Abstract
Mammary epithelial cells go through a series of developmental changes during pregnancy and lactation including proliferation, differentiation, secretion and apoptosis. HC11 mouse mammary epithelial cells, which undergo lactogen-induced differentiation in cell culture, were used to follow the changes in gene expression during this process. The expression profiles of over 20,000 genes were compared in HC11 cells undergoing lactogenic differentiation to non-differentiated cells using DNA microarray analysis. Greater than two fold changes were detected in 998 genes in the differentiated cells versus growth controls. Several genes including CTGF/CCN2 exhibited greater than five-fold increase. Validation of the gene expression pattern for more than twenty genes was performed. The results indicate the involvement of numerous genes and pathways in the differentiation of mouse mammary epithelial cells in culture and they identify genetic pathways associated with specific transcriptional regulation. In addition, the expression of a subset of genes regulated by lactogenic differentiation in HC11 cells, including CTGF/CCN2 and osteopontin, was examined in mouse mammary glands revealing expression during pregnancy and lactation that declined during involution of the glands. To probe the mechanism by which epidermal growth factor (EGF), a known inhibitor of lactogenic differentiation in HC11 cells, blocks lactogenesis, the HC11 cells stimulated with lactogenic hormone in the presence of EGF were profiled. This data revealed EGF regulation of a specific subset of genes including important cell cycle regulators. The studies confirm the value of expression profiling in defining gene transcription associated with differentiation of mammary epithelial cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-009-0047-5) contains supplementary material, which is available to authorized users.
Keywords: CTGF/CCN2, Lactogenic differentiation, Dexamethasone
Introduction
The mammary gland undergoes periodic cycles of growth, differentiation and regression throughout adult life. Normal mammary gland development is dependent on signals from growth factors, mammotrophic hormones, and tissue stroma, and the dysregulation of these process can lead to mammary epithelial hyperplasia and tumorigenesis. The HC11 cell line was derived from the COMMA1D cells, which originated from the mammary gland tissue of a pregnant BALB/c mouse (Danielson et al. 1984). HC11 cells are an excellent model system for studying differentiation states of mammary epithelial cells. These cells retain important characteristics of normal mammary epithelial cells including the synthesis of the milk protein ß-casein in vitro upon treatment with lactogenic hormones (Ball et al. 1988). HC11 cells also exhibit normal ductal morphogenesis when injected into the cleared fat pad of syngeneic mice (Humphreys and Rosen 1997). A crucial event in the mammary epithelial cells is the activation of the epidermal growth factor (EGF) receptor during the growth phase. In HC11 cells, activation of the EGF receptor promotes growth and is required for the establishment of competence to respond to the lactogenic hormones. Following EGF removal, HC11 cells differentiate in vitro in response to the synergistic actions of insulin, glucocorticoids, and prolactin (DIP) (Ball et al. 1988; Taverna et al. 1991). However, the inclusion of EGF in the lactogenic hormone mix blocks differentiation (Hynes et al. 1990; Marte et al. 1995b; Merlo et al. 1996; Peterson and Haldosen 1998). The regulatory mechanisms for signal transduction pathways controlling development of the mammary gland epithelium and the molecular switch from proliferation to differentiation have been studied in recent years. Prolactin stimulation results in the phosphorylation and nuclear translocation of Stat5 and Stat5 binds to the β-casein promotor initiating its transcription (Ali 1998; Gouilleux et al. 1994; Han et al. 1997; Marte et al. 1995b; Stocklin et al. 1996). The activation of β-casein transcription is enhanced by the glucocorticoid receptor (Nagaiah et al. 1981; Stocklin et al. 1996). While these studies addressed the requirements for milk protein transcription other changes contributing to lactogenic differentiation are less well understood.
HC11 mammary epithelial cell differentiation was examined using two-dimensional gels and proteomic techniques to decipher differences between the protein patterns of growing versus differentiating cells (Desrivieres et al. 2003). This study identified approximately sixty proteins whose expression levels changed following four days of differentiation. These proteins include cytoskeletal components, molecular chaperones and regulators of protein folding and stability, calcium-binding proteins, and components of RNA-processing pathways. More recently a proteomic study of nuclear protein changes in HC11 cells undergoing differentiation revealed differential expression of several transcription factors that have the potential to regulate mammary differentiation or development as well as proteasome subunits, hnRNPs and HSPs (Desrivieres et al. 2007). The profile of induced proteins provided insights into the mechanistic differentiation of HC11 mammary epithelial cells and the regulation of this process. Gene expression profiling using DNA microarrays allows the concurrent analysis of thousands of genes and has been applied to provide insight into the regulatory and functional pathways involved in mouse mammary gland development (Master et al. 2002; Clarkson and Watson 2003; Gass et al. 2003; Rudolph et al. 2003). These studies measure transcription in all cells in the mammary gland including stroma and immune cells. In the study reported here, oligonucleotide microarrays containing approximately 20,000 genes were used to examine the gene expression patterns during lactogenic differentiation of the HC11 mammary epithelial cell line. The study confirms that HC11 cells undergoing lactogenic differentiation in culture recapitulate the process in the mammary gland. In addition, the results identify potential signaling pathways not previously associated with mammary differentiation. Among the newly identified molecules contributing to lactogenic differention the rapid elevation of CTGF/CCN2 and osteopontin are highly significant as these proteins are known to play a role in tissue remodeling and the pathology of breast cancer.
Materials and methods
Cell preparation HC11 mouse mammary epithelial cells were cultured in RPMI 1640 medium containing 10% fetal calf serum, 5 µg/ml Insulin, 10 mM Hepes and 10 ng/ml epidermal growth factor (EGF) (Cerrito et al. 2004). For differentiation the cells were maintained in T75 flasks for 4 days after reaching confluence, then exposed to media without EGF for 24 h. The cells were then incubated in DIP differentiation media, serum-containing RPMI with dexamethasone (10−6 M), insulin (5 µg/ml) and prolactin (5 µg/ml) for 72 h and undifferentiated HC11 cells were maintained in growth media and these served as a control. In another set of experiments, control cells were incubated in DIP differentiation media for 72 h, while experimental cells were incubated in DIP differentiation media except that EGF was present. The cells were scraped from the flasks for RNA extraction.
RNA preparation RNA was extracted using Trizol reagent (Invitrogen) and RNeasy maxi kit (Qiagen). RNA samples were concentrated to greater than 1 mg/ml by centrifugation on a MicroCon 100 filter unit at 500 g. The concentration and 260/280 OD ratio of the RNA was determined by spectrophotometry and RNA was stored at −80°C. Alternatively, the RNA was purified to recover mRNA using Oligtex mRNA kit.
Labeling, hybridization and analysis Mouse (Development) Oligo Microarrays were purchased from Agilent Technologies. The content on this microarray was derived from the National Institute on Aging/National Institute of Health cDNA mouse clone set that includes genes from sequences of stem cells and very early-stage embryo cDNA libraries. It contains 20,371 60-mer oligonucleotides representing over 20,000 known mouse genes. Detailed process for labeling and hybridization were according to manufacturer’s manual. Briefly, fluorescent cRNA was synthesized using low RNA input fluorescent linear amplification kit from Agilent Technologies. 500 ng of total RNA was used in each experiment. In the differentiation experiments, five slides were used (repetitions N = 5), DIP treated RNA was labeled with cyanine 5-CTP, and control RNA was labeled with cyanine 3-CTP. In differentiation blocking experiments, six slides were used (N = 6), EGF treated DIP RNA was labeled with cyanine 5-CTP, while DIP treated RNA was labeled with cyanine 3-CTP. Amplified cRNA was purified using Qiagen’s RNeasy mini spin columns. Hybridization was carried out using In situ Hybridization Kit Plus from Agilent Technologies, 750 ng of cyanine 3 and cyanine 5 labeled cRNA was used in each hybridization. Hybridization continued at 60°C for over 17 h. Post hybridization washes includes 6 X SSC, 0.005% Triton X-102 (10 min), and 0.1 X SSC, 0.005% Triton X-102 (5 min) followed by drying with nitrogen gas and immediate scanning. Scanning was performed by GenePix 4000A scanner (Axon instruments, Inc., Foster city, CA) with Axon GenePix image acquisition and analysis software. Analysis of gene expression was performed using BRB-Array Tools Version 3 which was developed by Biometrics Research Branch, NCI (http://linus.nci.nih.gov/BRB-ArrayTools.html).In addition, a smaller study using oligonucleotides arrays representing 3800 genes (Atlas Glass Mouse 3.8 Microarrays from Clontech) (N = 4) was performed. Fluorescent labeling of RNAs was performed by using an Atlas Glass fluorescent labeling kit (Clontech Laboratories) according to manufacturer’s protocol. Synthesized first-strand cDNAs from RNA of HC11 cells with and without differentiation were labeled with fluorescent dyes, Cy3 and Cy5 (Amersham Pharmacia Biotech), respectively. The reciprocal labeling was used during the experiment, i.e. differentiation group was labeled with Cy3 two times, and Cy5 two times; and the control group was labeled with Cy5 two times, and Cy3 two times. The quality of the labeling and the amount of each probe used were determined by absorbance measurement for Cy3 and Cy5 probes in a Beckman DU-600 scanner. Hybridization of the microarrays was carried out in a hybridization solution for 16 h at 50°C. The slides were washed 3 times. The microarray slides were scanned and analyzed by using a GenePix 4000B scanner in both Cy3 and Cy5 channels. The control value was obtained from four experiments and used to determine the differentiation induced gene up- or down-regulations. The average of the Cy3 and Cy5 signals from nine house-keeping genes were used to obtain a ratio value for normalization of the individual signals. Much of the information obtained from the differentiation experiments analyzed on the smaller oligonucleotide based arrays, Clontech 3.8 K arrays, were contained within the larger 20280 gene set data. However, Supplemental file 3 contains the list of genes detected as differentially expressed during lactogenic differentiation of HC11 cells on 3.8 K arrays.
Statistical Analysis of Microarray Normalization and analysis of the gene expression profiles were performed as follows: the spot was excluded if red and green intensity was below 30. Each array was normalized (centered) using the median over entire array. Intensity ratios (and inverse ratios) greater than 64 were truncated. Genes were excluded under any of the following conditions: less than 20 % of expression data had at least a 1.5 -fold change in either direction from gene’s median value or if the percentage of data that was missing or filtered out exceeded 50%.In DIP versus control experiments, there are 10813 genes that passed filtering criteria in total of 20280 genes. And the first 2479 genes are significant at the nominal 0.05 level of the paired T-test. In EGF plus DIP versus DIP experiments, there are 1386 genes passed filtering criteria in total of 20280 genes. And the first 1129 genes are significant at the nominal 0.05 level of the paired T-test.
Generation of probes In order to generate DNA hybridization probes, the accession number of interested gene was used to obtain the mRNA sequence and primers were designed for RT-PCR to produce a DNA fragment several hundred basepairs in length. RT-PCR products were inserted into a pCR2.1 TA cloning vector (Invitrogen) and the identities of candidate clones were confirmed by DNA sequence. Double strand DNA plasmid inserts were isolated by restriction endonuclease digestion of the pCR2.1 plasmid and the released DNA fragment was gel purified for use as a hybridization probe. The fragments cloned and amplified include: PIP, position 77 to 477 in cDNA (accession number NM_008843); KLF9 (Bteb1), position 932 to 1262 in cDNA (accession number NM_010638); SGK position 92 to 592 in cDNA(accession number NM_011361); AP3 position 92 to 592 in cDNA (accession number NM_009681); Spi-C position 362 to 862 in cDNA(accession number NM_011461). The probes and primers for CTGF/CCN2, β-casein, and actin have been described (Wang et al. 2008).
Northern blot and Realtime PCR For northern blot experiments, HC11 cells were treated identically to the methods in the microarray experiment, and then exposed to DIP differentiation media for 0, 12, 24, 48, 72 and 96 h, respectively. RNA was extracted for northern blot by using Trizol reagent. In some experiments, the same RNA preparations used in Agilent Microarrays were utilized. RNA from mouse mammary gland at distinct stages of pregnancy and lactation were prepared as described (Wang et al. 2008). Northern blotting was performed as described (Cerrito et al. 2004; Galbaugh et al. 2006). Beta-actin probe was hybridized to the same membrane and scanned on Packard beta scanner for normalization purposes. Realtime PCR was performed using SYBR green PCR kits and a 7500 RealTime PCR instrument (Applied Biosystems). Primers include: actin forward 5’ TTACTGCTCTGGCTCCTAGCA, reverse 5’ GACTCATCGTACTCCTGCTTGC; CTGF/CCN2 primers amplified a 500 bp fragment in the mouse CTGF 3’UTR, forward 5’ACAGTTGTTCATTAGCGCAC and reverse: 5’CTATGGTGTTTGGAGTTTGA.
Expression vector construction The open reading frame of identified genes was tagged with the myc epitope tag by PCR and inserted into pCR3.1 vector (Invitrogen) as described (Wang et al. 2008). The clones were verified by DNA sequencing prior to use in expression studies.
Results
Lactogenic differentiation-induced gene expression in HC11 cells To follow the global changes in gene transcription in HC11 mouse mammary epithelial cells undergoing lactogenic differentiation, we employed an oligonucleotide based microarray technique. The experiments were performed with RNA extracted from HC11 cells maintained in either growth media or HC11 cells exposed to lactogenic differentiation media containing dexamethasone, insulin and prolactin (DIP). HC11 cells undergoing lactogenic differentiation have a unique phenotypic appearance as clusters of enlarged cells in dome-like structures, referred to as mammospheres, appear in the culture (Fig. 1c). Total RNA was purified from the cells and fluorescent cRNA was synthesized. In the differentiation experiments (N = 5), RNA from DIP treated cells was labeled with cyanine 5-CTP, and RNA from the control cells was labeled with cyanine 3-CTP. Differences in gene expression were detected by hybridization of labeled RNA to slide-based oligonucleotide microarrays. The NIA mouse development oligonucleotide microarrays containing 20280 genes (produced by Agilent Technologies) were employed for the study. Hybridization reactions were analyzed using an Axon microarray reader. Normalization and statistical analysis and the analysis of gene expression using BRB-Array Tools Version 3 was performed as described in the Materials and Methods Section. In the experiments comparing HC11 cells induced to differentiate with HC11 control cells (DIP versus control), there were 10813 genes that passed filtering criteria from a total of 20280 genes on the NIA microarrays. The changes in expression of 998 genes met the 2-fold change criteria. Table 1 shows the partial list of genes exhibiting increased expression during lactogenic differentiation of HC11 cells and Supplemental File 1 contains the information on the 998 genes that met the 2-fold change at the p value 0.05. In the case studies carried out using the NIA 20280 gene arrays fewer genes exhibited a significant reduction in expression than an increase in expression during the 72 h differentiation period. Table 2 contains a partial list from the 20 genes exhibiting a minimum two-fold decrease.
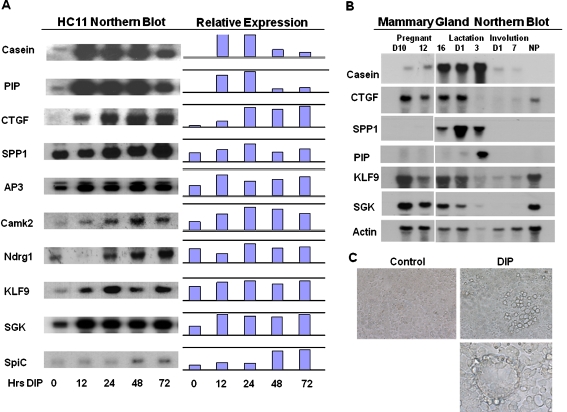
Fig. 1.
Northern blotting of genes exhibiting elevated expression in HC11 cells undergoing lactogenic differentiation. a. HC11 mouse mammary epithelial cells were cultured in complete medium with EGF for 4 days post confluence followed by incubation in the media without EGF for 24 h. The cells were then stimulated with differentiation media containing dexamethasone (10−6 M), insulin(5 µg/ml) and prolactin (5 µg/ml) for 0, 12, 24, 48, and 72 h. RNA was extracted and used for northern blots. The probes are described in Materials and Methods. The fold changes of gene expression of each probe normalized to β-actin is shown. b. Northern blotting of genes exhibiting elevated expression in HC11 cells hybridized to RNA from mouse mammary glands. The probes used in part A were hybridized to northern blots of RNA extracted form mouse mammary glands at various stages of pregnancy and lactation. The RNAs are from pregnancy days 10, 12 and 16; lactation days 1 and 3; involution days 1 and 7; and NP represents RNA from non-pregnant mammary glands of adult female mice. The blots were hybridized to β-actin as a loading control. C. HC11 cells undergoing lactogenic differentiation. Control = untreated cells, DIP = cells exposed to DIP for 5 days. Top panel: cells at magnification 200X; bottom panel: cells at magnification of 500X
Table 1.
Elevation of gene expression following stimulation of HC11 cells with DIP
| mean of log ratio | GB acc | Description |
|---|---|---|
| 17.067 | NM_010217.1 | connective tissue growth factor |
| 13.447 | NM_138314.1 | protein expressed in non-metastatic cells 7 |
| 11.15 | XM_150141.2 | LOC234574 |
| 9.52 | NM_009263.1 | secreted phosphoprotein 1 |
| 7.69 | BC009155.1 | microsomal glutathione S-transferase 1 |
| 7.64 | NM_022032.1 | p53 apoptosis effector related to Pmp22 |
| 6.99 | NM_011326.1 | sodium channel, nonvoltage-gated 1 gamma |
| 6.44 | NM_009681.1 | adaptor-related protein complex AP-3, sigma 1 subunit |
| 6.34 | NM_029083.1 | RIKEN cDNA 5830413E08 gene |
| 5.92 | BC036990.1 | metallothionein 1 |
| 5.85 | XM_123496.1 | Tcfcp2-related transcriptional repressor 1 |
| 5.78 | NM_007621.1 | carbonyl reductase 2 |
| 5.77 | NM_138578.1 | Bcl2-like |
| 5.76 | NM_012519.1 | calcium/calmodulin-dependent protein kinase II, delta |
| 5.72 | NM_009976.1 | cystatin C |
| 5.42 | NM_010286.1 | glucocorticoid-induced leucine zipper |
| 5.15 | NM_009128.1 | stearoyl-Coenzyme A desaturase 2 |
| 5.14 | NM_025610.1 | RIKEN cDNA 2410004D18 gene |
| 5.13 | NM_011361.1 | serum/glucocorticoid regulated kinase |
| 4.88 | AK009928.1 | RIKEN cDNA 2310051E17 gene |
| 4.82 | NM_011313.1 | S100 calcium binding protein A6 (calcyclin) |
| 4.79 | AF434663.1 | immunoglobulin superfamily, member 4 |
| 4.75 | NM_010730.1 | annexin A1 |
| 4.60 | NM_026217.1 | autophagy 12-like (S. cerevisiae) |
| 4.60 | NM_025436.1 | sterol-C4-methyl oxidase-like |
| 4.52 | NM_010884.1 | N-myc downstream regulated 1 |
| 4.48 | XM_177182.1 | expressed sequence C85344 |
| 4.44 | NM_011400.1 | solute carrier family 2 (facilitated glucose transporter) |
| 4.41 | NM_011803.1 | core promoter element binding protein |
| 4.41 | NM_010907.1 | nuclear factor of kappa light chain gene enhancer |
| 4.34 | NM_013470.1 | annexin A3 |
| 4.33 | NM_145942.1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| 4.21 | AL080093.1 | expressed sequence AU067636 |
| 4.16 | XM_132485.1 | RAS-related C3 botulinum substrate 1 |
| 4.11 | NM_010762.1 | myelin and lymphocyte protein, T-cell differentiation |
| 4.10 | NM_009983.2 | cathepsin D |
| 4.08 | AF378830.1 | prostaglandin-endoperoxide synthase 2 |
| 4.05 | NM_145977.1 | RIKEN cDNA 2210413P12 gene |
| 4.01 | NM_146120.1 | gelsolin |
| 3.98 | BC020081.1 | RIKEN cDNA 2310016C08 gene |
| 3.95 | BC019207.1 | low density lipoprotein receptor |
| 3.95 | NM_053623.1 | expressed sequence AU018108 |
| 3.93 | NM_019971.1 | platelet-derived growth factor, C polypeptide |
| 3.92 | NM_013492.1 | clusterin |
| 3.89 | NM_008084.1 | glyceraldehyde-3-phosphate dehydrogenase |
| 3.87 | BC038392.1 | ATPase, H+ transporting, lysosomal 70kD, V1 |
| 3.85 | NM_033079.1 | DNA segment, Chr 6, Miriam Meisler 5, expressed |
| 3.84 | BC037116.1 | adducin 3 (gamma) |
Regulation of gene expression in HC11 cells undergoing lactogenic differentiation. To determine the effect of lactogenic differentiation on gene transcription in HC11 cells oligonucleotide microarrays representing 20280 genes (NIA 20 k arrays from Agilent Technologies) were hybridized with RNA from DIP treated HC11 cells. The RNA was labeled with cyanine 5-CTP and control RNA was labeled with cyanine 3-CTP. Fluorescent cRNA was synthesized and hybridized in the differentiation experiments (N = 5) as described in Materials and Methods. Analysis of gene expression was performed using BRB-Array Tools Version 3. In The HC11-DIP versus HC11-control RNA experiments, there are 10813 genes that passed filtering criteria in total of 20280 genes. The first 2479 genes were significant at the nominal 0.05 level of the paired T-test, and 998 genes met the 2-fold change criteria. Table 1 shows the first 50 genes with elevated expression in cells undergoing lactogenic differentiation
Table 2.
Genes exhibiting decreased expression following stimulation of HC11 Cells with DIP
| Mean of log ratio | GB acc | Description |
|---|---|---|
| 0.66 | NM_007671.1 | cyclin-dependent kinase inhibitor 2C (p18) |
| 0.59 | NM_009335 | transcription factor AP-2, gamma |
| 0.56 | NM_007631.1 | cyclin D1 |
| 0.56 | Mm.11987 | ESTs, Moderately similar to POL2_MOUSE |
| 0.53 | Mm.173695 | ESTs, Moderately similar to cofactor forSp1 |
| 0.47 | AF316985.1 | toll-like receptor 1 |
| 0.44 | NM_021355.1 | fibromodulin |
| 0.44 | M_008086.1 | growth arrest specific 1 |
| 0.37 | C012724.1 | insulin-like growth factor binding protein 2 |
Decreased gene expression in HC11 cells undergoing lactogenic differentiation. RNA from HC11 cells undergoing lactogenic differentiation and HC11 control cells was analyzed for changes in gene transcription using oligonucleotide microarrays as described in the legend of Table 1. The results in Table 2 show the genes with significantly decreased expression in HC11 cells undergoing lactogenic differentiation
Elevated expression of hormone-regulated genes, genes associated with cell cycle control or cell survival and tissue reorganization during lactogenic differentiation of HC11 cells The genes induced during lactogenesis included transcriptional regulatory factors, proteins involved in growth control and survival, tissue re-organization and proteins important for milk production. Several previous studies performed on mouse mammary gland tissue reported alterations in the expression of genes that impact numerous pathways (Clarkson and Watson 2003) (Gass et al. 2003) (Anderson et al. 2007; Rudolph et al. 2007) (Stein et al. 2007). The majority of the studies examined changes in gene expression using RNA that was derived from the multiple cell types making up the mammary glands. In the present study the expression changes are restricted to genes expressed in epithelial cells. Therefore, while our findings confirm many of the findings of previous studies, all the changes reported are restricted to epithelial cells. Hence, some of the differences between HC11 cells and mammary tissue may be attributed to the lack of stromal or immune cell components in our analysis. Some examples of the functional divisions represented among DIP-regulated genes are included in Table 3. Many of the changes relate to milk protein production, but transcription factors and regulators, genes for growth control and tissue reorganization are well represented as are genes regulated by glucocorticoids and prolactin. Genes exhibiting significant changes in expression as a result of DIP-stimulation of HC11 cells were selected for analysis by northern blot to confirm the results obtained by microarray (Fig. 1a). The use of northern blotting for this aspect of the study allowed confirmation of the size of the mRNA in addition to the amount of RNA. Northern blots of HC11 cell RNA from cells stimulated with DIP for 0 to 72 h were hybridized with specific probes as well as a probe for actin. Following hybridization the results were quantified using a β-scanner and the fold changes in expression were obtained by normalization to β-actin expression. The normalized data revealed a positive correlation between the fold changes obtained in the microarray and the northern blot (Fig. 1). The expression of target genes was also analyzed by real-time PCR and the results support those obtained by northern blot. (data not shown). These data clearly demonstrated the significant increase in CTGF/CCN2 and SPP1/osteopontin gene expression following lactogenic stimulation of HC11 cells.To ascertain if the genes detected in the array analysis were similarly regulated in the mouse mammary gland in vivo RNA from pregnant and lactating mouse mammary glands was analyzed. Northern blots containing RNA from non pregnant, pregnant, lactating and involuting mouse mammary gland tissue were hybridized to probes encoding genes differentially expressed in DIP-stimulated HC11 cells. The results in Fig. 1b indicate that a number of the genes exhibit modest increases in expression during pregnancy and lactation. A number of the genes tested in this way, excluding β-casein, were also expressed in non-pregnant mammary glands but little or no expression of the specifc RNAs was detected during postlactation involution of the mammary gland.CTGF/CCN2 can be induced by dexamethasone in chondrocytes and renal epithelia. Hence, further studies explored the steroid regulation of CTGF/CCN2 expression in HC11 cells. While treatment of HC11 cells with dexamethasone induced expression of CTGF, neither estrogen nor progesterone produced the same result and CTGF/CCN2 expression was not dependent on TGFβ in these cells (Wang et al. 2008). The realtime PCR results in Fig. 2 demonstrate that the glucocorticoid receptor antagonist RU486 efficiently blocked CTGF/CCN2 expression in response to dexamethasone confirming the function of the steroid hormone in the HC11 cells. Our additional studies demonstrated that CTGF/CCN2 is required for HC11 lactogenic differentiation and that ectopic expression of CTGF/CCN2 enhances differentiation in HC11 cells as well as in primary mouse mammary epithelial cell cultures (Wang et al. 2008).
Table 3.
Functional division of lactogenic differentiation-regulated genes
| A. Up-Regulated genes | |
|---|---|
| Prolactin Regulated | |
| NM_008843 | prolactin induced protein |
| NM_009972 | casein beta |
| Dexamethasone Regulated | |
| NM_011361 | serum/glucocorticoid regulated kinase |
| NM_010286 | glucocorticoid-induced leucine zipper |
| NM_010217 | CTGF/CCN2 |
| NM_010730.1 | annexin A1 |
| Milk production: including carbohydrate and lipid metabolism | |
| NM_019739.1 | Fox O1 |
| AF378830.1 | prostaglandin-endoperoxide synthase 2 |
| NM_013467 | alcohol dehydrogenase family 1, subfamily |
| NM_007607 | carbonic anhydrase 4 |
| NM_007621.1 | carbonyl reductase 2 |
| NM_009128.1 | stearoyl-Coenzyme A desaturase 2 |
| NM_025436.1 | sterol-C4-methyl oxidase-like |
| NM_145942.1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 |
| NM_008084.1 | glyceraldehyde-3-phosphate dehydrogenase |
| BC038392.1 | ATPase, H+ transporting, lysosomal |
| BC019207.1 | low density lipoprotein receptor |
| NM_019703.1 | phosphofructokinase, platelet |
| NM_134469.1 | farnesyl diphosphate synthetase |
| NM_013820.1 | hexokinase 2 |
| NM_010687.1 | like-glycosyltransferase |
| NM_009270.1 | squalene epoxidase |
| U12330.1 | sterol regulatory element binding factor |
| NM_028803.1 | glucan (1,4-alpha-), branching enzyme 1 |
| NM_007408.1 | adipose differentiation related protein |
| XM_131456.2 | solute carrier family 31, member 1 |
| NM_011400.1 | solute carrier family 2glucose transporter) |
| NM_008489 | lipopolysaccharide binding protein |
| Apoptosis | |
| NM_022032.1 | Perp, p53 apoptosis effector |
| NM_138578.1 | Bcl2-like |
| NM_026217.1 | autophagy 12-like |
| U05989.1 | Par-4 |
| NM_019745.1 | programmed cell death 10 |
| NM_013492.1 | clusterin |
| Cytoskeleton, extracellular matrix, membrane structure | |
| NM_146120.1 | gelsolin |
| NM_009071.1 | Rho-associated coiled-coil kinase 1 |
| XM_132485.1 | RAS-related C3 botulinum substrate |
| BC037116.1 | adducin 3 (gamma) |
| NM_013470.1 | annexin A3 |
| NM_011313.1 | S100 calcium binding protein A6 (calcyclin) |
| BC013248.1 | kinesin family member 5B |
| XM_130768.1 | laminin, alpha 5 |
| NM_130448.1 | protocadherin 18 |
| NM_007404.1 | ADAM-9 (meltrin gamma) |
| D67076.1 | ADAMTS-1 |
| NM_009263.1 | secreted phosphoprotein/ostepontin |
| NM_010217 | CTGF/CCN2 |
| NM_010728.1 | lysyl oxidase |
| NM_009983.2 | cathepsin D |
| NM_144554.1 | claudin 4 |
| BC038280.1 | caveolin, caveolae protein, 22 kDa |
| NM_016900. | caveolin 2 |
| Transcription Factors and regulators | |
| NM_011461 | Spi-C ETS transcription factor |
| XM_123496.1 | Tcfcp2-related transcriptional repressor 1 |
| NM_011803.1 | CPEBP |
| NM_057211.1 | Kruppel-like factor 9 |
| NM_019739.1 | Fox O1 |
| NM_010907.1 | inhibitor of NFkB |
| Growth Control and Signaling | |
| NM_007669.1 | cdk inhibitor 1A (P21) |
| NM_007635.1 | cyclin G2 |
| NM_008960.1 | phosphatase and tensin homolog |
| NM_010515.1 | insulin-like growth factor 2 receptor |
| NM_021099.2 | kit oncogene |
| NM_007918.1 | eukaryotic translation initiation factor 4E binding protein 1 |
| NM_009721 | ATPase, Na+/K+transporting, beta 1 poly |
| BC009155.1 | microsomal glutathione Stransferase 1 |
| NM_010884.1 | N-myc downstream regulated 1 |
| NM_019971.1 | PDGF, C polypeptide |
| NM_011057 | PDGF, B polypeptide |
| NM_012671.1 | transforming growth factor alpha |
| NM_011338 | small inducible cytokine A9 |
| NM_011944 | MAPKK |
| NM_012519.1 | calcium/calmodulin-dependent protein kinase II, delta |
| AF378830.1 | prostaglandin-endoperoxide synthase 2 |
| BC038069.1 | IGFBP5 |
| BC002092.1 | v-maf oncogene family, protein G |
The functional division of genes transcriptionally regulated in HC11 cells undergoing lactogenic differentiation. The list of genes that exhibit elevated expression in HC11 cells undergoing lactogenic differentiation are categorized in functional divisions. The list does not include all genes with 2-fold or greater increase and some genes are listed in more than one category
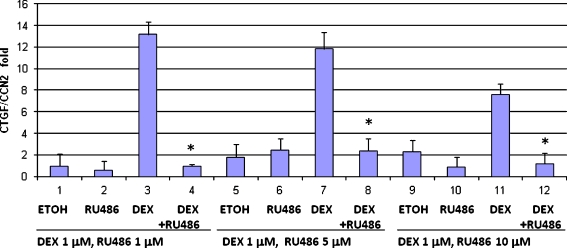
Fig. 2.
CTGF/CCN2 expression is regulated by dexamethasone in HC11 cells. HC11 mammary epithelial cells were grown to confluence and stimulated with dexamethasone (DEX)(1 μM) in serum-containing media in the presence of insulin. RU486 at varying concentrations or vehicle (ethanol) was added to cells alone or in combination with dexamethasone. RNA was isolated and levels of CTGF and actin RNA were determined by realtime PCR. The results indicate the induction of CTGF/CCN2 normalized to actin. *These Dex+Ru486 values represent statistically significant difference (p-value .001) from the Dex alone condition
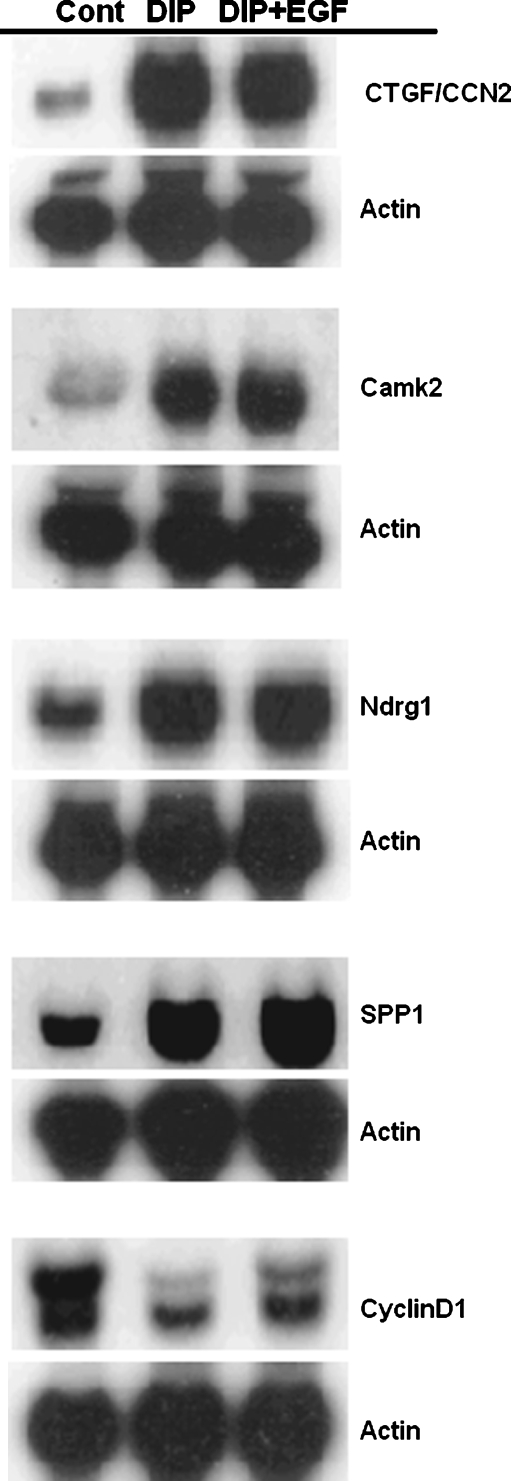
Regulation of gene expression by EGF during lactogenic differentiation in HC11 cells EGF, or other potent mitogens, are required for the proliferation and development of “competency” of HC11 cells to differentiate (Marte et al. 1995a; Merlo et al. 1994; Taverna et al. 1991). However, the inclusion of EGF, other mitogenic growth factors or expression of transforming oncogenes in HC11 cells blocks lactogenic differentiation (Cerrito et al. 2004; Hynes et al. 1990; Marte et al. 1995a; Merlo et al. 1996; Peterson and Haldosen 1998). To examine the effect of EGF on the DIP-induced expression pattern of the HC11 cells another microarray experiment was performed. For this study RNA was extracted from HC11 cells induced to differentiate with DIP and from HC11 cells stimulated with DIP containing 10 ng/ml EGF. Total RNA was purified and fluorescent cRNA was synthesized as described above. RNA from DIP plus EGF treated cells was labeled with cyanine 5-CTP, and RNA from DIP-treated cells was labeled with cyanine 3-CTP. Hybridization was carried out and the results were analyzed as described above. The analysis of gene expression revealed that in EGF plus DIP versus DIP experiments 1129 genes are significant at the nominal 0.05 level of the paired T-test, and 128 genes met the 2-fold change criteria. Table 4 shows the partial list of genes for which expression was regulated by EGF in the presence of DIP. The remainder of the data is contained in Supplemental file 2. Many of the genes expressed at higher level in the EGF plus DIP-stimulated cells are known targets of EGF signal transduction pathways or are associated with malignant growth, for example cyclin D1 and osteopontin. Confirmation of the array results for a number of genes was performed by northern blotting. RNA from undifferentiated HC11 cells or cells stimulated with either DIP or DIP plus EGF was hybridized to probes for several differentially expressed genes. The results are shown in Fig. 3. The genes that are transcriptionally activated by DIP were generally unaffected by the inclusion of EGF in the differentiation media. However, it was clear that the expression of cyclin D1 decreased during differentiation but it increased with inclusion of EGF in the differentiation media. This finding reflects the increase in the level of cyclin D1 in the microarray analysis.
Table 4.
Partial list of genes transcriptionally regulated by EGF (DIP + EGF) versus DIP
| Expression elevated in DIP+EGF | ||
|---|---|---|
| Mean log ratio | GB acc | Description |
| 2.63 | NM_009263.1 | secreted phosphoprotein 1 |
| 2.54 | NM_016980.1 | ribosomal protein L5 |
| 2.52 | AF378830.1 | prostaglandin-endoperoxide synthase 2 |
| 2.46 | NM_008972.1 | prothymosin alpha |
| 2.35 | NM_009121.1 | spermidine/spermine N1-acetyl transferase |
| 2.33 | NM_019682.1 | dynein, cytoplasmic, light chain 1 |
| 2.32 | BC006739.1 | catenin beta |
| 2.31 | NM_133777.2 | ubiquitin-conjugating enzyme E2S |
| 2.30 | NM_023372.1 | ribosomal protein L38 (Rpl38), transcript variant 2 |
| 2.29 | NM_007631.1 | cyclin D1 |
| 2.28 | BC010581.1 | stathmin 1 |
| 2.28 | XM_134967.2 | eukaryotic translation elongation factor 1 alpha 1 |
| 2.27 | NM_031165.1 | heat shock protein 8 |
| 2.27 | NM_022891.1 | ribosomal protein L23 |
| 2.26 | NM_008305.1 | perlecan (heparan sulfate proteoglycan 2) |
| 2.26 | NM_025592.2 | ribosomal protein L35 |
| 2.26 | NM_009608.1 | actin, alpha, cardiac |
| 2.26 | BC034257.1 | epithelial membrane protein 1 |
| 2.25 | NM_019703.1 | phosphofructokinase, platelet |
| 2.25 | XM_178595.1 | chromobox homolog 3 (Drosophila HP1 gamma) |
| 2.23 | NM_133834.1 | expressed sequence AA407306, |
| 2.23 | NM_009255.1 | serine (or cysteine) proteinase inhibitor, clade E, |
| 2.22 | BC010726.1 | phospholipase A2, group VII (platelet-activating factor) |
| 2.22 | NM_013261.1 | peroxisome proliferative activated receptor, gamma, |
| 2.22 | XM_129211.2 | phosphoserine aminotransferase |
| 2.21 | NM_010480.1 | heat shock protein, 1 |
| 2.21 | NM_025919.1 | ribosomal protein L11 (Rpl11) |
| 2.20 | XM_128846.2 | similar to 60 S RIBOSOMAL PROTEIN L17 (L23) |
| 2.20 | BC012508.1 | SET translocation |
| 2.20 | NM_145142.1 | HNK-1 sulfotransferase |
| 2.19 | NM_009076.1 | hypothetical pseudogene ribosomal protein L12 |
| 2.19 | NM_007961.1 | ets variant gene 6 (TEL oncogene) |
| 2.17 | AK021352.1 | RIKEN cDNA D730048J04 gene (day10 lact mamm gland) |
| 2.17 | NM_028044.1 | calponin 3, acidic |
| 2.16 | BC003308.1 | casein kinase 1, delta |
| 2.15 | NM_011400.1 | solute carrier family 2 (facilitated glucose transporter), |
| 2.14 | NM_008774.1 | poly A binding protein, cytoplasmic 1 |
| 2.14 | XM_135387.1 | similar to 40 S RIBOSOMAL PROTEIN S19 |
| 2.14 | BC002046.1 | ephrin A1 |
| 2.13 | NM_011218.1 | protein tyrosine phosphatase, receptor type, Sigma |
| 2.13 | XM_126364.1 | similar to ATP synthase, H + transporting, mitochondrial |
| Expression lower in presence of EGF | ||
| 0.56 | NM_002747 | MAPK4 |
| 0.59 | L25602.1 | BMP-2 |
| 0.59 | mannose-6-phosphate receptor transporter | |
| 0.59 | NM_011224.1 | muscle glycogen phosphorylase |
| 0.59 | NM_011500.1 | striatin |
| 0.58 | NM_021454.1 | cdc42 effector protein |
| 0.57 | NM_010086.1 | ADAM24 |
| 0.56 | NM_008372.2 | interleukin 7 receptor |
| 0.56 | U44955.1 | connexin 3 |
| 0.55 | NM_020496.1 | T-box 20 |
| 0.50 | BC033410.1 | eukaryotic translation initiation factor 4E |
Genes exhibiting elevated or decreased gene expression in HC11 cells undergoing lactogenic differentiation in the presence of EGF compared to HC11 lactogenic controls. RNA from HC11 cells undergoing lactogenic differentiation in the presence or absence of 10 ng/ml EGF was analyzed for changes in gene transcription using oligonucleotide microarrays as described in Materials and Methods. The results include genes exhibiting a greater than two-fold increase or significant decrease in the presence of EGF
Fig. 3.
Northern blot results of selected on RNA from HC11 mouse mammary epithelial cells undergoing lactogenic differentiation in the presence and absence of EGF. HC11 cells were grown as described in Fig. 1A and then incubated in differentiation media containing dexamethasone (10−6 M), insulin (5 µg/ml) and prolactin (5 µg/ml) with or without EGF (10 ng/ml) for 0 or 72 h. RNAs were extracted and used for Northern blot with the probes indicated. The blots were hybridized to β-actin as a loading control
Discussion
Several microarray studies have examined transcriptional control in mouse mammary epithelial cells undergoing lactogenic differentiation (Kelly et al. 2002; Clarkson and Watson 2003; Rudolph et al. 2003; Rudolph et al. 2007). In addition, proteomics analyses have been used to examine changes at the level of the protein expression in differentiating HC11 cells (Desrivieres et al. 2003) (Desrivieres et al. 2007). Our study analyzed mammary epithelial cell lactogenic differentiation and developed a global expression profile of HC11 cells undergoing differentiation using oligonucleotide microarrays. In addition, the changes in gene expression that occur as a result of mitogenic stimulation of HC11 cells undergoing lactogenic differentiation were analyzed. To qualify the accuracy of the microarray data both northern blot and real-time PCR techniques were used, and results showed very good correspondence. While numerous genes were regulated during differentiation (Table 1), the analysis of expression in different functional pathways (Table 3) revealed some changes not previously observed either by proteomic studies of HC11 cells or by the analysis of the lactational switch in the mouse mammary gland.
A number of proteins that regulate aspects of the extracellular matrix were expressed in differentiating HC11 cells. Two members of the small integrin binding protein family, connective tissue growth factor (CTGF/CCN2) and secreted phosphoprotein (SPP1/osteopontin) were transcriptionally activated during lactational differentiation of HC11 cells suggesting that these proteins may play a role in extracellular matrix accumulation necessary for competence to undergo differentiation. CTGF/CCN2, a CCN protein, is a cysteine-rich protein that can modulate numerous cellular responses including proliferation, chemotaxis, adhesion, migration, and extracellular matrix production (Perbal 2004; Brigstock 2002; Brigstock 2003; Leask and Abraham 2006). CTGF/CCN2 promotes endothelial cell growth, migration, adhesion, and survival in vitro, and CTGF action in angiogenesis is mediated at least partly through interactions with integrins (Ivkovic et al. 2003; Gao and Brigstock 2004; Gao and Brigstock 2006). Appropriate integrin signaling is required for lactogenic differentiation in vivo (Li et al. 2005; Naylor et al. 2005). The expression of integrins is regulated during lactogenesis in mouse mammary gland (Kelly et al. 2002; Clarkson and Watson 2003; Rudolph et al. 2003; Rudolph et al. 2007). Profibrotic activity of CTGF/CCN2 is related to high levels in many fibrotic lesions, and CTGF/CCN2 is induced by and acts as a co-factor for transforming growth factor β in the induction of fibrogenesis (Perbal 2004). In our study, CTGF/CCN2 is highly up-regulated in differentiated HC11 cells, suggesting that it might play an important role in matrix signals required for differentiation. However, expression of CTGF/CCN2 in HC11 cells does not require TGFβ, its expression is induced by dexamethasone (Wang et al. 2008). Moreover, ectopic expression of CTGF/CCN2 enhances HC11 lactogenic differentiation and depletion of CTGF inhibits differentiation (Wang et al. 2008).
Secreted phosphoprotein1(SPP1)/ osteopontin is a secreted glycoprotein that is rich in aspartate and sialic acid residues (Craig et al. 1988) and contains functional domains for calcium-binding, phosphorylation, glycosylation, and extra-cellular matrix adhesion (Kazanecki et al. 2007). Osteopontin is associated with cartiledge and bone but has multiple molecular functions that mediate cell adhesion, chemotaxis, macrophage-directed interleukin-10 suppression, stress-dependent angiogenesis, prevention of apoptosis, and anchorage-independent growth of tumor cells (Mi et al. 2004). A substantial body of data has recently linked osteopontin with the regulation of metastatic spread by tumor cells (Hayashi et al. 2007). A number of genes that are transcriptionally activated in HC11 cells undergoing lactogenic differentiation are also regulated during skeletogenesis. A recent study identified a number of proteins that are elevated in HC11 lactogenic differentiation as Runx2-regulated in developing skeleton including osteopontin, CTGF/CCN2, and CCCL9 (Hecht et al. 2007). Like CTGF expression, osteopontin levels are high in breast tumors that metastasize to the bone (Kang et al. 2003) (Carlinfante et al. 2003). If there is a connection between the expression of these small secreted proteins and the ability of metastatic breast tumors to target the bone environment remains to be conclusively determined, but they are readily transcriptionally activated in HC11 cells. The secreted osteopontin protein is also widely detected in plasma, urine and bile, and a recent study demonstrated that it is present in human milk (Nagatomo et al. 2004). A variety of stimuli, including phorbol 12-myristate 13-acetate, 1,25-dihydroxyvitamin D, basic fibroblast growth factor, tumor necrosis factor-α, interleukin-1, interferonγ and lipopolysaccharide, elevated osteopontin expression. However, whether osteopontin expression in HC11 cells is due to dexamethasone, prolactin, or insulin is not established, but osteopontin expression was enhanced by the addition of EGF to the lactogenic hormone mix. Other extracellular matrix proteins and proteins involved in tissue reorganization are induced during lactogenic differentiation of HC11 cells (Table 3). The list includes structural proteins as well as proteases. In addition, an increase in expression of Rac1 and ROCK were observed, relevant because a role for Rac1 has been proposed in mammary epithelial cell differentiation (Akhtar and Streuli 2006).
The regulation of key growth and cell cycle regulators plays an important role in the HC11 differentiation progression. Relative expression of cyclin D1 declines along with that of the p18INK4. HC11 cells undergo growth arrest during differentiation in part regulated by the decrease in cyclin D1. There is a concurrent increase in p21CIP suggesting it is the potent cdk inhibitor contributing to control of growth arrest in HC11 cells in this system. While cyclin G2 increased following DIP-stimulation it is likely that this is the result of glucocorticoid-mediated event (Vedoy and Sogayar 2002). Our analysis detected alterations in the regulation of IGFBP2 and IGFBP5 in HC11 cells undergoing lactogenic differentiation in agreement with a previously reported finding (Phillips et al. 2003).
Lactogenic differentiation induced multiple gene transcription events, while EGF blocked a number of these processes. For example, during lactogenic differentiation, the transcription element binding proteins (KLF4, KLF9) were induced, but were inhibited by EGF. KLF9 has been implicated as a transactivator of progesterone – and estrogen-dependent transcription in endometrial epithelium, another hormonal responsive tissue. (Velarde et al. 2007; Zhang et al. 2003). This provided, in part, an explanation of the large scale gene expression changes during lactogenic differentiation that were blocked by EGF. Numerous ribosomal protein transcripts were induced in both differentiation and EGF blockage process, suggesting that these genes are not a target of the mitogen-induced inhibition. In conclusion, our data provide further insight into the signal transduction pathways and gene interactions in the HC11 mouse mammary epithelial cell line during lactogenic differentiation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Dr. David Salomon for commentary and advice. The work was supported by grants from the Congressionally Directed Medical Research Fund (DAMD17-01-0264), NIH (R01CA90908) and USMCI to M. L. Cutler.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-009-0047-5) contains supplementary material, which is available to authorized users.
References
- Akhtar N, Streuli CH (2006) Rac1 links integrin-mediated adhesion to the control of lactational differentiation in mammary epithelia. J Cell Biol 173:781–793. doi:10.1083/jcb.200601059 [DOI] [PMC free article] [PubMed]
- Ali S (1998) Prolactin receptor regulates Stat5 tyrosine phosphporylation and nuclear translocation by two separate pathways. J Biol Chem 273:7709–7716. doi:10.1074/jbc.273.13.7709 [DOI] [PubMed]
- Anderson SM, Rudolph MC, McManaman JL, Neville MC (2007) Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis!. Breast Cancer Res 9:204. doi:10.1186/bcr1653 [DOI] [PMC free article] [PubMed]
- Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B (1988) Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J 7:2089–2095 [DOI] [PMC free article] [PubMed]
- Brigstock DR (2002) Regulation of angiogenesis and endothelial cell function by connective tissue growth factor (CTGF) and cysteine-rich 61 (CYR61). Angiogenesis 5:153–165. doi:10.1023/A:1023823803510 [DOI] [PubMed]
- Brigstock DR (2003) The CCN family: a new stimulus package. J Endocrinol 178:169–175. doi:10.1677/joe.0.1780169 [DOI] [PubMed]
- Carlinfante G, Vassiliou D, Svensson O, Wendel M, Heinegard D, Andersson G (2003) Differential expression of osteopontin and bone sialoprotein in bone metastasis of breast and prostate carcinoma. Clin Exp Metastasis 20:437–444. doi:10.1023/A:1025419708343 [DOI] [PubMed]
- Cerrito MG, Galbaugh T, Wang W, Chopp T, Salomon D, Cutler ML (2004) Dominant negative Ras enhances lactogenic hormone-induced differentiation by blocking activation of the Raf-Mek-Erk signal transduction pathway. J Cell Physiol 201:244–258. doi:10.1002/jcp.20077 [DOI] [PMC free article] [PubMed]
- Clarkson RW, Watson CJ (2003) Microarray analysis of the involution switch. J Mammary Gland Biol Neoplasia 8:309–319. doi:10.1023/B:JOMG.0000010031.53310.92 [DOI] [PubMed]
- Craig AM, Nemir M, Mukherjee BB, Chambers AF, Denhardt DT (1988) Identification of the major phosphoprotein secreted by many rodent cell lines as 2ar/osteopontin: enhanced expression in H-ras-transformed 3 T3 cells. Biochem Biophys Res Commun 157:166–173. doi:10.1016/S0006-291X(88)80028-7 [DOI] [PubMed]
- Danielson K, Oborn C, Durbam E, Butel J, Medina D (1984) Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci USA 81:3756–3760. doi:10.1073/pnas.81.12.3756 [DOI] [PMC free article] [PubMed]
- Desrivieres S, Kuhn K, Muller J, Glaser M, Laria NC, Korder J, Sonnentag M, Neumann T, Schwarz J, Schafer J et al (2007) Comparison of the nuclear proteomes of mammary epithelial cells at different stages of functional differentiation. Proteomics 7:2019–2037. doi:10.1002/pmic.200600994 [DOI] [PubMed]
- Desrivieres S, Prinz T, Castro-Palomino Laria N, Meyer M, Boehm G, Bauer U, Schafer J, Neumann T, Shemanko C, Groner B (2003) Comparative proteomic analysis of proliferating and functionally differentiated mammary epithelial cells. Mol Cell Proteomics 2:1039–1054. doi:10.1074/mcp.M300032-MCP200 [DOI] [PubMed]
- Galbaugh T, Cerrito MG, Jose CC, Cutler ML (2006) EGF-induced activation of Akt results in mTOR-dependent p70S6 kinase phosphorylation and inhibition of HC11 cell lactogenic differentiation. BMC Cell Biol 7:34. doi:10.1186/1471-2121-7-34 [DOI] [PMC free article] [PubMed]
- Gao R, Brigstock DR (2004) Connective tissue growth factor (CCN2) induces adhesion of rat activated hepatic stellate cells by binding of its C-terminal domain to integrin alpha(v) beta(3) and heparan sulfate proteoglycan. J Biol Chem 279:8848–8855. doi:10.1074/jbc.M313204200 [DOI] [PubMed]
- Gao R, Brigstock DR (2006) A novel integrin alpha5beta1 binding domain in module 4 of connective tissue growth factor (CCN2/CTGF) promotes adhesion and migration of activated pancreatic stellate cells. Gut 55:856–862. doi:10.1136/gut.2005.079178 [DOI] [PMC free article] [PubMed]
- Gass S, Harris J, Ormandy C, Brisken C (2003) Using gene expression arrays to elucidate transcriptional profiles underlying prolactin function. J Mammary Gland Biol Neoplasia 8:269–285. doi:10.1023/B:JOMG.0000010029.85796.63 [DOI] [PubMed]
- Gouilleux F, Wakeo H, Mundt M, Groner B (1994) Prolactin induces phosphorylation of Tyr 694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J 13:4361–4369 [DOI] [PMC free article] [PubMed]
- Han Y, Watling D, Rogers N, Stark G (1997) Jak2 and Stat5, but not Jak1 and Stat1, are required for prolactin-induced beta-lactoglobulin transcription. Mol Endocrinol 11:1180–1188. doi:10.1210/me.11.8.1180 [DOI] [PubMed]
- Hayashi C, Rittling S, Hayata T, Amagasa T, Denhardt D, Ezura Y, Nakashima K, Noda M (2007) Serum osteopontin, an enhancer of tumor metastasis to bone, promotes B16 melanoma cell migration. J Cell Biochem 101:979–986. doi:10.1002/jcb.21298 [DOI] [PubMed]
- Hecht J, Seitz V, Urban M, Wagner F, Robinson PN, Stiege A, Dieterich C, Kornak U, Wilkening U, Brieske N et al (2007) Detection of novel skeletogenesis target genes by comprehensive analysis of a Runx2(-/-) mouse model. Gene Expr Patterns 7:102–112. doi:10.1016/j.modgep.2006.05.014 [DOI] [PubMed]
- Humphreys R, Rosen J (1997) Stably transfected HC11 cells provide and in vitro and in vivio model system for studying Wnt gene expression. Cell Growth Differ 8:839–849 [PubMed]
- Hynes NE, Taverna D, Harwerth IM, Ciardiello F, Salomon DS, Yamamoto T, Groner B (1990) Epidermal growth factor receptor, but not c-erbB-2, activation prevents lactogenic hormone induction of the beta-casein gene in mouse mammary epithelial cells. Mol Cell Biol 10:4027–4034 [DOI] [PMC free article] [PubMed]
- Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM (2003) Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130:2779–2791. doi:10.1242/dev.00505 [DOI] [PMC free article] [PubMed]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3:537–549. doi:10.1016/S1535-6108(03)00132-6 [DOI] [PubMed]
- Kazanecki CC, Uzwiak DJ, Denhardt DT (2007) Control of osteopontin signaling and function by post-translational phosphorylation and protein folding. J Cell Biochem 102:912–924. doi:10.1002/jcb.21558 [DOI] [PubMed]
- Kelly PA, Bachelot A, Kedzia C, Hennighausen L, Ormandy CJ, Kopchick JJ, Binart N (2002) The role of prolactin and growth hormone in mammary gland development. Mol Cell Endocrinol 197:127–131. doi:10.1016/S0303-7207(02)00286-1 [DOI] [PubMed]
- Leask A, Abraham DJ (2006) All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci 119:4803–4810. doi:10.1242/jcs.03270 [DOI] [PubMed]
- Li N, Zhang Y, Naylor MJ, Schatzmann F, Maurer F, Wintermantel T, Schuetz G, Mueller U, Streuli CH, Hynes NE (2005) Beta1 integrins regulate mammary gland proliferation and maintain the integrity of mammary alveoli. EMBO J 24:1942–1953. doi:10.1038/sj.emboj.7600674 [DOI] [PMC free article] [PubMed]
- Marte B, Jeschke M, Grause-Porta D, Taverna D, Hofer P, Groner B, Yarden Y, Hynes N (1995a) Neu differentiaiton factor/heregulin modulates growth and differentitaion of HC11 mammary epithelial cells. Mol Endocrinol 9:14–23. doi:10.1210/me.9.1.14 [DOI] [PubMed]
- Marte BM, Jeschke M, Graus-Porta D, Taverna D, Hofer P, Groner B, Yarden Y, Hynes NE (1995b) Neu differentiation factor/heregulin modulates growth and differentiation of HC11 mammary epithelial cells. Mol Endocrinol 9:14–23. doi:10.1210/me.9.1.14 [DOI] [PubMed]
- Master SR, Hartman JL, D’Cruz CM, Moody SE, Keiper EA, Ha SI, Cox JD, Belka GK, Chodosh LA (2002) Functional microarray analysis of mammary organogenesis reveals a developmental role in adaptive thermogenesis. Mol Endocrinol 16:1185–1203. doi:10.1210/me.16.6.1185 [DOI] [PubMed]
- Merlo G, Grause-Porta D, Cella N, Marte B, Taverna D, Hynes N (1996) Growth, differentiation and survival of HC11 mammary epithelial cells: diverse effects of receptor tyrosine kinase-activating peptide growth factors. Eur J Cell Biol 70:97–105 [PubMed]
- Merlo G, Venesio T, Taverna D, Marte B, Callahan R, Hynes N (1994) Growth suppression of normal mammary epithelial cells by wild type p53. Oncogene 9:443–453 [PubMed]
- Mi Z, Guo H, Wai PY, Gao C, Wei J, Kuo PC (2004) Differential osteopontin expression in phenotypically distinct subclones of murine breast cancer cells mediates metastatic behavior. J Biol Chem 279:46659–46667. doi:10.1074/jbc.M407952200 [DOI] [PubMed]
- Nagaiah K, Bolander FF Jr, Nicholas KR, Takemoto T, Topper YJ (1981) Prolactin-induced accumulation of casein mRNA in mouse mammary explants: a selective role of glucocorticoid. Biochem Biophys Res Commun 98:380–387. doi:10.1016/0006-291X(81)90851-2 [DOI] [PubMed]
- Nagatomo T, Ohga S, Takada H, Nomura A, Hikino S, Imura M, Ohshima K, Hara T (2004) Microarray analysis of human milk cells: persistent high expression of osteopontin during the lactation period. Clin Exp Immunol 138:47–53. doi:10.1111/j.1365-2249.2004.02549.x [DOI] [PMC free article] [PubMed]
- Naylor MJ, Li N, Cheung J, Lowe ET, Lambert E, Marlow R, Wang P, Schatzmann F, Wintermantel T, Schuetz G et al (2005) Ablation of beta1 integrin in mammary epithelium reveals a key role for integrin in glandular morphogenesis and differentiation. J Cell Biol 171:717–728. doi:10.1083/jcb.200503144 [DOI] [PMC free article] [PubMed]
- Perbal B (2004) CCN proteins: multifunctional signalling regulators. Lancet 363:62–64. doi:10.1016/S0140-6736(03)15172-0 [DOI] [PubMed]
- Peterson H, Haldosen L (1998) EGF modulates expression of Stat5 in mammary epithelial cells. Exp Cell Res 243:347–358. doi:10.1006/excr.1998.4160 [DOI] [PubMed]
- Phillips K, Park MA, Quarrie LH, Boutinaud M, Lochrie JD, Flint DJ, Allan GJ, Beattie J (2003) Hormonal control of IGF-binding protein (IGFBP)-5 and IGFBP-2 secretion during differentiation of the HC11 mouse mammary epithelial cell line. J Mol Endocrinol 31:197–208. doi:10.1677/jme.0.0310197 [DOI] [PubMed]
- Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC (2003) Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia 8:287–307. doi:10.1023/B:JOMG.0000010030.73983.57 [DOI] [PubMed]
- Rudolph MC, McManaman JL, Phang T, Russell T, Kominsky DJ, Serkova NJ, Stein T, Anderson SM, Neville MC (2007) Metabolic regulation in the lactating mammary gland: a lipid synthesizing machine. Physiol Genomics 28:323–336. doi:10.1152/physiolgenomics.00020.2006 [DOI] [PubMed]
- Stein T, Salomonis N, Gusterson BA (2007) Mammary gland involution as a multi-step process. J Mammary Gland Biol Neoplasia 12:25–35. doi:10.1007/s10911-007-9035-7 [DOI] [PubMed]
- Stocklin E, Wissler M, Gouilleux F, Groner B (1996) Functional interactions between Stat5 and the glucocorticoid receptor. Nature 383:726–728. doi:10.1038/383726a0 [DOI] [PubMed]
- Taverna D, Groner B, Hynes NE (1991) Epidermal growth factor receptor, platelet-derived growth factor receptor, and c-erbB-2 receptor activation all promote growth but have distinctive effects upon mouse mammary epithelial cell differentiation. Cell Growth Differ 2:145–154 [PubMed]
- Vedoy CG, Sogayar MC (2002) Isolation and characterization of genes associated with the anti-tumor activity of glucocorticoids. Brain Res Mol Brain Res 106:57–69. doi:10.1016/S0169-328X(02)00410-2 [DOI] [PubMed]
- Velarde MC, Zeng Z, McQuown JR, Simmen FA, Simmen RC (2007) Krupple-like Factor 9 is a negative regulator of ligand dependent estrogen receptor signaling in Ishikawa endometrial carcinoma cells. Mol Endocrinol 21:2988–3001 [DOI] [PubMed]
- Wang W, Morrison B, Galbaugh T, Jose CC, Kenney N, Cutler ML (2008) Glucocorticoid induced expression of connective tissue growth factor contributes to lactogenic differentiation of mouse mammary epithelial cells. J Cell Physiol 214:38–46. doi:10.1002/jcp.21159 [DOI] [PubMed]
- Zhang XL, Zhang D, Michel FJ, Blum JL, Simmen FA, Simmen RC (2003) Selective interactions of Kruppel-like factor 9/basic transcription element-binding protein with progesterone receptor isoforms A and B determine transcriptional activity of progesterone-responsive genes in endometrial epithelial cells. J Biol Chem 278:21474–21482. doi:10.1074/jbc.M212098200 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.