Abstract
The development of the pancreas depends on epithelial-mesenchymal interactions. Fibroblast growth factors (FGFs) and their receptors (FGFRs 1–4) have been identified as mediators of epithelial-mesenchymal interactions in different organs. We show here that FGFR-2 IIIb and its ligands FGF-1, FGF-7, and FGF-10 are expressed throughout pancreatic development. We also show that in mesenchyme-free cultures of embryonic pancreatic epithelium FGF-1, FGF-7, and FGF-10 stimulate the growth, morphogenesis, and cytodifferentiation of the exocrine cells of the pancreas. The role of FGFs signaling through FGFR-2 IIIb was further investigated by inhibiting FGFR-2 IIIb signaling in organocultures of pancreatic explants (epithelium + mesenchyme) by using either antisense FGFR-2 IIIb oligonucleotides or a soluble recombinant FGFR-2 IIIb protein. Abrogation of FGFR-2 IIIb signaling resulted in a considerable reduction in the size of the explants and in a 2-fold reduction of the development of the exocrine cells. These results demonstrate that FGFs signaling through FGFR-2 IIIb play an important role in the development of the exocrine pancreas.
Keywords: epithelial-mesenchymal interactions
The development of the pancreas as well as that of many other organs depends on epithelial-mesenchymal interactions (1, 2). Although the differentiation of the endocrine tissue of the pancreas can occur in the absence of mesenchyme (3, 4), the growth, morphogenesis, and cytodifferentiation of the exocrine tissue entirely depends on mesenchymal signals (3–5). Despite many attempts to characterize such signals (6), the nature of the mesenchymal factors that govern the development of the exocrine tissue of the pancreas remain largely unknown.
The fibroblast growth factor (FGF) family consists of several structurally related proteins (to date 18 members are known) (7) that display a variety of biological effects, including regulation of cell proliferation, migration, and differentiation (8). There are four FGF receptor (FGFR) genes, designated FGFR-1–FGFR-4, which encode a cytoplasmic tyrosine-kinase domain, and an extracellular region composed of two or three Ig-like domains (designated as loops I, II and III). Ligand specificity is determined by the C-terminal portion of loop III. In the case of FGFRs 1–3, alternative exon usage exists at the level of loop III, giving rise to FGFR IIIb (here abbreviated FGFR2b) and FGFRIIIc isoforms (9).
Recently, the implication of FGFs in organogenesis has been demonstrated in transgenic mice expressing a soluble dominant negative form of FGFR2b (dnFGFR2b), which is thought to interfere with multiple FGFs, including FGF-1, FGF-3, FGF-7, and FGF-10 (10). Those mice presented agenesis of many organs, including lung, thyroid, and salivary glands, whereas other organs like the thymus, stomach, and pancreas presented a considerable reduction of size and/or malformations. To the best of our knowledge besides the very summarized description of the pancreas of mice expressing the dnFGFR2b (10), the role of FGFs in the development of the pancreas had not been analyzed.
In the present study, we have investigated the possible implication of FGFs signaling through FGFR2b in mediating epithelial-mesenchymal interactions in the developing pancreas. For this purpose, we have analyzed the expression of FGFR2b ligands during pancreatic development and used a mesenchyme-free pancreatic epithelial culture system to investigate the effect of FGFs on the growth morphogenesis and cytodifferentiation of the embryonic rat pancreas. We have further extended our analysis by following the in vitro development of the pancreas after inhibition of FGFR2b signaling.
We show that three ligands of FGFR2b (FGF-1, FGF-7, and FGF-10) are expressed during pancreatic development and stimulate the growth, morphogenesis, and cytodifferentiation of the exocrine pancreas. Furthermore abrogation of FGFR2b signaling in pancreatic organocultures results in a considerable reduction of the development of the exocrine pancreas.
MATERIALS AND METHODS
Organ Culture.
Pancreatic epithelium was prepared from embryonic day (E) 11.5 rat embryos as described and grown into three-dimensional collagen gels (4, 11). In some experiments, the pancreatic rudiments were covered with 200 μl of Matrigel (Becton Dickinson). The culture medium consisted of RPMI medium 1640 without serum and supplemented or not with 50 ng/ml of either recombinant human FGF-1 (Boehringer Mannheim), recombinant human FGF-7 (Becton Dickinson), or recombinant human FGF-10. FGF-10 was produced as described (12) and purified by affinity chromatography using Ni-NTA agarose (Qiagen, Hilden, Germany). Heparin (Sigma) at a concentration of 50 μg/ml was added to the rudiments cultured in the presence of FGF-1 or FGF-10.
Cultures were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2. Medium and growth factors were replaced every 48 hr. At the indicated times, the pancreatic epithelia were photographed and fixed for immunohistology as described below.
Immunohistology and Quantitative Analysis.
The pancreatic epithelia were fixed and analyzed as described (4, 11). The antisera used in this study were used at the following dilutions: guinea pig anti-porcine insulin (Dako) 1:500; mouse monoclonal anti-porcine glucagon (Sigma) 1:2,000; rabbit anti-human amylase (Sigma) 1:2,000; mouse monoclonal anti-human insulin (Sigma) 1:2,000, and mouse monoclonal anti-human mitotic proteins 1:200 (clone mpm-2, Dako).
The fluorescent secondary antibodies were: fluorescein anti-guinea pig antibodies (Dako) 1:500, and the following secondary antibodies from Jackson ImmunoResearch: fluorescein anti-rabbit antibodies 1:200; fluorescein anti-mouse antibodies 1:200; Texas-red anti-rabbit antibodies 1:500; and Texas-red anti-mouse antibodies 1:200.
DNA Content and Amylase Activity.
Pancreatic rudiments were harvested and stored frozen. Their DNA content was measured by using the Hoechst dye procedure (13). Their amylase activity was determined from pancreatic rudiment extracts by using a Diagnostics Amylase test from Sigma. Statistical differences were determined by using a Student’s t test for independent samples.
Reverse Transcriptase–PCR Analyses.
Total RNA was extracted from pancreatic rudiments and reverse-transcribed as described (14). The oligonucleotides used for amplification were: FGF-1 (forward) 5′-GGTGCTCCCTATTTTTGGTTGAC-3′; FGF-1 (reverse) 5′-TACACATGGCTCCCCTCTTTGTA-3′; FGF-7 (forward) 5′-ATCCTGCCAACTCTGCTCTACAGA-3′; FGF-7 (reverse) 5′-CTTCCCTTTGACAGGAATCCCCTT-3′; FGF-10 (forward) 5′-TGTTGCTCTTCTTGGTGTCTTCC-3′; FGF-10 (reverse) 5′-GACCTTGCCGTTCTTTTCAATCT-3′; cyclophilin (forward) 5′-ATGGTCAACCCCACCGTGTT-3′; and cyclophilin (reverse) 5′-CGTGTGAAGTCACCACCCT-3′.
Typically 30 cycles of amplification were performed. Amplification parameters included a 1-min denaturation step at 94°C, a 1-min annealing step at 57°C, and a 30-sec extension step at 72°C. The products of amplification were separated on a 2% agarose gel and photographed.
Northern Blot.
Northern blot analysis was performed as described (14). The probes used in this study were a 290-bp fragment corresponding to the FGFR2b cDNA and a 280-bp fragment from mouse glyceraldehyde-3-phosphate dehydrogenase cDNA. Membranes were washed three times for 15 min at 65°C in 0.5 × SSC containing 0.1% SDS. Autoradiography was performed at −70°C with Fujii RX film.
Abrogation of FGFR2b Signaling Pathway.
Two different approaches were used. (i) The use of antisense oligonucleotides specific to rat FGFR2b sequence involved: (a) antisense FGFR ON-I, 5′-AGGCAGACTGGTTGGCCTG-3; such an oligonucleotide has been described as being able to block FGFR expression in the developing rat lung (15, 16), and sense FGFR ON-I: 5′-CAGGCCAACCAGTCTGCCT-3′; (b) antisense FGFR ON-II, 5′-GCGCATTGGCATGCAGTGCCC-3′ consisting of base pairs 2826–2844; sense FGFR ON-II, 5′-GGGCACTGCACTCCAATGCGC-3. The phosphorotioate oligodeoxynucleotides, purified by HPLC, were prepared by Genset (Paris).
(ii) Competition using the extracellular domain of mouse FGFR2b was used. The cDNA encoding the extracellular domain of FGFR2b containing a DNA fragment encoding an E-tag (GAPVPYPDPLEPR) and a hexameric His tag at the 3′ end was constructed into the baculovirus expression vector, BacPAK6 (CLONTECH). Insect cells, High Five cells, were transfected with the recombinant baculovirus. Recombinant soluble FGFR2b was purified from the culture medium by affinity chromatography with Ni-NTA agarose (Qiagen) and used in pancreatic organocultures at a final concentration of 5 μg/ml.
RESULTS
Expression of FGF-1, FGF-7, FGF-10, and Their Receptor FGFR2b in the Developing Rat Pancreas.
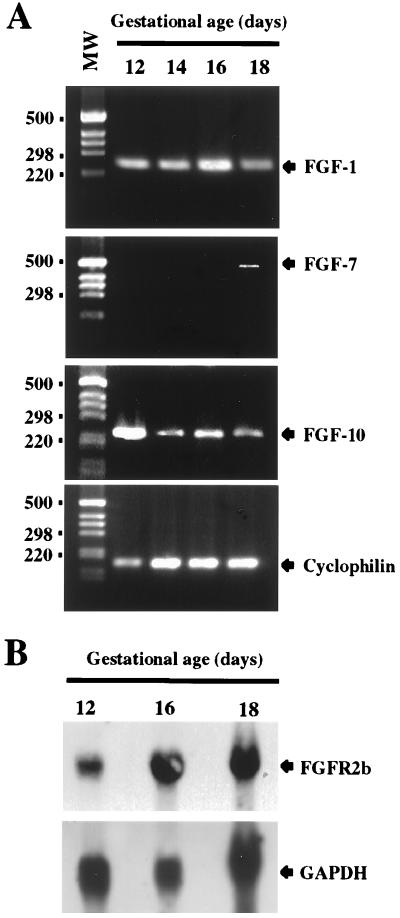
The expression of FGF-1, FGF-7, and FGF-10 mRNAs was studied by reverse transcriptase–PCR in the E12 rat pancreas. As shown in Fig. 1A, although the expression of FGF-1 and FGF-10 could be detected as early as E12 and remained relatively constant throughout pancreatic development, the expression of FGF-7 could not be detected until E18.
Figure 1.
Expression of FGF-1, FGF-7, and FGF-10 and their receptor FGFR2b during pancreas development. (A) Reverse transcriptase–PCR was carried out on pancreatic cDNAs from the indicated stages of development by using appropriate primers for each FGF. Thirty-five cycles of amplification were used for the amplification of FGFs transcripts. Cyclophilin amplification (30 cycles) was used to control the quality of the cDNAs. (B) Expression of FGFR2b mRNA in the developing rat pancreas. FGFR2b expression was analyzed by Northern blot. Total RNA from pancreases at different stages of development were hybridized successively with FGFR2b and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes.
The temporal expression of mRNAs coding for FGFR2b, which acts as a receptor for FGF-1, FGF-7, and FGF-10 was analyzed next. As illustrated (Fig. 1B), the expression of FGFR2b mRNAs was already detectable in the rat pancreas at E12 and remained stable during fetal life.
Cultures of Pancreatic Epithelium from E11.5 Rats.
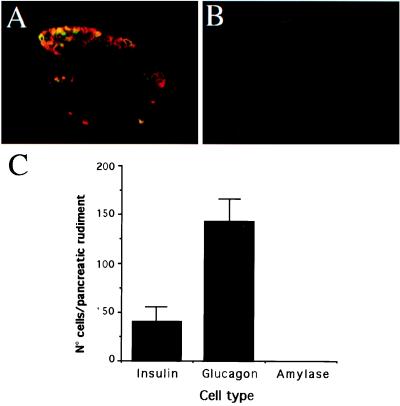
We next studied the effects of FGFs on the in vitro development of the rat pancreatic epithelium. Those experiments were performed on E11.5 pancreatic epithelia. At that stage, the pancreatic epithelium appears as a unlobulated small bulge, allowing a total depletion of mesenchymal cells. Our immunohistochemical analysis indicates that at that stage the pancreatic rudiment is mainly composed of glucagon-expressing cells and a few insulin-expressing cells (Fig. 2). The large majority of the insulin-expressing cells coexpressed glucagon (Fig. 2A). No amylase-expressing cells could be detected at this stage (Fig. 2B). The pancreatic epithelia next were cultured for 7 days in the absence of serum to avoid proliferation of the few mesenchymal cells that would remain attached to the epithelium. Under those conditions, after 7 days in culture, the number of both glucagon- and insulin-expressing cells did increase, reaching 338 ± 65 and 250 ± 50 cells per rudiment, respectively. Few amylase-expressing cells (25 ± 6 cells per rudiment) also developed after 7 days in culture.
Figure 2.
Immunohistological and quantitative analysis of E11.5 rat pancreatic rudiments. (A) Immunostaining with anti-glucagon (red fluorescence) and anti-insulin (green fluorescence) antibodies. (B) The lack of positive staining for amylase staining indicates the absence of development of acinar cells. (C) Quantification of the number of insulin-, glucagon-, and amylase-expressing cells in the E11.5 rat pancreas. Each data point represents the mean ± SEM of 15 pancreatic rudiments.
Effects of FGF-1, FGF-7, and FGF-10 on the Embryonic Pancreatic Epithelium.
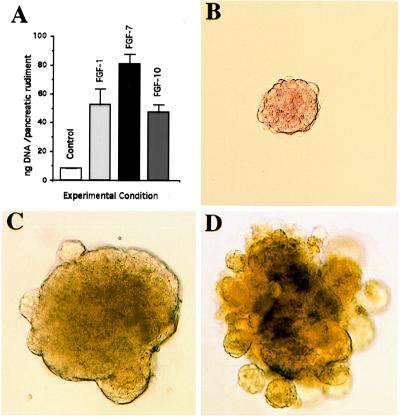
The mitogenic effects of FGF-1, FGF-7, and FGF-10 were studied next. For that purpose, E11.5 mesenchyme-free pancreatic epithelia were cultured in collagen gels in the absence of serum and with, or without, added recombinant FGFs. As indicated in Fig. 3A, after 7 days of culture, the DNA contents of the rudiments treated with all three FGFs were considerably higher than those of the control rudiments. Moreover, FGF treatment induced morphological changes. Indeed, in Matrigel, known to be permissive in different models for morphogenesis, morphogenesis did not occur in the absence of FGFs (Fig. 3B), but FGF-1 induced the formation of a cystic structure displaying some big buds on the surface of the rudiment (Fig. 3C), and in the presence of FGF-7 (Fig. 3D) or FGF-10 (data not shown) long buds giving a branching appearance develop. Such a branching was confirmed by histological analysis (data not shown).
Figure 3.
Mitogenic and morphogenic effects of FGF-1, FGF-7, and FGF-10. (A) DNA content of E11.5 mesenchyme-free pancreatic epithelia cultured during 7 days in the absence or presence of FGF-1, FGF-7, or FGF-10 (50 ng/ml). Each data point represents the mean ± SEM of 10 pancreatic rudiments. (B-D) Effects of FGF-1 and FGF-7 on pancreatic morphogenesis in Matrigel (B) absence of exogenous FGF, (C) FGF-1, 50 ng/ml, and (D) FGF-7, 50 ng/ml.
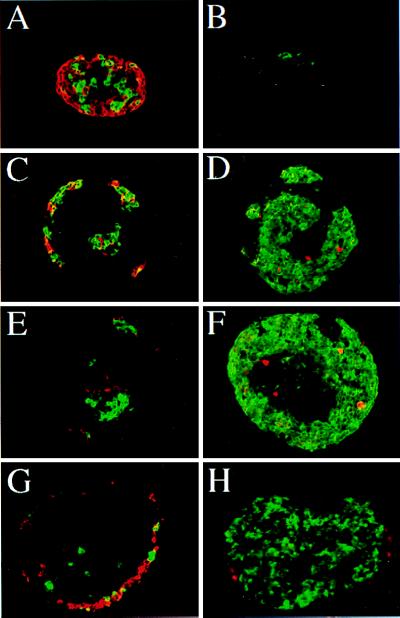
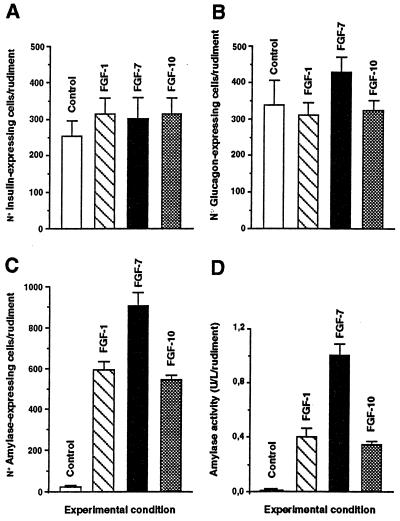
The effects of FGF-1, FGF-7, and FGF-10 on cytodifferentiation were studied next. After a 7-day culture period, cells staining positively for the endocrine hormones insulin and glucagon were detectable both in controls (Fig. 4A) and FGF-treated rudiments (Fig. 4 C, E, and G). No significant quantitative difference in the numbers of insulin- and glucagon-expressing cells was found between the rudiments treated or not with the different FGFs (Fig. 5 A and B). On the contrary, FGFs induce an increase in the number of cells staining positive for amylase, an exocrine-specific product (Fig. 4, compare B to D, F, and H). Indeed, although 25 ± 6 amylase-expressing cells/rudiment develop in the absence of FGF treatment, FGF-1, FGF-7, and FGF-10 induce the development of 600 ± 40, 900 ± 150 and 570 ± 25 amylase-expressing cells per rudiment, respectively (Fig. 5C). Such an increase in the number of amylase-expressing cells could be explained, at least in part, by their proliferation. Indeed, our immunohistological analysis revealed that in FGF-treated cultures, some amylase-expressing cells stained positive for MPM-2 (Fig. 4 D and F), a marker of cells in M phase (17). Finally, this increase in the number of amylase-expressing cells upon FGF treatment was paralleled by an increase in amylase activity (Fig. 5D).
Figure 4.
Immunohistological analysis of pancreatic epithelium grown in vitro in the presence of FGFs. E11.5 mesenchyme-free pancreatic epithelia were grown in collagen gels for 7 days in the absence (A and B) or presence of 50 ng/ml of either FGF-1 (C and D), FGF-7 (E and F), or FGF-10 (G and H). The development of the endocrine tissue (A, C, E, and G) was evaluated after anti-glucagon (red fluorescence) and anti-insulin (green fluorescence) staining. The development of the exocrine tissue (B, D, F, and H) was evaluated after anti-amylase (green fluorescence) staining. The effect of FGF-1 and FGF-7 on acinar cell proliferation (B, D, and F) was analyzed by staining with the antimitotic proteins, mpm2 antibody (red fluorescence). Magnifications: A and B, ×600; C-H, ×400.
Figure 5.
Quantitative analysis of the development of E11.5 mesenchyme-free pancreatic epithelia grown in vitro for 7 days in the absence or presence of FGFs. The absolute number of insulin- (A), glucagon- (B), and amylase-expressing cells (C) was quantified. (D) Quantification of amylase activity in pancreatic epithelia after 7 days in culture in the absence or presence of FGFs. Each data point represents the mean ± SEM of at least 12 pancreatic rudiments.
Effect of the Abrogation of FGFR2b Signaling Pathway in Cultured Embryonic Pancreatic Rudiments.
We next designed in vitro experiments to inhibit the effects of endogenously produced FGFs. Those experiments were performed on E11.5 embryonic pancreatic rudiments (mesenchyme + epithelium). For this purpose, different approaches were used.
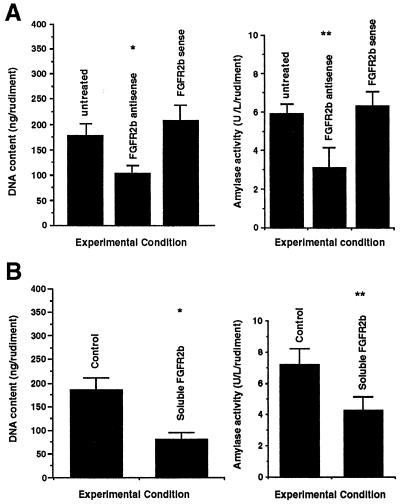
We first used an antisense oligonucleotide specific to FGFR2b. Treatment of pancreatic rudiments with such an oligonucleotide, which had been used previously with success to inhibit FGFR2b expression in the lung (15, 16), induced a 2-fold decrease in both DNA content and amylase activity (Fig. 6A). Moreover the immunohistological study showed that although the number of insulin and glucagon cells was not significantly modified, the number of amylase cells was clearly reduced (data not shown). Identical results were obtained with antisense FGFR ON-II. No significant effect was seen when the rudiments were treated with sense oligonucleotides (Fig. 6A).
Figure 6.
Effects of the abrogation of FGFR2b signaling pathway on the development in vitro of E11.5 pancreatic rudiments (epithelium + mesenchyme). (A) E11.5 pancreatic rudiments were cultured in serum-free medium in the presence of antisense and sense FGFR2b oligonucleotides or in the absence of oligonucleotides. After 7 days in culture, the DNA content and amylase activity per rudiment were analyzed. As shown, treatment with antisense FGR2b oligonucleotides provokes a 2-fold decrease in both DNA (∗, P < 0.01)and amylase activity (∗∗, P < 0.01). At least five rudiments were used for each experiment. The concentration of oligonucleotides was 30 mM. (B) Similar results were obtained when pancreatic rudiments were cultured in the presence of recombinant soluble FGFR2b.
Recombinant protein corresponding to the extracellular ligand-binding domain of human FGFR2b next were used to block the effects of signals transduced via FGFR2b. This recombinant protein is able to bind FGF-1, FGF-7, and FGF-10 (unpublished observation). As shown in Fig. 6B, when E11.5 rat pancreatic rudiments (epithelim + mesenchyme) were cultured for 7 days in the presence of this FGFR2b fragment (5 μg/ml), a significant reduction in both DNA content and amylase activity was noticed.
DISCUSSION
The present study provides experimental evidence indicating that signals transduced via FGFR2b are important for the growth, morphogenesis, and cytodifferentiation of the exocrine component of the pancreas.
We show that in the rat pancreas FGFR2b is expressed as early as E12, its expression remaining fairly constant throughout pancreatic development. This finding is consistent with previous reports showing that this splice variant of FGFR2 is expressed in the pancreatic epithelium during embryonic life (18, 19). Our results also show that mRNAs encoding three known ligands of FGFR2b, FGF-1, FGF-7, and FGF-10 are expressed in the developing pancreas. It previously had been shown that FGF-7 was expressed late during pancreatic development (20). Our present data confirm this point. To our knowledge, the expression of FGF-1 and FGF-10 in the embryonic pancreas had not been studied. We demonstrate here that those two ligands of FGFR2b are expressed as early as E12 in the developing pancreas. Both FGF-1 and FGF-10 thus could interact with FGFR2b during early pancreatic development.
To establish a functional role for FGFR2b ligands during pancreatic development, their effects were tested on cultures of E11.5 mesenchyme-free pancreatic epithelium. Our results clearly show that FGF-1, FGF-7, and FGF-10 induce the proliferation of the cells of the embryonic pancreatic epithelium. These results were not entirely unexpected because in the lung, for example, those ligands have been shown to stimulate epithelial cell proliferation (12, 21, 22). Moreover, transgenic mice overexpressing FGF-7 under the control of the apolipoprotein promoter display hyperplasia in numerous organs including kidney, liver, and pancreas (23).
Our results also demonstrate that FGFs are implicated in the morphogenesis of the exocrine tissue. Indeed, although the pancreatic epithelium remains round in the absence of FGFs, it displayed numerous outgrowths upon treatment with FGFs. Such morphogenic effects of FGFs were found when the epithelium was grown in Matrigel, but not in collagen, the former matrix being known as permissive in different models for budding (12, 21). An interesting difference was found in the type of outgrowths induced by FGF-7 and FGF-10 on one side and FGF-1 on the other side. Although both FGF-7 and FGF-10 cause bud-like branching, FGF-1 induces dilated cyst structures with poor budding. Those different effects could be explained by the fact that at the concentrations used in the present study, although FGF-7 and FGF-10 do not interact with other FGFR family members than FGFR2b (24), FGF-1 can interact with FGFR2b as well as FGFR1 and FGFR4 (25), those receptors being expressed in the developing pancreas (26, 27).
In addition to their effects on the proliferation and morphogenesis of the embryonic pancreatic epithelium, FGF-1, FGF-7, and FGF-10 strongly modified the cytodifferentiation program of the pancreatic epithelium. Indeed, although they do not modify the final number of endocrine cells differentiated from the mesenchyme-free pancreatic rudiments, they induce the development of the exocrine tissue. The fact that the three FGFs do not increase the development of the endocrine tissue is in apparent contradiction with data demonstrating that overexpression of FGF-7 under the control of the apolipoprotein promoter in transgenic mice provokes, among other effects, an increase in the insulin expression by pancreatic ductal epithelial cells, suggesting that FGF-7 may induce endocrine cell differentiation (23). However it must be remembered that (i) in the present work the effects of FGFs were studied during embryonic life, whereas in the work of Nguyen et al. (23) the adult pancreas was studied; and (ii) in the present study, the pancreatic rudiments developed in the absence of mesenchyme. Because the mesenchyme is the site of synthesis of numerous cytokines that can influence pancreatic development, it cannot be excluded that the effect of FGF-7 on endocrine differentiation seen in vivo (23) but not in vitro (the present study) could be indirect or could result from the interactions between FGF-7 and other factors produced by the mesenchyme.
We show here that FGF-1, FGF-7, or FGF-10 induce the development of embryonic pancreatic epithelial cells into acinar exocrine cells expressing amylase. It is well known that the proper development of the exocrine component of the pancreas depends on mesenchymal factors (3–5). Such factors previously were unknown. In light of our results, it can be postulated that FGFs, in particular FGF-1, FGF-7, and FGF-10, represent such factors. Although in our model, FGF-7 was very potent in promoting exocrine cytodifferentiation, it is unlikely that in vivo this factor acts as the principal inducer of exocrine development. Indeed, FGF-7 is not expressed in the pancreas before E18 (ref. 20 and the present study), whereas amylase-expressing cells are detected at E16. Thus, FGF-1 and FGF-10 could promote the formation of the exocrine tissue during early development, whereas all three FGFs tested, which are expressed at E18, could be implicated in later events. This redundancy in ligands of FGFR2b could explain the fact that no obvious pancreatic phenotype was found in FGF-7- or FGF-10-deficient mice (28–30).
In other tissues, such as the lung, a role for FGF-1 and FGF-7 in cytodifferentiation had been demonstrated (22). In the pancreas, the spectacular development of the exocrine tissue in the rudiments cultured in the presence of FGFs is probably the combination of the effects of FGFs both on the differentiation of precursor cells into acinar cells and on the proliferation of such differentiated cells, a large number of amylase-expressing cells being found to express the mitotic antigen mpm-2 upon FGF treatment.
In the present study, an almost 2-fold decrease of the amylase activity and DNA content was observed when FGFR2b activity was inhibited. Such data are in good agreement with the ones recently found in transgenic mice expressing a soluble dominant-negative FGFR under the control of the metallothionein promoter, allowing the expression of the transgene relatively early during mice development (10). Such transgenic mice displayed developmental abnormalities in multiple organs. Their pancreas showed a greatly reduced number of morphologically aberrant acinar cells. Thus, both in the present study as well as in the transgenic mice expressing a soluble dominant negative FGFR, the growth of the pancreas and the cytodifferentiation of the acinar cells were affected but not completely abrogated. There are different possibilities to explain the fact that inhibition of FGFR2b signaling does not totally abrogate exocrine development. One obvious explanation is a technical one: signal transduction mediated by FGFR2b would not be totally abolished in our experiments. Another explanation is that additional soluble factors produced by the mesenchyme also could be implicated in the development of the exocrine pancreas. Indeed, in the present study, although FGF-1, FGF-7, and FGF-10 strongly enhance the development of the pancreatic epithelium depleted of its surrounding mesenchyme into exocrine tissue in term of induction of amylase activity, such effects of FGFs remain below the ones obtained when the differentiation occurs in the presence of mesenchyme (compare absolute values in Figs. 5D and 6). Such factors produced by the mesenchyme could be other members of the FGF family, acting via other FGFR family members. We have observed that FGF-2, which has a very low affinity for FGFR2b (25) and whose biological effects are essentially transduced via FGFR1, FGFR2c, and FGFR4, is able to stimulate both the proliferation (a 7-fold increase on DNA content/rudiment upon treatment with FGF-2) and the development of the exocrine tissue, the number of amylase-expressing cells in FGF-2 treated pancreatic rudiments being 15-fold higher than in control rudiments (data not shown). Other factors implicated in epithelial-mesenchymal signaling like hepatocyte growth factor, platelet-derived growth factor, or epidermal growth factor also could be involved in the development of the exocrine pancreas, their receptors being expressed in the pancreas in development (27).
Taken together, we have demonstrated in the present study that FGFR2b and its ligands FGF-1, FGF-7, and FGF-10 must be considered as crucial mediators of the epithelial-mesenchymal interactions, which promote the development of the exocrine component of the pancreas.
Acknowledgments
O. Dubois and S. Mahe are acknowledged for technical assistance. This work was supported by grants from the Juvenile Diabetes Foundation International (198207 to R.S.).
ABBREVIATIONS
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- E(n)
embryonic day
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Golosow N, Grobstein C. Dev Biol. 1962;4:242–255. doi: 10.1016/0012-1606(62)90042-8. [DOI] [PubMed] [Google Scholar]
- 2.Kallman F, Grobstein C. J Cell Biol. 1968;20:399–413. doi: 10.1083/jcb.20.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gittes G, Galante P, Hanahan D, Rutter W, Debas H. Development (Cambridge, UK) 1996;122:439–447. doi: 10.1242/dev.122.2.439. [DOI] [PubMed] [Google Scholar]
- 4.Miralles F, Czernichow P, Scharfmann R. Development (Cambridge, UK) 1998;125:1017–1024. doi: 10.1242/dev.125.6.1017. [DOI] [PubMed] [Google Scholar]
- 5.Pictet R, Rutter W. In: Handbook of Physiology: The Endocrine Pancreas. Steiner D, Freinkel N, editors. Vol. 1. Baltimore: Williams & Wilkins; 1972. pp. 25–66. [Google Scholar]
- 6.Ronzio R, Rutter W. Dev Biol. 1973;30:307–320. doi: 10.1016/0012-1606(73)90091-2. [DOI] [PubMed] [Google Scholar]
- 7.Ohbayashi N, Hoshikawa M, Kimura S, Yamasaki M, Fukui S, Itoh N. J Biol Chem. 1998;273:18161–18164. doi: 10.1074/jbc.273.29.18161. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi T, Rossant J. Curr Opin Genet Dev. 1995;5:485–491. doi: 10.1016/0959-437x(95)90053-j. [DOI] [PubMed] [Google Scholar]
- 9.Miki T, Bottaro D, Flemnig T, Smith C, Burgess W, Chan A, Aaronson S. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celli G, LaRochelle W, Mackem S, Sharp R, Merlino G. EMBO J. 1998;17:1642–1655. doi: 10.1093/emboj/17.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miralles F, Battelino T, Czernichow P, Scharfmann R. J Cell Biol. 1998;143:827–836. doi: 10.1083/jcb.143.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan B. Development (Cambridge, UK) 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 13.Labarca C, Paigen K. Anal Biochem. 1980;102:344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- 14.Atouf F, Czernichow P, Scharfmann R. J Biol Chem. 1997;272:1929–1934. doi: 10.1074/jbc.272.3.1929. [DOI] [PubMed] [Google Scholar]
- 15.Post M, Souza P, Liu J, Tseu I, Wang J, Kuliszewski M, Tanswell A. Development (Cambridge, UK) 1996;122:3107–3115. doi: 10.1242/dev.122.10.3107. [DOI] [PubMed] [Google Scholar]
- 16.Ohmichi H, Koshimizu U, Matsumoto K. Development (Cambridge, UK) 1998;125:1315–1324. doi: 10.1242/dev.125.7.1315. [DOI] [PubMed] [Google Scholar]
- 17.Westendorf J, Rao P, Gerace L. Proc Natl Acad Sci USA. 1994;91:714–718. doi: 10.1073/pnas.91.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finch P, Cunha G, Rubin J, Wong J, Ron D. Dev Dyn. 1995;203:223–240. doi: 10.1002/aja.1002030210. [DOI] [PubMed] [Google Scholar]
- 19.Orr-Urtreger A, Bedford M, Burakowa T, Arman E, Zimmer Y, Yayon A, Givol D, Lonai P. Dev Biol. 1993;158:475–486. doi: 10.1006/dbio.1993.1205. [DOI] [PubMed] [Google Scholar]
- 20.Mason I, Fuller-Pace F, Smith R, Dickson C. Mech Dev. 1994;45:15–30. doi: 10.1016/0925-4773(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 21.Nogawa H, Ito T. Development (Cambridge, UK) 1995;121:1015–1022. doi: 10.1242/dev.121.4.1015. [DOI] [PubMed] [Google Scholar]
- 22.Cardoso W, Itoh A, Nogawa H, Mason I, Brody J. Dev Dyn. 1997;208:398–405. doi: 10.1002/(SICI)1097-0177(199703)208:3<398::AID-AJA10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen H, Danilenko D, Bucay N, DeRose M, Van G, Thomason A, Simonet W. Oncogene. 1996;12:2109–2119. [PubMed] [Google Scholar]
- 24.Igarashi M, Finch P, Aaronson S. J Biol Chem. 1998;273:13230–13235. doi: 10.1074/jbc.273.21.13230. [DOI] [PubMed] [Google Scholar]
- 25.Ornitz D, Xu J, Colvin J, McEwen D, McArthur C, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 26.LeBras S, Miralles F, Basmaciogullari A, Czernichow P, Scharfmann R. Diabetes. 1998;47:1236–1242. [PubMed] [Google Scholar]
- 27.LeBras S, Czernichow P, Scharfmann R. Diabetologia. 1998;41:1471–1481. doi: 10.1007/s001250051094. [DOI] [PubMed] [Google Scholar]
- 28.Guo L, Degentstein L, Fuchs E. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]
- 29.Min H, Danilenko D, Scully S, Bolon B, Ring B, Tarpley J, DeRose M, Simonet W. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]