalian brain, new neurons born in the SVZ migrate anteriorly into the olfactory bulb (OB), where they mature [34-36]. In post-natal mice, a second germinal zone exists; the subgranular zone (SGZ) of dentate gyrus in the hippocampus [36,37]. Our in situ results showed that during brain development Neil3 was highly expressed in cells found in places rich in neural progenitors such as the VZ and the SVZ. The expression decreased during embryonic development and at P0, only a few cells around the lateral ventricles and in the dentate gyrus of the hippocampus were positive for Neil3 expression. We have recently described the distribution of Neil3 during postnatal development and found Neil3 expression in the SVZ, the rostral migratory stream (RMS), the dentate gyrus of the hippocampal formation and the Purkinje cells of the cerebellum in P3 mice. The expression of Neil3 decreased dramatically in postnatal mice so that in 1-month-old mice only a few cells in the SVZ and in layer V of neocortex were detected. In 1-year-old mice Neil3 was detected in layer V of neocortex only [19]. In the present study no Neil3 expression was detected in brains from 5-week-old mice when using quantitative real-time PCR. The discrepancy between our results and the previously reported data could be due to a lower sensitivity of real-time PCR compared to in situ hybridization. Thus, the limited number of Neil3-positive cells detected in 1-month-old mice by in situ hybridization is probably below the detection limit of the real-time PCR. Altogether, previous data and results presented here suggest that Neil3 is highly expressed in regions where neurogenesis occurs during embryogenesis and to a lesser extent in neonatal animals [19]. Hippocampal Neil3 expression disappeared in 1-month-old mice but could still be observed in the SVZ [19]. This may suggest the existence of different progenitors in the two germinal areas. Since Neil3 appears to be enhanced in proliferating cells, the decline in Neil3 expression during development could be due to a decline in cell proliferation.
Neil3 knockout mice were generated a few years ago and these mice are viable and remain apparently healthy into adulthood with no overt phenotype [18]. A putative role in lymphocytes and/or other immune cells has been proposed since Neil3 is highly expressed in lymphatic cells and tissues and the knockout mice have a slightly reduced number of blood cells [18]. The expression pattern described in this paper warrants a closer examination of the distribution/phenotype of neural progenitor cells in the embryo and adult Neil3 knockout mice. Studies using the Neil3 knockout mice model that involve insults to the brain such as ischemic stroke would be central in revealing whether Neil3 has a role in the regeneration of damaged tissue.
In a recent study, NEIL3 was shown to be overexpressed in primary melanomas giving rise to metastasis [20]. NEIL3 was therefore suggested to be associated with the progression of primary melanoma to distant metastasis [20]. Interestingly, we found increased NEIL3 expression in 16 of 18 cancer tissues compared to normal tissues. Intriguingly, NEIL3 has also recently been suggested to be a host factor for HIV and thus, a target for antiviral medicines [38]. Taken together all these observations and the fact that no robust DNA glycosylase activity has been detected [16,17], indicate that NEIL3 is not a typical DNA glycosylase but rather a protein involved in processes where extensive proliferation is a hallmark such as brain development and possibly tumor development/progression.
Conclusion
We have characterized the expression pattern of Neil 3 during embryonic development. Our results clearly showed that the expression of Neil3 appeared to be tightly regulated both temporally and spatially. High expression was detected in the oocytes and preimplantation stages of development and later during organogenesis. In the embryonic brain, Neil3 was detected in sites rich in neural stem/progenitor cells. Finally, tumor tissues were shown to express higher amounts of NEIL3 compared to their normal counterparts, except for testis and pancreas. These results call for a closer examination of the brains of Neil3 knock-out mice not only during development and aging, but also under detrimental conditions such as ischemic stress.
Methods
Northern Blot Analysis
A mouse embryo MTN blot purchased from Clontech (catalog no. 7763-1) was probed for Neil3 expression. Northern blot hybridization was carried out using ExpressHyb solution (Clontech) and probes were labeled using the Rediprime DNA labeling system (Amersham Biosciences), both according to the manufacturer's protocol. The full-length murine cDNA was used as a probe.
Real-time Quantitative PCR
Messenger RNA was isolated from the brains of embryos and newborn mice (mixed background: 50% 129Sv and 50% C57BL/6) using the Dynabeads mRNA DIRECT Kit (Dynal). The RNA was treated with TurboDNase (Applied Biosystems) and cDNA synthesized using the High-Capacity cDNA Reverse Transcription kit (Applied Biosytems). The expression of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA was used as endogenous control. Primers used: mouse Gapdh, forward 5'-TCG TCC CGT AGA CAA AAT GGT-3'-, reverse 5'-CGC CCA ATA CGG CCA AA-3'. Mouse Neil3, forward 5'-GGG CAA CAT CAT CAA AAA TGA A-3', reverse 5'-CTG TTT GTC TGA TAG TTG ACA CAC CTT-3'. Quantitative real-time PCR was performed in 20-μl reactions containing 20 ng of cDNA using the Power SYBR Green PCR master mix and the Step One Plus real-time PCR system (both from Applied Biosystems) according to the system and kit instructions. Relative gene expression was calculated using the comparative CT method and primers were designed using the Primer Express software version 2.0 (Applied Biosystems).
To investigate more specifically the expression of Neil3 in the developing mouse brain, a Mouse Developmental Tissue qPCR array (MDRT101; OriGene Technologies) was used. Neil3 primers used were the same as mentioned above. The array contained normalized cDNA prepared from various brain regions at five developmental stages. The cDNAs were normalized against Gapdh. Relative gene expression was calculated using the comparative CT method. The data presented are relative Neil3 mRNA levels.
For measurement of NEIL3 mRNA expression in normal and tumor tissues, we used a Disease Tissue qPCR array (CSRT303; OriGene Technologies). This array consisted of normalized cDNA prepared from pathologist-verified human tumor tissues obtained from 18 different tissues. The cDNAs were normalized against β-actin. Clinical information associated with each of these samples can be found in Additional file 1. Primers used: Human NEIL3, forward 5'-GGT CTC CAC CCA GCT GTT AAA G-3', reverse 5'-CAC GTA TCA TTT TCA TGA GGT GAT G-3'. Two parallel experiments were run and the raw data is presented in the additional file 1. Relative gene expression was calculated using the comparative CT method. The data presented are relative NEIL3 mRNA levels.
In situ hybridization
For characterization of Neil3 gene expression in the brain, wild type (C57BL6/CBA F1 strain) embryos and newborn mice were used. Coronal cryosections were prepared and in situ hybridization was performed as previously described [39]. A plasmid containing full-length mNeil3 (clone 4945750; Invitrogen,) was linearized with appropriate enzymes before sense and antisense riboprobes were synthesized using a DIG RNA labeling kit (SP6/T7) (catalog no. 1175025; Roche).
Authors' contributions
GAH, performed Northern Blot analysis, took care of the mice and removed brains from mice for RNA isolation, isolated RNA, discussed the results and contributed to the editing of the manuscript. CGN, performed real-time RT-PCR. DBD, performed in situ hybridization. CGC, took care of the mice and helped removed brains from mice for RNA isolation. SK, contributed to the planning of the experiment and provided financial support. MB, contributed to the planning of the experiment, discussed results, contributed to the editing of the manuscript and provided financial support. LL, conceived and supervised the experiments, performed the electronic Northern blot, discussed results and prepared the manuscript.
Supplementary Material
Raw data of NEIL3 mRNA expression in normal and tumor tissues. The data provided are the measurement of NEIL3 mRNA expression in normal and tumor tissues, we used a Disease Tissue qPCR array (CSRT303; OriGene Technologies). This array consisted of normalized cDNA prepared from pathologist-verified human tumor tissues obtained from 18 different tissues. The cDNA concentrations were normalized to β-actin. Raw data from two parallel experiments and information about the samples examined are shown in this file.
Acknowledgments
Acknowledgements
We are grateful to Dr. Mattias Backman (University of Umeå, Sweden) for helpful technical assistance. This work was supported through the Norwegian Center for Stem Cell Research by the Research Council of Norway and The European Union Program DNA Repair.
Contributor Information
Gunn A Hildrestrand, Email: gunn.hildrestrand@rr-research.no.
Christine G Neurauter, Email: christine.gran@rr-research.no.
Dzung B Diep, Email: dzung.diep@umb.no.
Cesilie G Castellanos, Email: cesilie.castellanos@veths.no.
Stefan Krauss, Email: stefan.krauss@rr-research.no.
Magnar Bjørås, Email: magnar.bjoras@rr-research.no.
Luisa Luna, Email: luisa.luna@rr-research.no.
References
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Baute J, Depicker A. Base excision repair and its role in maintaining genome stability. Crit Rev Biochem Mol Biol. 2008;43:239–276. doi: 10.1080/10409230802309905. [DOI] [PubMed] [Google Scholar]
- Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Gamkrelidze M, Mamamtavrishvili N, Bejitashvili N, Sanikidze T, Ratiani L. Role of oxidative stress in pathogenesis of atherosclerosis. Georgian Med News. 2008;163:54–57. [PubMed] [Google Scholar]
- Martin LJ. DNA damage and repair: relevance to mechanisms of neurodegeneration. J Neuropathol Exp Neurol. 2008;67:377–387. doi: 10.1097/NEN.0b013e31816ff780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I, Grotewiel MS. Oxidative damage and age-related functional declines. Mech Ageing Dev. 2006;127:411–423. doi: 10.1016/j.mad.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Ide H, Kotera M. Human DNA glycosylases involved in the repair of oxidatively damaged DNA. Biol Pharm Bull. 2004;27:480–485. doi: 10.1248/bpb.27.480. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, Horst GT van der, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2003;2:581–591. doi: 10.1016/S1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- Krokeide SZ, Bolstad N, Laerdahl JK, Bjoras M, Luna L. Expression and purification of NEIL3, a human DNA glycosylase homolog. Protein Expr Purif. 2009;65:160–164. doi: 10.1016/j.pep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Takao M, Oohata Y, Kitadokoro K, Kobayashi K, Iwai S, Yasui A, Yonei S, Zhang QM. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells. 2009;14:261–270. doi: 10.1111/j.1365-2443.2008.01271.x. [DOI] [PubMed] [Google Scholar]
- Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic Tissue-Specific Expression of Mouse Neil3 for Endonuclease VIII-Like Protein. J Biochem. 2005;138:763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- Rolseth V, Runden-Pran E, Luna L, McMurray C, Bjoras M, Ottersen OP. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair (Amst) 2008;7:1578–1588. doi: 10.1016/j.dnarep.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, Oord JJ van den, Spatz A, Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27:565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/S1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–144. doi: 10.1016/S1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wang S, Cowan CA, Chipperfield H, Powers RD. Gene expression in the preimplantation embryo: in-vitro developmental changes. Reprod Biomed Online. 2005;10:607–616. doi: 10.1016/s1472-6483(10)61668-9. [DOI] [PubMed] [Google Scholar]
- Gillies K, Price DJ. Cell migration and subplate loss in explant cultures of murine cerebral cortex. Neuroreport. 1993;4:911–914. doi: 10.1097/00001756-199307000-00018. [DOI] [PubMed] [Google Scholar]
- Levers TE, Edgar JM, Price DJ. The fates of cells generated at the end of neurogenesis in developing mouse cortex. J Neurobiol. 2001;48:265–277. doi: 10.1002/neu.1056. [DOI] [PubMed] [Google Scholar]
- Rao MS, Jacobson M. Developmental Neurobiology. 4. Kluwer Academic/Plenum Publishers; 2005. [Google Scholar]
- Tan SS, Kalloniatis M, Sturm K, Tam PP, Reese BE, Faulkner-Jones B. Separate progenitors for radial and tangential cell dispersion during development of the cerebral neocortex. Neuron. 1998;21:295–304. doi: 10.1016/S0896-6273(00)80539-5. [DOI] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Merkle FT, Flames N, Tramontin AD, Garcia-Verdugo JM, varez-Buylla A. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Appolloni I, Calzolari F. Radial glia and neural stem cells. Cell Tissue Res. 2008;331:165–178. doi: 10.1007/s00441-007-0481-8. [DOI] [PubMed] [Google Scholar]
- Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc Natl Acad Sci USA. 1993;90:2074–2077. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Garcia-Verdugo JM. Neurogenesis in adult subventricular zone. J Neurosci. 2002;22:629–634. doi: 10.1523/JNEUROSCI.22-03-00629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Kang E, Liu CY, Ming GL, Song H. Development of neural stem cell in the adult brain. Curr Opin Neurobiol. 2008;18:108–115. doi: 10.1016/j.conb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Machon O, Bout CJ van den, Backman M, Rosok O, Caubit X, Fromm SH, Geronimo B, Krauss S. Forebrain-specific promoter/enhancer D6 derived from the mouse Dach1 gene controls expression in neural stem cells. Neuroscience. 2002;112:951–966. doi: 10.1016/S0306-4522(02)00053-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data of NEIL3 mRNA expression in normal and tumor tissues. The data provided are the measurement of NEIL3 mRNA expression in normal and tumor tissues, we used a Disease Tissue qPCR array (CSRT303; OriGene Technologies). This array consisted of normalized cDNA prepared from pathologist-verified human tumor tissues obtained from 18 different tissues. The cDNA concentrations were normalized to β-actin. Raw data from two parallel experiments and information about the samples examined are shown in this file.