Abstract
Two important cytokines mediating inflammation are tumor necrosis factor α (TNFα) and IL-1β, both of which require conversion to soluble forms by converting enzymes. The importance of TNFα-converting enzyme and IL-1β-converting enzyme in the production of circulating TNFα and IL-1β in response to systemic challenges has been demonstrated by the use of specific converting enzyme inhibitors. Many inflammatory responses, however, are not systemic but instead are localized. In these situations release and/or activation of cytokines may be different from that seen in response to a systemic stimulus, particularly because associations of various cell populations in these foci allows for the exposure of procytokines to the proteolytic enzymes produced by activated neutrophils, neutrophil elastase (NE), proteinase 3 (PR3), and cathepsin G (Cat G). To investigate the possibility of alternative processing of TNFα and/or IL-1β by neutrophil-derived proteinases, immunoreactive TNFα and IL-1β release from lipopolysaccharide-stimulated THP-1 cells was measured in the presence of activated human neutrophils. Under these conditions, TNFα and IL-1β release was augmented 2- to 5-fold. In the presence of a specific inhibitor of NE and PR3, enhanced release of both cytokines was largely abolished; however, in the presence of a NE and Cat G selective inhibitor, secretory leucocyte proteinase inhibitor, reduction of the enhanced release was minimal. This finding suggested that the augmented release was attributable to PR3 but not NE nor Cat G. Use of purified enzymes confirmed this conclusion. These results indicate that there may be alternative pathways for the production of these two proinflammatory cytokines, particularly in the context of local inflammatory processes.
Keywords: neutrophil elastase, cytokine processing, proteinase inhibitors
Tumor necrosis factor α (TNFα) and IL-1β are two of the more important proinflammatory cytokines that have been shown to play major roles in the pathogenesis of both systemic diseases, such as sepsis (1–4), and diseases with significant, localized tissue destruction, such as rheumatoid arthritis (5). Although there is considerable overlap in the biology of these two cytokines, the mechanisms of their release, regulation, and activation are significantly different. The precursor of TNFα is synthesized and initially expressed as a 26-kDa transmembrane protein. Thereafter it is cleaved from the membrane by a metalloproteinase disintegrin, TNFα converting enzyme (TACE) (6, 7).
In contrast, although IL-1β is synthesized as an immature precursor (pro-IL-1β), this form lacks biological activity (8, 9). Conversion to the 17-kDa active form depends on cleavage by a membrane-associated cysteine proteinase, caspase-1, or IL-1β converting enzyme (ICE), which also may be important for transport of the mature protein across the cytoplasmic membrane (10–12). However, the release of pro-IL-1β from activated monocytes also has been described (13, 14).
Although inhibitors of TACE and ICE ameliorate inflammatory responses to systemically administered stimuli, many inflammatory responses are not systemic in nature but are instead highly localized and involve intimate interactions between many cell types, both resident at the inflamed site and contained in the infiltrate. It therefore seems inevitable that cytokines found in these microenvironments will be exposed to various proteinases released by the inflammatory cells. Keratinocytes have been shown to produce, but not process, pro-IL-1β (15), further suggesting an obligatory requirement for extracellular processing in some inflammatory situations. Furthermore, it has been reported that ICE-deficient mice still can generate mature IL-1β in response to a local inflammatory stimulus (16), indicating that alternative processing of this cytokine can occur in vivo and that the release and/or activation of cytokines in local sites may have considerably different mechanisms from those encountered systemically. The enzymes likely to play a major role in these processes are those contained within the azurophilic granules of neutrophils, namely neutrophil elastase (NE), cathepsin G (Cat G), and proteinase 3 (PR3) (reviewed in ref. 17).
Both TNFα and IL-1β are sensitive to processing by proteases other than their specific converting enzymes. The neutrophil-derived proteinases, NE and Cat G, as well as members of the matrix metalloproteinase family, granzyme A and human mast cell chymase, have been shown to cleave recombinant pro-IL-1β in vitro (9, 18–21). Serine protease inhibitors were found to inhibit secretion of TNFα but had no effect on levels of TNFα mRNA (22). Moreover, an alternative processing mechanism for the release of membrane-bound TNFα has been described by using an in vitro translated precursor of TNFα. Cleavage was attributed to PR3 based on the sensitivity of this activity to various serine proteinase inhibitors (23). These studies support the concept that alternative pathways for the processing of TNFα and IL-1β might be manifested in certain inflammatory conditions.
We were interested in whether neutrophil-derived proteinases, and in particular PR3, could play amplifying roles in the generation of bioactive cytokines and whether these activities could be detected in a cellular system that might mimic conditions within an inflammatory site. PR3 originally was described as a neutral serine proteinase distinct from NE and Cat G found in the azurophilic granules of human neutrophils (17). It was further characterized as being capable of degrading elastin in vitro and of inducing emphysema in hamsters (24, 25). Despite its characterization as an elastase-like enzyme, however, the physiological role of PR3 has remained speculative.
The ability of activated human neutrophils to augment the release of TNFα from lipopolysaccharide (LPS)-activated THP-1, a human monocytic cell line, initially was investigated. Under these conditions, cytokine release took place in the presence of maximally inhibitory concentrations of a TACE-specific inhibitor, and therefore, we assessed whether release was abolished in the presence of an NE/PR3-specific inhibitor. It became apparent, however, during the course of these studies that release IL-1β was similarly augmented under these same conditions. We therefore used purified enzymes in place of neutrophils to elucidate the underlying mechanism(s) involved in the enhanced release of these cytokines. The implications of these results in defining a potential new role for PR3 in the generation of inflammatory responses are presented.
MATERIALS AND METHODS
THP-1 Cell Line.
The human promonocytic leukemia cell line, THP-1 (26), was obtained from the American Type Culture Collection and was cultured in RPMI medium 1640 (BioWhittaker) supplemented with 10% FCS (Irvine Scientific), 5 × 10−5 M 2-mercaptoethanol, and 2 mM l-glutamine in an atmosphere of 5% CO2 at 37°C.
Enzymes and Inhibitors.
Purified human PR3 and Cat G were obtained from Athens Research & Technology (Athens, GA). NE was obtained from Elastin Products (Owensville, MI). Recombinant human secretory leukocyte proteinase inhibitor (SLPI) was purchased from R & D Systems. Acetyl-Tyr-Val-Ala-Asp-aldehyde (YVAD-CHO) was purchased from Bachem. GI-129471/GW9471 (GI-1) and CE-2072 were synthesized at Cortech, Denver (see Table 1). Inhibitors were stored as 10 mM solutions in DMSO at −20°C. For use in cellular assays, inhibitors were freshly diluted at the time of use in RPMI or assay medium (see below). Dose–response curves were generated for GI-1 and YVAD-CHO with LPS-stimulated THP-1 cells to assess maximally inhibitory concentrations for use in the assays (data not shown). Unless otherwise indicated, all inhibitors were used at a final concentration of 10 μM.
Table 1.
Specificities of inhibitors used in the current study
| Inhibitor | Target enzyme | Ki, nM | Ref(s). |
|---|---|---|---|
| CE-2072 | NE | 0.025 | This paper |
| PR3 | 5.0 | This paper | |
| Cat G | >50,000 | This paper | |
| SLPI | NE | 0.1 | 31 |
| PR3 | ≫2,500 | This paper | |
| No inhibition | 32, 33 | ||
| Cat G | 3.5 | 31 | |
| GI-1 | TACE | 5 to 13 | 6 |
| YVAD-CHO | ICE | 0.76 | 10 |
Measurement of Enzyme and Inhibitory Activity.
PR3. The assays were performed as described (25, 27). The conditions for the reaction were 5 nM PR3 and 0.625 mM substrate (Boc-Ala-O-nitrophenyl; Bachem). Residual activity of the enzyme was measured after preincubation with different concentrations of inhibitors for 10–30 min at 25°C in 0.1 M Hepes buffer, containing 0.1 M NaCl, 10 mM CaCl2, 0.005% Triton X-100, and 5% DMSO, pH 7.5. The formation of the p-nitrophenol product was monitored at 410 nm in a diode array spectrophotometer. The data were processed by using a software program (enzfit, Elsevier-Biosoft, Cambridge, U.K.) to generate IC50 values. IC50 approximated to Ki under the experimental conditions.
NE.
Initial conditions of the reaction were 0.05 M sodium phosphate (pH 7.5), 0.1 M NaCl, 0.005% Triton X-100, 5% DMSO, and 5 nM NE. The reaction was begun with the simultaneous addition of CE-2072 and 50 mM N-methoxysuccinyl-K(2-picolinoyl)-A-P-V-p-nitroanilide (NA) substrate (Bachem). The appearance of p-NA product was monitored over 15–30 min at 410 nm in a diode array spectrophotometer. Steady-state velocities were measured and processed by using enzfit to generate IC50 values. From these data, the Ki values were calculated by using the equation: Ki = IC50/[1+([S]/Km)]. In the case of SLPI, the inhibitor (0.25 to 2.5 M) and NE were preincubated for 10 min, and the residual enzyme activity was measured by using 0.5 mM N-methoxysuccinyl-A-A-P-V-p-NA (Sigma).
Cat G.
Enzyme was diluted in 0.1 M Hepes, 0.1 M NaCl, 10 mM CaCl2, and 0.005% Triton X-100, pH 7.5. The general protocol for NE was followed. Reaction was commenced by addition of enzyme to the cuvette already containing buffer, inhibitor, and 0.5 mM N-succinyl-A-A-P-F-p-NA (Sigma).
Stimulation of THP-1 Cells in the Presence of PR3 and NE.
THP-1 cells were harvested by centrifugation and washed once in RPMI. The cells were resuspended in RPMI and added to wells at a final concentration of 2 × 106 cells/ml. Cells were preincubated for 15 min at 37°C with inhibitors and enzymes before activation. Purified NE and PR3 were diluted in Hanks’ balanced salt solution and added to wells at a final concentration of 10 μg/ml. LPS (serotype O127.88; Sigma) was preincubated in FCS at 1 mg/ml for 30 min at 37°C then serially diluted in RPMI and added to wells at a final concentration of 1 μg/ml. The final concentration of serum in the wells was 0.1%. Cells were incubated with LPS for 4 h at 37°C in 5% CO2 after which time the plates were centrifuged at 500 × g for 5 min and the supernatants were aspirated and stored at −70°C. THP-1 cell viability at the end of these experiments was verified by trypan blue exclusion.
Coincubation of THP-1 and Purified Human Neutrophils.
THP-1 cells were harvested by centrifugation, washed once in assay medium (RPMI lacking phenol red, plus 5% dextrose), and added to wells at a final concentration of 106 cell/ml in assay medium. Neutrophils were isolated from healthy, nonsmoking volunteers. Blood was collected into EDTA vacutainers, and separation of neutrophils was carried out within 1 h of collection. Blood was layered onto separation medium (one-step Polymorph, Accurate Chemicals) in sterile polystyrene tubes, which then were centrifuged for 30 min at 500 × g at room temperature. The neutrophil layer was aspirated and diluted by addition of 1 vol of 0.45% sterile saline to restore osmolarity. The cells then were washed twice in assay medium. This procedure routinely yielded a preparation of greater than 95% neutrophils as assessed by light microscopy. Cells were resuspended in assay medium and were used at final concentration of 4 × 106 cells/ml. THP-1, neutrophils, or the combination of both were preincubated in wells for 15 min at 37°C with inhibitors before activation. LPS was preincubated in alpha-1 proteinase inhibitor-deficient serum (obtained through the Rocky Mountain Alpha-1 Anti-trypsin Registry) at 1 mg/ml for 30 min at 37°C, then diluted in RPMI and added to wells at a final concentration of 1 μg/ml. FNLP (N-formyl-Nle-Leu-Phe; Sigma) was diluted from a stock concentration of 10 mM in DMSO to a final concentration in the wells of 10 μM. All wells of the assay were adjusted to contain the same final concentrations of DMSO (typically 0.1–0.2%). Cultures were set up in duplicate and were incubated with LPS/FNLP for 4 h at 37°C in 5% CO2 after which time the plates were centrifuged at 500 × g for 5 min and the supernatants were aspirated and stored at −70°C. To confirm activation and degranulation of neutrophils, aliquots of culture supernatants were removed after 2 h of culture and assayed for NE activity by using the substrate N-methoxysuccinyl-K(2-picolinoyl)-A-P-V-p-NA.
Measurement of Cytokine Levels.
TNFα and IL-1β levels in cell culture supernatants were measured by ELISA (PerSeptive Diagnostics, Cambridge, MA for TNFα; R & D Systems for IL-1β). Data are expressed as the mean cytokine level ± SD by using values from duplicate tissue culture wells with each of these wells run in duplicate in ELISA (n = 4).
RESULTS
Augmentation of TNFα by Coincubation of Stimulated THP-1 Monocytes and Purified Human Neutrophils.
To investigate the levels of TNFα that could be induced from coincubation of major cell types infiltrating inflammatory sites, we chose to look at the effect of mixing monocytic THP-1 cells, which release high levels of TNFα and IL-1β after stimulation with LPS (28, 29), with isolated human neutrophils. THP-1 was used as a source of homogeneous monocytic cells in preference to monocytes purified from human blood to reduce the variability in cytokine production from individual donors. Cells were mixed at 4:1 neutrophil/monocyte ratio in an attempt to approximate conditions in an acute inflammatory site.
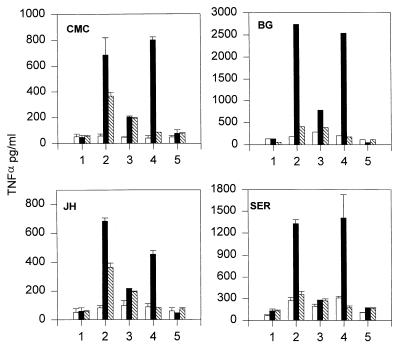
When cells were stimulated with LPS and FNLP, increased release of TNFα was observed when neutrophils and THP-1 were coincubated compared with either cell population stimulated alone (Fig. 1, position 2). Of particular note, however, was the augmentation of TNFα levels observed when the two stimulated cell populations were incubated together in the presence of GI-1, the TACE-specific inhibitor (Fig. 1, position 4). Under these circumstances, the levels were increased approximately 5-fold over the sum of the two cell populations stimulated separately and 10-fold over the level of TNFα produced from stimulated THP-1 alone in the presence of GI-1. When CE-2072, a specific serine elastase inhibitor (see Table 1), was included with the mixed cells, the enhancement was greatly reduced or abolished entirely (Fig. 1, position 3) and when both CE-2072 and GI-1 were used in combination, levels of TNFα were reduced to background (Fig. 1, position 5). These data indicate that a mechanism distinct from that attributable to TACE was involved in the release of TNFα.
Figure 1.
Enhancement of TNFα release from coincubation of stimulated THP-1 cells and human neutrophils. Representative data are shown from two separate experiments by using two neutrophil donors per day. Initials in top left hand corner of graphs refer to individual donors. One × 106 THP-1 cells (hatched bars) and 4 × 106 neutrophils isolated from individual donors were added to tissue culture wells either separately (open bars) or as a mixture of both populations (closed bars). Numbers along the horizontal axes indicate different conditions. Cells were incubated for 4 h at 37°C either with no further additives (position 1), with 1 μg/ml LPS plus 10-5 M FNLP (position 2), 10 μM CE-2072 (position 3), 10 μM GI-1 (position 4), or 10 μM CE-2072 plus 10 μM GI-1 (position 5). Supernatants were recovered and assayed for TNFα by ELISA. For each donor data represent mean TNFα levels ± SD from two tissue culture wells with each well run in duplicate in ELISA (n = four observations per donor).
Augmentation of IL-1β Release by Coincubation of Stimulated THP-1 Monocytes and Human Neutrophils.
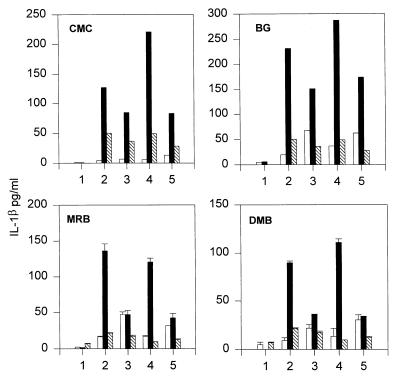
When levels of IL-1β were measured in similar experiments to those illustrated in Fig. 1, it was apparent that these also were increased in stimulated cocultures compared with either cell population activated in isolation (Fig. 2, position 2) and that CE-2072 could reduce the levels in cocultures to various degrees (Fig. 2, position 3). Levels of IL-1β in the cocultures also were sometimes increased in the presence of GI-1 (Fig. 2, position 4), which could have been the result of the increased levels of TNFα in these cultures (see Fig. 1). Alternatively, because IL-1β has been reported to be readily degraded by matrix metalloproteinases (30), GI-1 might have prevented this degradation. Nevertheless, the fact that the increase in IL-1β levels appeared to mirror those of the TNFα levels suggested that both TNFα and IL-1β release were augmented by a neutrophil-dependent mechanism.
Figure 2.
Enhancement of IL-1β release from coincubation of stimulated THP-1 cells and human neutrophils. Data shown are from two separate experiments by using neutrophils isolated from four different donors. Experiments were set up as described for Fig. 1, and supernatants were assayed for the presence of IL-1β by ELISA.
Inhibition Profile of CE-2072.
In the above experiments, neutrophil activation and degranulation were confirmed by measuring the presence of NE activity in the culture supernatants (data not shown). Therefore, it seemed likely that augmentation of levels of both TNFα and IL-1β seen in these cocultures could be attributed to the action of an enzyme released by activated neutrophils. Because a role for PR3 in the cleavage of TNFα has been reported (23), this enzyme could be responsible for the release of this cytokine, if not IL-1β as well, in the cocultures. Examination of the inhibition profile of CE-2072 (Table 1) indicated that it could inhibit purified NE and PR3 completely but had no activity against Cat G (Ki > 50 μM).
Augmentation of Cytokine Release in Cocultures of Activated THP-1 and PMN Is Not Inhibited By SLPI.
Because Cat G was ruled out as a mechanism of TNFα and IL-1β augmentation in the above experiments, we next wanted to distinguish between the activities of NE and PR3. Activated THP-1 and neutrophils were cocultured in the presence of SLPI, which inhibits NE and Cat G (31), but not PR3 (32, 33) (see Table 1).
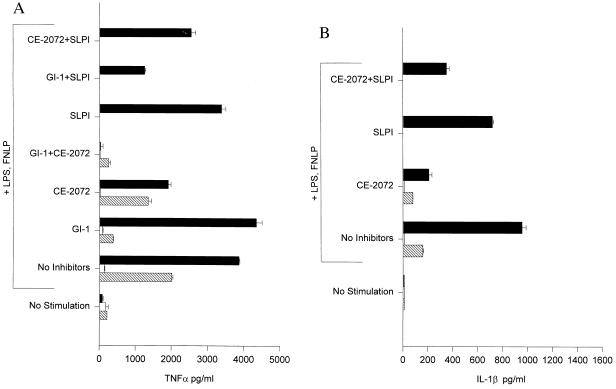
Fig. 3A shows a representative experiment in which a modest increase (2-fold) in TNFα levels was observed upon coculture of stimulated THP-1 with neutrophils compared with THP-1 stimulated alone. Inclusion of CE-2072 reduced TNFα levels in the cocultures essentially to those of THP-1 alone (a 52% reduction in levels) whereas in the presence of 2.5 μM SLPI, only a 12% reduction in levels occurred. Furthermore, whereas addition of both GI-1 and CE-2072 abolished TNFα levels to background, the combination of GI-1 and SLPI still allowed a greater than 100-fold increase over background levels, indicating that a substantial proportion of TNFα release from THP-1 in the presence of activated neutrophils was TACE, NE, and Cat G independent.
Figure 3.
SLPI does not inhibit augmented release of TNFα (A) nor IL-1β (B) from coincubation of stimulated THP-1 cells and human neutrophils. Representative data are shown by using neutrophils isolated from a single donor (BG). The experimental design was as described in Figs. 1 and 2 except that inhibitors were preincubated with cells for 15 min at 37°C before the addition of LPS and FNLP. SLPI was added at a final concentration of 2.5 μM. Levels of TNFα and IL-1β in the presence of SLPI alone and of SLPI plus CE-2072 were not measured for neutrophils nor for THP-1cells cultured separately.
In the same experiment, IL-1β production in the stimulated cocultures was increased 6-fold over stimulated THP-1 alone (Fig. 3B). CE-2072 greatly reduced this enhancement in the cocultures as did the combination of both CE-2072 and SLPI, whereas SLPI alone caused only a modest decrease in levels (25% reduction). Augmented IL-1β release in the cocultures therefore also substantially depended on a SLPI-insensitive but CE-2072-sensitive enzyme.
Augmentation of TNFα and IL-1β Release by Purified PR3 But Not NE.
The preceding data implicated PR3 in the augmentation of both TNFα and IL-1β production in the monocyte-neutrophil-stimulated cocultures. We therefore turned to the use of the purified enzyme to establish whether the effects of the activated neutrophils on LPS-stimulated THP-1 could be reproduced by PR3.
It was found (Fig. 4) that PR3 increased TNFα levels produced by LPS-stimulated THP-1 cells approximately 6-fold whereas NE caused only a modest increase. In the additional presence of GI-1, however, neither of these levels was substantially reduced. Thus although NE could not produce exaggerated levels of TNFα, it could reverse the effect of GI-1. This finding is in agreement with the data shown in Fig. 3A in which the combination of SLPI and GI-1 caused a substantial decrease in TNFα production. In contrast, PR3 compensated for the inhibition of TACE by GI-1 by causing greatly increased TNFα production.
Figure 4.
Augmentation of TNFα release from LPS-stimulated THP-1 cells by PR3 but not NE. Two × 106 THP-1 cells were added to tissue culture wells and preincubated with enzymes and inhibitors for 15 min at 37°C before addition of LPS.
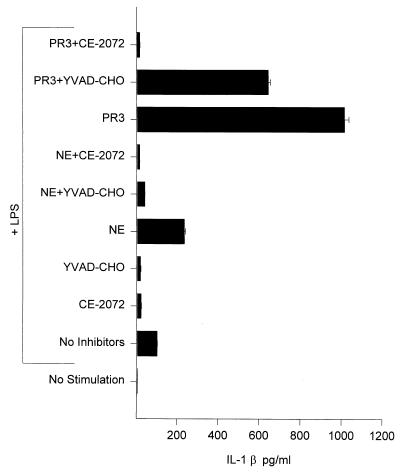
When the effects of PR3 and NE on IL-1β production were examined, it was found (Fig. 5) that PR3 caused a 10-fold increase in IL-1β production from LPS-stimulated THP-1 cells compared with levels produced by stimulated cells alone, whereas NE only caused a modest (2-fold) increase in IL-1β levels. The inclusion of the ICE-specific inhibitor, YVAD-CHO (see Table 1), revealed an even more dramatic difference in the ability of these two enzymes to augment IL-1β production. In the presence of this inhibitor, PR3 enhanced levels of IL-1β 30-fold over those of stimulated THP-1 alone. In contrast, NE caused no increase in IL-1β levels produced by stimulated THP-1 in the presence of YVAD-CHO.
Figure 5.
Augmentation of IL-1β release from stimulated THP-1 cells by PR3 but not NE. Experimental design was as described for Fig. 4. YVAD-CHO was added at a final concentration of 5 μM.
Selected supernatants from the above experiment were tested for the presence of biologically active IL-1β by using the IL-1β-sensitive cell line, HaCaT, and were found to be capable of inducing IL-8 production with levels corresponding to those of IL-1β measured by ELISA (data not shown).
DISCUSSION
The proinflammatory cytokines, TNFα and IL-1β, are released from cells by the action of specific proteinases, TACE and ICE, respectively, which convert the precursors of these molecules into soluble mature forms with biological activity (6, 10). In this report we show that the neutrophil-derived proteinase, PR3, appears to provide an alternative mechanism for the cleavage and release of biologically active forms of both of these cytokines. Specifically we show that purified PR3 is capable of augmenting the release of TNFα and IL-1β from LPS-stimulated THP-1, a human monocytic cell line, in the presence of maximally inhibitory concentrations of inhibitors (typically causing greater than 80% inhibition) specific for the enzymes TACE (GI-1) and ICE (YVAD-CHO). The augmentation of IL-1β by PR3 in the presence of YVAD-CHO was particularly dramatic, with levels increased 30-fold over those generated by stimulated THP-1 alone in the presence of this inhibitor. Although ICE is also involved in the secretion of mature IL-1β (4, 12), Thornberry and coworkers (10) have reported that pro-IL-1β accumulates outside the cell in the presence of YVAD-CHO, thus making it available to the action of extracellular PR3. That augmentation of TNFα and IL-1β by the purified enzyme is abolished in the presence of a strong inhibitor of PR3 (and NE), CE-2072, indicates that PR3 augments production of mature forms of these cytokines via its proteolytic activity. It was also apparent that CE-2072 caused a reduction in levels of TNFα released from LPS-stimulated THP-1 alone on some occasions. Whether this reflects the involvement of a serine elastase-like proteinase in the release of TNFα when TACE activity is not limited or the action of such a proteinase at another point in the LPS stimulation pathway upstream to TNFα release is currently unknown.
Augmentation of TNFα and IL-1β release first was observed in cocultures of LPS-activated monocytic THP-1 cells and activated neutrophils, suggesting that a neutrophil-derived mechanism was responsible for this enhancement as well as suggesting that this alternative mechanism might be operative at an inflammatory site. Because the presence of SLPI did not affect the levels of cytokines measured in these cocultures, this finding strongly suggested that PR3, but not NE nor Cat G, caused this enhanced release because only PR3 is insensitive to inhibition by SLPI (31–33). The involvement of Cat G was further ruled out because it is not inhibited by CE-2072, yet enhancement of both TNFα and IL-1β levels was largely abolished in the presence of this inhibitor. We therefore turned to use of purified enzymes to establish the mechanism of enhanced cytokine production.
Enhancement of both TNFα and IL-1β levels by PR3 could have been caused by a generalized physiological effect of the enzyme on the LPS stimulation pathway in THP-1 cells. However, we consider this unlikely because enhancement could not be abolished by the specific inhibitors of TACE and ICE. This observation indicates that PR3 acts independently of the action of these converting enzymes and most likely at the most distal points of these pathways. Furthermore PR3 previously has been shown to process an in vitro-translated precursor of TNFα to a biologically active form (23), and serine protease inhibitors have been found to inhibit secretion of TNFα with no effect on TNFα mRNA (22). We also have observed that PR3 can process recombinant pro-IL-1β to a biologically active form (unpublished observations). For these reasons we believe that PR3 augments release of these cytokines by alternative processing of their immature forms, although we have not formally eliminated the possibility that this enzyme could act at the level of gene regulation and expression or at other points in these pathways.
PR3 is colocalized with NE and Cat G in neutrophil granules and is coreleased by degranulation upon activation of these cells (17). NE and PR3 have been shown to have approximately the same substrate specificities with respect to small molecular weight peptide substrates (25, 34–36), and both enzymes also degrade elastin (24, 25). In addition, both enzymes share significant structural and sequence homology and, as a result, have been classified as serine elastases. Furthermore, with the exception of SLPI, both enzymes appear to be subject to the same antiproteinase regulatory mechanisms (32, 37, 38). NE, however, plays a well-documented role in degradation of matrix proteins to enable cell migration (39). In contrast, although PR3 has been shown to degrade a variety of extracellular matrix proteins (24, 25), preliminary data indicate that degradation of extracellular matrix proteins in vitro by activated human neutrophils is not attributable to PR3 (S. R. Simon, personal communication). In the present study, although NE could not produce enhanced levels of TNFα, it could reverse the effect of GI-1 but had no effect on IL-1β levels in the present of YVAD-CHO. Thus, although PR3 and NE share many characteristics, the apparent differing abilities of these proteinases to degrade tissue, as well as the data contained in this report, suggest that the physiological roles of these two enzymes may be quite different.
The physiological relevance of the ability of PR3 to process TNFα and IL-1β in vitro remains to be elucidated but is strengthened by the observations that SLPI-insensitive, but CE-2072-sensitive, enhancement of cytokine levels occurred in cocultures of stimulated monocytic cells and neutrophils under conditions that approximate a localized inflammatory focus. In addition to reports of the cleavage of biologically active TNFα in vitro by PR3 (this paper and ref. 23), serine proteinase inhibitors have been found to inhibit TNFα release in vivo (40). However, the importance of TACE as a pivotal enzyme in the development of inflammatory responses is indisputable given the ability of TACE inhibitors to dramatically reduce circulating TNFα levels in endotoxin-treated rats and mice (1–3).
The importance of ICE in inflammation is also well documented in studies reporting successful amelioration of inflammation in various animal models by inactivation of this enzyme, e.g., ICE inhibitors reduced systemic inflammation induced by endotoxin in mice (41) and ICE-deficient mice were resistant to endotoxic shock (42). In contrast, it was found that localized inflammation induced by turpentine application to the skin induced levels of the IL-1β-dependent cytokine, IL-6, in ICE-deficient mice that were comparable to those of wild-type mice, whereas no IL-6 was produced in IL-1β-deficient mice (16). Thus an ICE-independent mechanism could compensate for loss of ICE activity in this model of inflammation by providing an alternative processing pathway for IL-1β. Other enzymes have been reported to cleave pro-IL-1β in vitro to its active form (9, 18–21) but they have not been shown to have activity in a cellular environment.
In addition, other cytokines and regulatory molecules, such as IL-8, transforming growth factor (TGF) type α, and TGFβ, also can be processed extracellularly to mature or more active forms (43–49). These reports are manifestations of the same concept illustrated in the current study, namely that these molecules may depend on the extracellular environment for release or amplification of their activity. For some of these this processing event may be an obligatory step as in the case of TGFβ (46–48). For others, such as IL-1β and TNFα, as described in this report, extracellular activation may represent an alternative mechanism.
In conclusion, we suggest that the major role for PR3 in localized inflammatory sites may be to amplify the response via its action on cytokines secreted into the microenvironment and that its role in degradation of tissue may be secondary to this activity. Given the ability of PR3 to augment the production of TNFα and IL-1β in vitro, it seems imperative to approach the use of specific inhibitors in various inflammatory disorders with caution. Although application of ICE and TACE inhibitors to treat systemic inflammation may well be beneficial, their use for diseases with largely localized inflammation may be less effective and, given the data in this report, may even be detrimental.
Acknowledgments
We thank Dr. R. Sandhaus and Ms. D. Craft for alpha-1 proteinase inhibitor-deficient serum donation, and Drs. A. Gyorkos, L. Spruce, and the Cortech NE Project Team for synthesis of CE-2072.
ABBREVIATIONS
- Cat G
cathepsin G
- GI-1
GI-129471/GW9471
- NE
neutrophil elastase
- ICE
IL-1β converting enzyme
- NA
nitroanilide
- PR3
proteinase 3
- SLPI
secretory leucocyte proteinase inhibitor
- YVAD-CHO
acetyl-Tyr-Val-Ala-Asp-aldehyde
- TNFα
tumor necrosis factor α
- TACE
TNFα converting enzyme
- LPS
lipopolysaccharide
- FNLP
N-formyl-Nle-Leu-Phe
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.McGeehan G M, Becherer J D, Bast R C, Jr, Boyer C M, Champion B, Connolly K M, Conway J G, Furdon P, Karp S, Kidao S, et al. Nature (London) 1994;370:558–561. doi: 10.1038/370558a0. [DOI] [PubMed] [Google Scholar]
- 2.Mohler K M, Sleath P R, Fitzner J N, Cerretti D P, Alderson M, Kerwar S S, Torrance D S, Otten-Evans C, Greenstreet T, Weerawarna K, et al. Nature (London) 1994;370:218–220. doi: 10.1038/370218a0. [DOI] [PubMed] [Google Scholar]
- 3.Gearing A J H, Beckett P, Christodoulou M, Churchill M, Clements J, Davidson A H, Drummond A H, Galloway W A, Gilbert R, Gordon J L, et al. Nature (London) 1994;370:555–557. doi: 10.1038/370555a0. [DOI] [PubMed] [Google Scholar]
- 4.Dinarello C A. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 5.Feldmann M, Brenna F M, Maini R N. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 6.Moss M L, Jin S L, Milla M E, Bickett D M, Burkhart W, Carter H L, Chen W J, Clay W C, Didsbury J R, Hassler D, et al. Nature (London) 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 7.Black R A, Rauch C T, Kozlosky C J, Peschon J J, Slack J L, Wolfson M F, Castner B J, Stocking K L, Reddy P, Srinivasan S, et al. Nature (London) 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 8.Hazuda D, Webb R L, Simon P, Young P. J Biol Chem. 1989;264:1689–1693. [PubMed] [Google Scholar]
- 9.Black R A, Kronheim S R, Cantrell M, Deeley M C, March C J, Prickett K S, Wignall J, Conlon P J, Cosman D, Hopp T P, et al. J Biol Chem. 1988;263:9437–9442. [PubMed] [Google Scholar]
- 10.Thornberry N A, Bull H G, Calaycay J R, Chapman K T, Howard A D, Kostura M J, Miller D K, Molineaux S M, Weidner J R, Aunins J, et al. Nature (London) 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 11.Alnemri E S, Livingston D J, Nicholson D W, Salvesen G, Thornberry N A, Wong W E, Yuan J. Cell. 1996;87:171. doi: 10.1016/s0092-8674(00)81334-3. [DOI] [PubMed] [Google Scholar]
- 12.Singer I I, Scott S, Chin J, Bayne E K, Limjuco G, Weidner J, Miller D K, Chapman K, Kostura M J. J Exp Med. 1995;182:1447–1459. doi: 10.1084/jem.182.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazuda D J, Lee J C, Young P R. J Biol Chem. 1988;263:8473–8479. [PubMed] [Google Scholar]
- 14.Bomford R, Abdulla E, Hughes-Jenkins C, Simpkin D, Schmidt J. Immunology. 1987;62:543–549. [PMC free article] [PubMed] [Google Scholar]
- 15.Mizutani H, Black R, Kupper T S. J Clin Invest. 1991;87:1066–1071. doi: 10.1172/JCI115067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fantuzzi G, Ku G, Harding M W, Livingston D J, Sipe J D, Kuida K, Flavell R A, Dinarello C A. J Immunol. 1997;158:1818–1824. [PubMed] [Google Scholar]
- 17.Baggiolini M, Bretz U, Dewald B, Feigenson M E. Agents Actions. 1978;8:3–10. doi: 10.1007/BF01972395. [DOI] [PubMed] [Google Scholar]
- 18.Hazuda D J, Strickler J, Kueppers F, Simon P L, Young P R. J Biol Chem. 1990;265:6318–6322. [PubMed] [Google Scholar]
- 19.Mizutani H, Schechter N, Lazarus G, Black R A, Kupper T S. J Exp Med. 1991;174:821–825. doi: 10.1084/jem.174.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irmler M, Hertig S, MacDonald H R, Sadoul R, Becherer J D, Proudfoot A, Solari R, Tschopp J. J Exp Med. 1995;181:1917–1922. doi: 10.1084/jem.181.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schonbeck U, Mach F, Libby P. J Immunol. 1998;161:3340–3346. [PubMed] [Google Scholar]
- 22.Scuderi P, Dorr R T, Liddil J D, Finley P R, Meltzer P, Raitano A B, Rybski J. Eur J Immunol. 1989;19:939–942. doi: 10.1002/eji.1830190523. [DOI] [PubMed] [Google Scholar]
- 23.Robache-Gallea S, Morand V, Bruneau J M, Schoot B, Tagat E, Realo E, Chouaib S, Roman-Roman S. J Biol Chem. 1995;270:23688–23692. doi: 10.1074/jbc.270.40.23688. [DOI] [PubMed] [Google Scholar]
- 24.Kao R C, Wener N G, Skubitz K M, Gray B H, Hoidal J R. J Clin Invest. 1988;82:1963–1973. doi: 10.1172/JCI113816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fruh H, Kostoulas G, Michel B A, Baici A. Biol Chem. 1996;377:579–586. doi: 10.1515/bchm3.1996.377.9.579. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Int J Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 27.Hoidal J R, Rao N V, Gray B. Methods Enzymol. 1994;244:61–67. doi: 10.1016/0076-6879(94)44005-0. [DOI] [PubMed] [Google Scholar]
- 28.Molina J-M, Scadden D T, Byrn R, Dinarello C A, Groopman J E. J Clin Invest. 1989;84:733–737. doi: 10.1172/JCI114230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krakauer T, Oppenheim J J. Cell Immunol. 1983;80:223–229. doi: 10.1016/0008-8749(83)90111-9. [DOI] [PubMed] [Google Scholar]
- 30.Ito A, Mukaiyama A, Itoh Y, Nagase H, Thogersen I B, Enghild J J, Sasaguri Y, Mori Y. J Biol Chem. 1996;271:14657–14660. doi: 10.1074/jbc.271.25.14657. [DOI] [PubMed] [Google Scholar]
- 31.Steffens G J, Heinzel-Wieland R, Saunders D, Wolf B, Rudolphus A, Stolk J, Krarnps J A, Dijkman J A. In: Agents and Actions Supplements. Cheronis J C, Repine J E, editors. Vol. 42. Basel: Birkhauser; 1993. pp. 111–121. [DOI] [PubMed] [Google Scholar]
- 32.Rao N V, Wener N G, Marshall B C, Gray W R, Gray B H, Hoidal J R. J Biol Chem. 1991;266:9450–9548. [PubMed] [Google Scholar]
- 33.Rao N V, Marshall B C, Gray B H, Hoidal J R. Am J Resp Cell Mol Biol. 1993;8:612–616. doi: 10.1165/ajrcmb/8.6.612. [DOI] [PubMed] [Google Scholar]
- 34.Kam C M, Kerrigan J E, Dolman K M, Goldschmeding R, Von dem Borne A E G K, Powers J C. FEBS Lett. 1992;297:119–123. doi: 10.1016/0014-5793(92)80340-m. [DOI] [PubMed] [Google Scholar]
- 35.Brubaker M J, Groutas W C, Hoidal J R, Rao N V. Biochem Biophys Res Commun. 1992;188:1318–1324. doi: 10.1016/0006-291x(92)91375-z. [DOI] [PubMed] [Google Scholar]
- 36.Nakajima K, Powers J C. J Biol Chem. 1979;254:4027–4032. [PubMed] [Google Scholar]
- 37.Wiedow O, Schroder J-M, Gregory H, Young J A, Christopher E. J Biol Chem. 1990;265:14791–14795. [PubMed] [Google Scholar]
- 38.Wiedow O, Ludemann J, Utecht B. Biochem Biophys Res Commun. 1991;174:6–10. doi: 10.1016/0006-291x(91)90476-n. [DOI] [PubMed] [Google Scholar]
- 39.Owen C A, Campbell E J. Semin Cell Biol. 1995;6:367–376. doi: 10.1016/s1043-4682(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 40.Niehorster M, Tiegs G, Schade U F, Wendel A. Biochem Pharmacol. 1990;40:1601–1603. doi: 10.1016/0006-2952(90)90461-s. [DOI] [PubMed] [Google Scholar]
- 41.Fletcher D S, Agarwal L, Chapman K T, Chin J, Egger L A, Limjuco G, Luell S, MacIntyre D E, Peterson E P, Thornberry N A, et al. J Interferon Cytokine Res. 1995;15:243–248. doi: 10.1089/jir.1995.15.243. [DOI] [PubMed] [Google Scholar]
- 42.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J, et al. Cell. 1995;80:401–411. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 43.Padrines M, Wolf M, Walz A, Baggiolini M. FEBS Lett. 1994;352:231–235. doi: 10.1016/0014-5793(94)00952-x. [DOI] [PubMed] [Google Scholar]
- 44.Cappelluti E, Harris R B. Perspect Drug Discovery Res. 1994;2:353–361. [Google Scholar]
- 45.Mueller S G, Paterson A J, Kudlowm J E. Mol Cell Biol. 1990;10:4596–4602. doi: 10.1128/mcb.10.9.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonelli-Orlidge A, Saunders K, Smith S R, D’amore P A. Proc Natl Acad Sci USA. 1989;86:4544–4548. doi: 10.1073/pnas.86.12.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawrence D A, Pircher R, Jullien P. Biochem Biophys Res Commun. 1985;133:1026–1034. doi: 10.1016/0006-291x(85)91239-2. [DOI] [PubMed] [Google Scholar]
- 48.Csernok E, Szymkowiak C H, Mistry N, Daha M R, Gross W L, Kekow J. Clin Exp Immunol. 1996;105:104–111. doi: 10.1046/j.1365-2249.1996.d01-715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taipale J, Lohi J, Saarinen J, Kovanen P T, Keski-Oja J. J Biol Chem. 1995;270:4689–4696. doi: 10.1074/jbc.270.9.4689. [DOI] [PubMed] [Google Scholar]