The prevalence of CTX-M β-lactamases has reached a critical level, which highlights the need for study of their spread in developing countries.
Keywords: Antimicrobial resistance, bacteria, enteric infections, Escherichia coli, community, urinary tract infections, β-lactamase resistance, developing countries, Cambodia, research
Abstract
Despite the recent global spread of CTX-M β-lactamases in Escherichia coli isolates from community-acquired urinary tract infections (CA-UTIs), their dissemination has been little studied in developing countries. In a 2-year prospective study, we documented the prevalence of extended-spectrum β-lactamases (ESBLs) in E. coli that were responsible for CA-UTIs in Phnom-Penh, Cambodia. Ninety-three E. coli strains were included. We observed a high prevalence of resistance to amoxicillin (88.2% of strains), cotrimoxazole (75.3%), ciprofloxacin (67.7%), gentamicin (42.5%), and third-generation cephalosporins (37.7%). A total of 34 strains carried ESBLs, all of which were CTX-M type. CTX-M carriage was associated with resistance to fluoroquinolones and aminoglycosides. U using repetitive extragenic palindromic–PCR, we identified 4 clusters containing 9, 8, 3, and 2 strains. The prevalence of CTX-M β-lactamases has reached a critical level in Cambodia, which highlights the need for study of their spread in developing countries.
Escherichia coli is the bacterium most frequently isolated in community- and hospital-acquired urinary tract infections (CA-UTIs and HA-UTIs, respectively). Despite possessing the gene encoding cephalosporinase, ampC (1), wild strains of E. coli are susceptible to most β-lactams because of the absence of an efficient ampC promoter region. The extensive use of β-lactam antimicrobial drugs has led to the emergence of resistant strains worldwide. β-lactam resistance is mostly mediated through acquisition of β-lactamase genes located on mobile genetic elements such as plasmids or transposons. Most β-lactamases found in E. coli belong to Ambler class A and can be further divided into narrow-spectrum β-lactamases (e.g., TEM-1, TEM-2, and SHV-1) and extended-spectrum β-lactamases (ESBLs) (e.g., TEM-3, SHV-5, and CTX-M-like) (1–3). ESBLs confer resistance to extended-spectrum cephalosporins, widely used to treat E. coli infections.
CTX-M–type β-lactamases (CTX-Ms) are broad-spectrum β-lactamases derived from the chromosomally encoded β-lactamases of Kluyvera sp. (4–6). So far, >70 CTX-M types have been isolated (www.lahey.org/studies, updated October 2008); these have been divided into 5 clusters on the basis of amino acid sequence: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25. Native CTX-Ms are cefotaximases that usually hydrolyze cefotaxime rather than ceftazidime. However, point mutations can extend their target spectrum to ceftazidime. Thus, CTX-M-15 and CTX-M-27 are derived by a single Asp240Gly substitution from CTX-M-3 and CTX-M-14, respectively (7,8).
Until recently, most ESBLs in clinical samples came from a hospital environment, belonged to the TEM or SHV β-lactamase family, and were produced by Klebsiella spp., Enterobacter spp., and E. coli (9). Within the past few years, the nature of ESBL dissemination has changed: E. coli is now the most frequently isolated ESBL-carrying bacterium, and CTX-Ms have become the most frequently isolated ESBLs (10). Moreover, in the last few years CTX-M–type ESBLs have emerged within the community, particularly among E. coli isolated from UTIs (11–16). Risk factors for CTX-M carriage in the community are largely unknown. CTX-Ms have been isolated from patients with CA-UTIs who have neither had recent antimicrobial drug treatment nor been admitted to a hospital or long-term care facility (17–19). Resistance to fluoroquinolones, aminoglycosides, and cotrimoxazole is often associated with CTX-M production (16,17), thereby limiting the choice of effective antimicrobial drugs to carbapenems or colistin.
Some studies have shown high prevalence of ESBLs among various Enterobacteriaceae in hospitals in developing countries (20–22). However, little information is available regarding prevalence within the community. Few bacteriologic data are available for Cambodia, and no studies of resistance of Enterobacteriaceae to antimicrobial agents have been reported. We investigated E. coli CA-UTIs in a 2-year prospective study. Our aims were 1) to establish the prevalence of community-acquired urinary E. coli resistance to a wide range of antimicrobial drugs and 2) to characterize the mechanisms underlying E. coli resistance to β-lactams.
Methods
Clinical Isolates from Patient Samples
Our laboratory at the Institut Pasteur in Phnom Penh, Cambodia, is a community laboratory, involved in various biological analyses. We receive biological samples from outpatients and hospitals that do not have bacteriology laboratories. Included in the study were patients who visited our laboratory with a suspected UTI from January 2004 through December 2005. Currently hospitalized patients were excluded, yet previous hospitalization was not an exclusionary criterion. We collected basic clinical data for each patient, including age and gender, whether antimicrobial drugs had been taken within the month preceding the sample submission, UTI history, and recent hospital visit. A UTI was defined by >104 leukocytes/mL urine and >105 CFU/mL urine. We used CLED (cystine-lactose-electrolyte deficient) agar (Dynamic Pharma, Phnom-Penh, Cambodia) for culture. Bacteria were identified with the API20E identification gallery (bioMérieux, Marcy l’Etoile, France). Strains were named (CEC [Cambodian Escherichia coli]) and numbered independently from isolation date.
Antimicrobial Drug Susceptibility Testing and ESBL Confirmatory Testing
We determined antimicrobial drug susceptibility by the disk-diffusion method on Mueller-Hinton agar plates (Bio-Rad, Marnes-la-Coquette, France), as recommended by the French Society for Microbiology (www.sfm.asso.fr). We tested the following antimicrobial agents: amoxicillin, amoxicillin/clavulanic acid (coamoxiclav), ticarcillin, ticarcillin/clavulanic acid, piperacillin, piperacillin/tazobactam, cefalothin, cefoxitin, cefotaxime, ceftazidime, cefepime, imipenem, moxalactam, aztreonam, nalidixic acid, norfloxacin, ciprofloxacin, gentamicin, tobramycin, netilmicin, amikacin, nitroxolin, fosfomycin, and sulfamethoxazole/trimethoprim (cotrimoxazole). We detected ESBLs by using the double-disk synergy test (clavulanic acid and cefotaxime, ceftazidime, cefepime and aztreonam) performed on Mueller-Hinton media. We determined MICs for cefoxitin, cefotaxime, ceftazidime, and cefepime by using the agar-dilution method for ESBL-carrying strains and strains with decreased susceptibility to cefoxitin (as interpreted by the disk-diffusion method) as recommended by the French Society for Microbiology. For cefoxitin, strains were considered susceptible if MICs were <8 mg/L; intermediate susceptible if MICs were 16–32 mg/L, and resistant if MICs were >32 mg/L. For cefotaxime, strains were considered susceptible if MICs were <1 mg/L; intermediate susceptible if MICs were 2 mg/L, and resistant if MICs were >2 mg/L. For ceftazidime and cefepime, strains were considered susceptible if MICs were <4 mg/L, intermediate susceptible if MICs were 8 mg/L, and resistant if MICs were >8 mg/L.
PCR Amplifications
Template DNA was prepared by boiling. Briefly, 5 colonies were suspended thoroughly in 1 mL DNase- and RNase-free water and boiled for 10 min. After centrifugation, supernatant was used as template DNA. We amplified the ampC upstream region, blaTEM, blaSHV, blaCTX-M, blaVEB, blaOXA-1, and blaCMY by PCR, using specific oligodeoxynucleotides (Table 1). PCR was performed in a 25-µL mixture of 1× buffer (supplied with Taq polymerase), 2.5 mmol/L MgCl2, 2.5 U of FIREPol DNA polymerase (Solis BioDyne, Tartu, Estonia), 200 µmol/L of each deoxynucleoside triphosphate, and 25 pmol of each primer. The PCR mixture was subjected to a 5-min denaturation step at 94°C, followed by 30 cycles of 45 s at 94°C, 45 s at 55°C, and 60 s at 72°C, and a final elongation step of 5 min at 72°C. PCR products were separated by 100-V electrophoresis in a 2% agarose gel for 30 min, after which they were stained with ethidium bromide.
Table 1. Primers used to study CTX-M β-lactamases in Escherichia coli, Cambodia, 2004–2005.
| Gene detected | Primer name | Primer sequence (5′ → 3′) | Reference |
|---|---|---|---|
| blaTEM | C | TCG GGG AAA TGT GCG CG | (23) |
|
|
D |
TGC TTA ATC AGT GAG GCA CC |
|
| blaSHV | OS-5 | TTA TCT CCC TGT TAG CCA CC | (24) |
|
|
OS-6 |
GAT TTG CTG ATT TCG CTC GG |
|
| blaCTX-M | MA-1 | SCS ATG TGC AGY ACC AGT AA | (23) |
|
|
MA-2 |
CCG CRA TAT GRT TGG TGG TG |
|
| blaCTX-M group 9 | M9U | ATG GTG ACA AAG AGA GTG CA | (23) |
|
|
M9L |
CCC TTC GGC GAT GAT TCT C |
|
| blaCTX-M group 1 | M13U | GGT TAA AAA ATC ACT GCG TC | (23) |
|
|
M13L |
TTG GTG ACG ATT TTA GCC GC |
|
| blaampC | AmpC1 | AAT GGG TTT TCT ACG GTC TG | (25) |
|
|
AmpC2 |
GGG CAG CAA ATG TGG AGC AA |
|
| blaVEB | casF | CGA CTT CCA TTT CCC GAT GC | (26) |
|
|
casB |
GGA CTC TGC AAC AAA TAC GC |
|
| blaOXA-1 | OXA-1up | TAT CAA CTT CGC TAT TTT TTT A | (27) |
|
|
OXA-1low |
TTT AGT GTG TTT AGA ATG GTG A |
|
| blaCMY | CF1 | ATGATGAAAAAATCGTTATGC | (28) |
|
|
CF2 |
TTGTAGCTTTTCAAGAATGCGC |
|
| chuA | ChuA.1 | GAC GAA CCA ACG GTC AGG AT | (29) |
|
|
ChuA.2 |
TGC CGC CAG TAC CAA AGA CA |
|
| ygaA | YgaA.1 | TGA AGT GTC AGG AGA CGC TG | (29) |
|
|
YgaA.2 |
ATG GAG AAT GGG TTC CTC AAC |
|
| TspE4C2 | TspE4C2.1 | GAG TAA TGT CGG GGC ATT CA | (29) |
| TspE4C2.2 | CGC GCC AAC AAA GTA TTA GC |
PCR Product Sequencing
Amplification products were purified with Montage PCR Filter Units (Millipore, Billerica, MA, USA). Sequencing reactions were performed in a PTC-225 Peltier Thermal Cycler (MJ Research, Waltham, MA, USA) by using an ABI PRISM BigDye Terminator Cycle Sequencing Kit with AmpliTaq DNA polymerase (Applied Biosystems, Branchburg, NJ, USA), according to the manufacturer’s instructions. Each template was sequenced with the appropriate primer. Fluorescence-labeled fragments were purified from the unincorporated terminators with an ethanol precipitation protocol. The samples were resuspended in distilled water and subjected to electrophoresis in an ABI 3730xl sequencer (Applied Biosystems).
Phylogenetic Group Determination
To determine phylogenetic group (i.e., A, B1, B2, and D), we performed triplex PCR for all strains (n = 93) as described previously (29). We used chuA and yjaA genes and an E. coli DNA fragment, TSPE4.C2.
Repetitive Extragenic Palindromic PCR
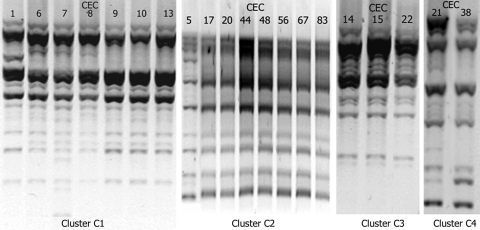
The clonality of ESBL-positive strains was assessed by repetitive extragenic palindromic (rep)–PCR. DNA was extracted by using the QIAGEN Mini kit (QIAGEN, Courtaboeuf, France). REP-PCRs were performed for all strains in the same batch with primers rep-1R and rep-2T, as described previously (28). The resulting products were separated by 70-V electrophoresis in a 1% agarose gel for 3 h, after which they were stained with ethidium bromide. Strains with suspected similar migration profiles were then migrated together with the ESBL-positive strains to assess their similarity. Photographs of the gels (Figure) have been harmonized to be more informative.
Figure.
Results of repetitive extragenic palindromic–PCR of Escherichia coli isolates belonging to the 4 clusters, Cambodia, 2004–2005. CEC, Cambodian E. coli.
Statistical Analysis
Data were analyzed by using Epi Info version 3.2 (www.cdc.gov/epiinfo). Risk factors for ESBL-producing E. coli were assessed by using univariate analysis with the χ2 or Fisher exact tests; however, we used analysis of variance to determine whether age had an association with ESBL-producing E. coli. Associations between ESBL type, co-resistance, mutations in ampC promoter regions, and phylogenetic groups were separately tested by using the χ2 or Fisher exact tests. The significance threshold was 0.05.
Results
Population Characteristics
Of the 861 urine samples, 194 were positive for UTI and 163 contained E. coli. Samples from hospitalized patients and samples isolated from the same patient in a short period were excluded from the study. Overall, 93 patients with E. coli–related CA-UTIs were recruited for the study, all of whom were living in Phnom Penh. Their mean age was 43 years (range 1–88 years, median 40 years), and the M:F ratio was 0.21. No pregnant women were included. Twenty-eight (30.1%) patients had a history of UTI (with no details available), 24 (25.8%) had taken antimicrobial drugs, and 9 (9.7%) had sought consultation at a hospital in the month preceding the urine sampling. Patients with a history of UTI had taken significantly more antimicrobial drugs than had persons without UTIs (p<0.0001).
Prevalence of Antimicrobial Drug Resistance in E. coli Strains
Disk-diffusion susceptibility testing indicated high prevalence of resistance to various antimicrobial agents: 88 (94.6%) strains were resistant to amoxicillin and ticarcillin, and 20 (21.5%) were immediate susceptible to cefoxitin. We observed synergy between clavulanic acid, oxyimino-cephalosporins, and aztreonam for 34 strains (36.6%). Decreased susceptibility to cefoxitin was significantly associated with ESBL expression (p<0.001). All strains (n = 93) were susceptible to imipenem. One strain (CEC93) showed a high level of resistance to cefoxitin, cefotaxime, and ceftazidime but not to cefepime; double-disk synergy test results were negative for clavulanate. We observed a substantial level of resistance to quinolones; 63 strains (67.7%) had an intermediate level of resistance or were fully resistant to nalidixic acid, norfloxacin, and ciprofloxacin. Quinolone resistance was strongly associated with ESBL expression (p<0.0001). Of the 34 ESBL-carrying strains, only 2 were susceptible to quinolones. Cotrimoxazole resistance was observed in 70 strains (75.3%) but was not significantly associated with ESBL expression (p = 0.06). Aminoglycoside resistance was also significantly associated with ESBL expression (p<0.01). Thirty-nine strains (41.9%) were resistant to at least 1 of the 4 tested aminoglycosides. The most frequently observed phenotypic profile included resistance to gentamicin, tobramycin, and netilmicin (37.6% of the strains). None of the strains were resistant to fosfomycin or nitrofurantoin (Table 2).
Table 2. Resistance to antimicrobial agents among ESBL-positive and ESBL-negative Escherichia coli, Cambodia, 2004–2005*.
| Antimicrobial agent | Resistance ratios, % (no. resistant strains) |
p value | ||
|---|---|---|---|---|
| ESBL+, n = 34 | ESBL–, n = 59 | Total, n = 93 | ||
| Fluoroquinolones | 94 (32) | 53 (31) | 68 (63) | <0.001 |
| Cotrimoxazole | 85 (29) | 69 (41) | 75 (70) | 0.06 |
| Aminoglycosides | 65 (22) | 27 (16) | 42 (39) | <0.01 |
| Co-amoxiclav | 94 (32) | 19 (11) | 46 (43) | <0.001 |
| Cefoxitin | 41 (13) | 11 (7) | 22 (20) | <0.001 |
| Nitrofurantoin | 0 | 0 | 0 | NS |
| Fosfomycin | 0 | 0 | 0 | NS |
*ESBL, extended-spectrum β-lactamase; NS, not significant.
Risk Factors for ESBL Carriage
Univariate analysis showed no risk factors for ESBL carriage. Carriage was not significantly associated with gender, age, previous hospitalization (within the past month), antimicrobial drug treatment (within the last month), or history of UTI.
β-Lactamase Characterization
Thirty-four strains were tested for ESBL identification. They were all positive for blaCTX-M and negative for blaVEB and blaSHV. We detected blaTEM in 26 (76.4%) of the ESBL-carrying strains. CTX-M-14 was the most frequently isolated ESBL (n = 15), followed by CTX-M-27 (n = 12) and CTX-M-15 (n = 5). One strain (CEC7) was carrying both blaCTX-M-14 and blaCTX-M-15. Strain CEC14 was carrying a blaCTX-M-14 variant, which differed from the parental enzyme by a single transversion (C825G) leading to a Ser273Arg amino acid change. blaTEM-1 was detected in 24 (96%) of the blaTEM-carrying strains. One strain (CEC23) had a DNA sequence that differed from TEM-1 by 4 nucleotides. Only 1 of these mutations resulted in an amino acid change (Met182Thr). Amino acid sequence comparison (www.lahey.org/studies/webt.asp) showed that this TEM had the same amino acid sequence as TEM-135.
Strain CEC93, exhibiting a phenotypic profile indicative of a high level of cephalosporinase expression, was positive for blaCMY. Sequencing confirmed the presence of blaCMY-2 in this strain. We detected blaOXA-1 in 4 CTX-M-15–producing strains: CEC15, CEC48, CEC68, and CEC89.
ampC Sequencing Results
Strains with different levels of resistance to cefoxitin, ranging from intermediate (cefoxitin MIC = 8 mg/L) to full resistance (cefoxitin MIC >8 mg/L), had different mutations within the region upstream from ampC (Table 3). Only 1 strain did not display any mutations. Most mutations occurred within the transcriptional attenuator region; the most frequently observed profiles were +22 C>T, +26 T>G, +27 A>T, +32 G>A, and +70 C>T (11 strains). The other strains each exhibited a different profile. Strain CEC57 had the profile –32 T>A with an additional nucleotide change, which significantly increases ampC expression (31). Strain CEC92 had the combination –42 C>T and –28 G>A, which also increases ampC expression (25).
Table 3. Mutations detected in the ampC promoter region of CTX-M β-lactam–resistant Escherichia coli*.
| Isolate | CEF MIC, mg/L | CTX-M | Phylogenetic group | Mutation at position† |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −42 | −32 | −28 | −18 | −1 | +22 | +26 | +27 | +32 | +58 | +70 | +81 | ||||
| E. coli K12 (U00096) | – | – | – | C | T | G | G | C | C | T | A | G | C | C | G |
| CEC1 | 16 | Yes | D | T | G | T | A | T | |||||||
| CEC3 | 32 | No | D | T | G | T | A | T | |||||||
| CEC4 | 32 | No | D | A | T | T | |||||||||
| CEC6 | 32 | Yes | D | T | G | T | A | T | |||||||
| CEC8 | 32 | Yes | D | T | G | T | A | T | |||||||
| CEC9 | 32 | Yes | D | T | G | T | A | T | |||||||
| CEC10 | 32 | Yes | D | T | G | T | A | T | |||||||
| CEC13 | 32 | Yes | D | T | G | T | A | T | |||||||
| CEC14 | 16 | Yes | D | T | |||||||||||
| CEC15 | 16 | Yes | D | T | |||||||||||
| CEC19 | 32 | No | D | T | G | T | A | T | |||||||
| CEC21 | 16 | Yes | D | A | T | A | |||||||||
| CEC22 | 16 | Yes | D | T | |||||||||||
| CEC31 | 16 | Yes | D | A | T | A | |||||||||
| CEC57 | 32 | No | D | A | T | G | T | A | T | ||||||
| CEC88 | 32 | No | B2 | A | A | ||||||||||
| CEC89 | 32 | Yes | D | T | G | T | A | T | |||||||
| CEC92 | >128 | Yes | D | T | A | T | T | ||||||||
| CEC93 | >128 | No | D | T | G | T | A | T | |||||||
*CEF, cefoxitin; CEC, Cambodian E. coli. †According to numbering of Jaurin and Grundström (30).
Phylogenetic Groups
Most of the strains studied belonged to groups B2 (53%; n = 49) and D (28%; n = 26), as do most pathogenic E. coli (32). Twelve strains belonged to group A (13%) and 6 to group B1 (6%). Among ESBL-carrying strains, 53% belonged to group B2 (n = 18), 38% to D (n = 13), 6% to A (n = 2), and 3% to B1 (n = 1). Resistance to cotrimoxazole was less prevalent in B2 strains than in strains from the other groups (p<0.05). Resistance to quinolones was more prevalent among group D strains than among strains from other groups (p<0.005). Furthermore, a strong association (p<0.0001) was found between decreased susceptibility to cefoxitin and group D strains. No association was shown between phylogenetic group and other characteristics such as ESBL carriage and aminoglycoside resistance. Among ESBL-carrying strains, none of the 4 groups were associated with 1 particular type of CTX-M.
Rep-PCR Findings
Twenty-one ESBL positive strains and the CMY-2–producing strain were divided into 4 clusters; the remaining strains were genetically unrelated (Appendix Table; Figure). Cluster C1 consisted of 8 strains with an identical ampC mutation profile, all belonging to phylogenetic group D. However, β-lactamase content was variable: 6 strains harbored CTX-M-27 and TEM-1, 1 strain harbored only CMY-2, and 1 harbored only CTX-M-15. Strain CEC7 (belonging to group B2, carrying CTX-M-14 and CTX-M-15, with no ampC mutations) exhibited a similar REP-PCR profile to that of strains in cluster C1. Cluster C2 consisted of 8 strains belonging to group B2 with no ampC mutations and carrying various β-lactamases; 6 strains produced CTX-M-14 (2 of which co-produced TEM-1), 1 produced CTX-M-27/TEM-1, and 1 produced CTX-M-15/TEM-1/OXA-1. Cluster C3 consisted of 3 strains from group D, all with the same ampC mutation profile, but with 3 distinct β-lactamase profiles (CTX-M-14 and TEM-1; CTX-M-14-like and TEM-1; and CTX-M-15 and TEM-1). Cluster C4 consisted of 2 strains from group D, each with the same ampC mutation profile but different β-lactamase profiles (CTX-M-14 and TEM-1; and CTX-M-27 only).
Discussion
We surveyed antimicrobial drug resistance in Cambodia. Our prospective study in this developing country focused on CTX-Ms CA-UTIs caused by E. coli. Although the patients included came from the community, antimicrobial drug resistance was prevalent among UTI-causing strains, particularly to β-lactams (including extended-spectrum cephalosporins). Our findings suggest that CTX-M-type β-lactamases are widespread in Cambodia. CTX-M production was significantly associated with resistance to quinolones and aminoglycosides and with decreased susceptibility to cefoxitin, leading to a high prevalence of multiresistant strains. The spread of CTX-M in the community has already been described through prospective studies in industrialized countries such as Canada (33), France (34), and the United Kingdom (12); however, we found higher prevalence of CTX-Ms in Cambodia than that reported in these previous studies.
We propose 3 possible explanations for the situation in Cambodia. First, although no reliable data were available, it is well known that many persons in the community self-medicate or obtain prescriptions for non-adapted drugs (drugs taken without any antibiotic susceptibility testing of the strain that causes the infections); these practices suggest that uncontrolled consumption of antimicrobial agents is likely to play a major role. This factor, together with the likely substandard quality of some drugs, undoubtedly contributes to the high prevalence of resistance. Second, hygiene in Cambodia is poor, and the population density in Phnom Penh is high. The saturated sewer system, particularly during the rainy season, likely facilitates efficient propagation and spread of bacteria within the community. Grenet et al. proposed that that poor hygiene in a French-Guyanese Indian community led to the spread of resistant bacteria despite low antibiotic pressure (35). Similarly, poor hygiene conditions may have led to the spread of resistant strains in the Cambodian community studied here. The TEM-type and SHV-type ESBL—until recently the predominant ESBL family subtypes—have never been implicated in the spread of ESBL in the community. Why CTX-M strains are the only ESBL types to spread in the community is not clear. CTX-M strains may have a particularly high capacity to disseminate or an ecologic advantage over other ESBLs and thus persist in the community, whereas other ESBL may be progressively eliminated with decreasing antibiotic pressure.
One limitation to our study is that patients at our institute are not necessarily representative of the Cambodian population. For most persons in Cambodia, antimicrobial agents and biologic analyses are expensive. Therefore, our patients may have been wealthier and may have taken more courses of antimicrobial drug treatment than other Cambodians.
REP-PCR yielded 4 clusters of strains, consistent with their identical ampC mutational profiles, yet with various contents of β-lactamases. Because all included patients were living in Phnom Penh, mediocre hygiene might have favored the diffusion of clones within an urban area, a phenomenon that had previously been observed in large-scale studies undertaken in the United Kingdom (12), Canada (36), Italy (37), and Brazil (15) but not in Hong Kong (13). In contrast to our results, clones identified in those studies harbored the same β-lactamases. Further investigation using multilocus sequence typing would be necessary to identify the molecular determinants of the CTX-M–carrying E. coli pandemic in Cambodia.
Branger et al. have observed that group D E. coli were more frequently resistant to quinolones (38) than were non-D E. coli; these findings are consistent with our results. Moreover, we found a strong association (p<0.001) between group D strains and decreased susceptibility to cefoxitin (secondary to the effect of mutations in the ampC promoter region). This association was also present when all strains were taken into account (p<0.001). However, although CTX-M carriage was more frequently observed for group D strains than for non-D strains, the association was not significant. Given that quinolone resistance and ampC hyperexpression involve several mutations, a possible explanation for this association with group D strains may be a stronger mutation capacity for these strains than for strains belonging to other groups. Further investigations will be required to explain this phenomenon.
Community-isolated ESBL-carrying strains are an emerging challenge for community practitioners and hospitals. Information is not readily available in either developing countries or in industrialized countries, and UTI treatment guidelines remain unchanged. In Cambodia, and probably in many other developing countries, resistant E. coli strains are endemic to the community. Investigating the current situation in Cambodia may improve our understanding of the situation in industrialized countries, where ESBLs are no longer uncommon in the community. According to our experience in Cambodia, measures should focus on improving hygiene and appropriate prescribing of antimicrobial agents. In conclusion, we suggest that this high prevalence of β-lactam resistance in Cambodia is due to the intrinsic capacity of CTX-M–encoding genes to disseminate through communities where hygiene and living conditions are poor and antimicrobial drug consumption is uncontrolled.
Supplementary Material
Characterization of the ESBL- or plasmidic ampC-type _-lactamase-carrying Escherichia coli strains (N = 35), Cambodia, 2004-2005*
Acknowledgments
This work was partially supported by grants from Université Pierre et Marie Curie–Paris and from the European Community (6th Programme Cadre de Recherche et de Développement Technologique Contract: LSHM-CT 2003-503335).
Biography
Dr Ruppé is a medical microbiologist at the Institut Pasteur du Cambodge in Phnom Penh, Cambodia. His main research interest is antimicrobial drug resistance in developing countries.
Footnotes
Suggested citation for this article: Ruppé E, Hem S, Lath S, Gautier V, Ariey F, Sarthou J-L, et al. CTX-M β-lactamases in Escherichia coli from community-acquired urinary tract infections, Cambodia. Emerg Infect Dis [serial on the Internet]. 2009 May [date cited]. Available from http://www.cdc.gov/EID/content/15/5/741.htm
References
- 1.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–31. 10.1098/rstb.1980.0049 [DOI] [PubMed] [Google Scholar]
- 2.Hall BG, Barlow M. Revised Ambler classification of β-lactamases. J Antimicrob Chemother. 2005;55:1050–1. 10.1093/jac/dki130 [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humeniuk C, Arlet G, Gautier V, Grimont P, Labia R, Philippon A. Beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob Agents Chemother. 2002;46:3045–9. 10.1128/AAC.46.9.3045-3049.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Kampfer P, Nordmann P. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2002;46:4038–40. 10.1128/AAC.46.12.4038-4040.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson AB, Silverman M, Boyd DA, McGeer A, Willey BM, Pong-Porter V, et al. Identification of a progenitor of the CTX-M-9 group of extended-spectrum beta-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob Agents Chemother. 2005;49:2112–5. 10.1128/AAC.49.5.2112-2115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnet R, Recule C, Baraduc R, Chanal C, Sirot D, De Champs C, et al. Effect of D240G substitution in a novel ESBL CTX-M-27. J Antimicrob Chemother. 2003;52:29–35. 10.1093/jac/dkg256 [DOI] [PubMed] [Google Scholar]
- 8.Karim A, Poirel L, Nagarajan S, Nordmann P. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol Lett. 2001;201:237–41. [DOI] [PubMed] [Google Scholar]
- 9.Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–51. 10.1128/CMR.14.4.933-951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9:466–75. 10.1016/j.mib.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 11.Arpin C, Coulange L, Dubois V, Andre C, Fischer I, Fourmaux S, et al. Extended-spectrum-beta-lactamase-producing Enterobacteriaceae strains in various types of private health care centers. Antimicrob Agents Chemother. 2007;51:3440–4. 10.1128/AAC.01431-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother. 2004;54:735–43. 10.1093/jac/dkh424 [DOI] [PubMed] [Google Scholar]
- 13.Ho PL, Poon WW, Loke SL, Leung MS, Chow KH, Wong RC, et al. Community emergence of CTX-M type extended-spectrum beta-lactamases among urinary Escherichia coli from women. J Antimicrob Chemother. 2007;60:140–4. 10.1093/jac/dkm144 [DOI] [PubMed] [Google Scholar]
- 14.Lartigue MF, Zinsius C, Wenger A, Bille J, Poirel L, Nordmann P. Extended-spectrum beta-lactamases of the CTX-M type now in Switzerland. Antimicrob Agents Chemother. 2007;51:2855–60. 10.1128/AAC.01614-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minarini LA, Gales AC, Palazzo IC, Darini AL. Prevalence of community-occurring extended spectrum beta-lactamase–producing Enterobacteriaceae in Brazil. Curr Microbiol. 2007;54:335–41. 10.1007/s00284-006-0307-z [DOI] [PubMed] [Google Scholar]
- 16.Mendonca N, Leitao J, Manageiro V, Ferreira E, Canica M. Spread of extended-spectrum beta-lactamase CTX-M–producing Escherichia coli clinical isolates in community and nosocomial environments in Portugal. Antimicrob Agents Chemother. 2007;51:1946–55. 10.1128/AAC.01412-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calbo E, Romani V, Xercavins M, Gomez L, Vidal CG, Quintana S, et al. Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum beta-lactamases. J Antimicrob Chemother. 2006;57:780–3. 10.1093/jac/dkl035 [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Bano J, Navarro MD, Romero L, Martinez-Martinez L, Muniain MA, Perea EJ, et al. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase–producing Escherichia coli in nonhospitalized patients. J Clin Microbiol. 2004;42:1089–94. 10.1128/JCM.42.3.1089-1094.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colodner R, Rock W, Chazan B, Keller N, Guy N, Sakran W, et al. Risk factors for the development of extended-spectrum beta-lactamase–producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis. 2004;23:163–7. 10.1007/s10096-003-1084-2 [DOI] [PubMed] [Google Scholar]
- 20.Jitsurong S, Yodsawat J. Prevalence of extended-spectrum beta-lactamases (ESBLs) produced in blood isolates of gram-negative bacteria in a teaching hospital in southern Thailand. Southeast Asian J Trop Med Public Health. 2006;37:131–5. [PubMed] [Google Scholar]
- 21.Kusum M, Wongwanich S, Dhiraputra C, Pongpech P, Naenna P. Occurrence of extended-spectrum beta-lactamase in clinical isolates of Klebsiella pneumoniae in a university hospital, Thailand. J Med Assoc Thai. 2004;87:1029–33. [PubMed] [Google Scholar]
- 22.Chanawong A, M’Zali FH, Heritage J, Lulitanond A, Hawkey PM. SHV-12, SHV-5, SHV-2a and VEB-1 extended-spectrum beta-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J Antimicrob Chemother. 2001;48:839–52. 10.1093/jac/48.6.839 [DOI] [PubMed] [Google Scholar]
- 23.Saladin M, Cao VT, Lambert T, Donay JL, Herrmann JL, Ould-Hocine Z, et al. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol Lett. 2002;209:161–8. [DOI] [PubMed] [Google Scholar]
- 24.Arlet G, Rouveau M, Philippon A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum beta-lactamase. FEMS Microbiol Lett. 1997;152:163–7. [DOI] [PubMed] [Google Scholar]
- 25.Caroff N, Espaze E, Berard I, Richet H, Reynaud A. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum beta-lactamase production. FEMS Microbiol Lett. 1999;173:459–65. [DOI] [PubMed] [Google Scholar]
- 26.Poirel L, Naas T, Guibert M, Chaibi EB, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999;43:573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavollay M, Mamlouk K, Frank T, Akpabie A, Burghoffer B, Ben Redjeb S, et al. Clonal dissemination of a CTX-M-15 beta-lactamase–producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob Agents Chemother. 2006;50:2433–8. 10.1128/AAC.00150-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckert C, Gautier V, Saladin-Allard M, Hidri N, Verdet C, Ould-Hocine Z, et al. Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother. 2004;48:1249–55. 10.1128/AAC.48.4.1249-1255.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8. 10.1128/AEM.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaurin B, Grundström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of beta-lactamases of the penicillinase type. Proc Natl Acad Sci U S A. 1981;78:4897–901. 10.1073/pnas.78.8.4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caroff N, Espaze E, Gautreau D, Richet H, Reynaud A. Analysis of the effects of –42 and –32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing ampC. J Antimicrob Chemother. 2000;45:783–8. 10.1093/jac/45.6.783 [DOI] [PubMed] [Google Scholar]
- 32.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitout JD, Hanson ND, Church DL, Laupland KB. Population-based laboratory surveillance for Escherichia coli–producing extended-spectrum beta-lactamases: importance of community isolates with blaCTX-M genes. Clin Infect Dis. 2004;38:1736–41. 10.1086/421094 [DOI] [PubMed] [Google Scholar]
- 34.Arpin C, Dubois V, Coulange L, Andre C, Fischer I, Noury P, et al. Extended-spectrum beta-lactamase–producing Enterobacteriaceae in community and private health care centers. Antimicrob Agents Chemother. 2003;47:3506–14. 10.1128/AAC.47.11.3506-3514.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grenet K, Guillemot D, Jarlier V, Moreau B, Dubourdieu S, Ruimy R, et al. Antibacterial resistance, Wayampis Amerindians, French Guyana. Emerg Infect Dis. 2004;10:1150–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitout JD, Church DL, Gregson DB, Chow BL, McCracken M, Mulvey MR, et al. Molecular epidemiology of CTX-M–producing Escherichia coli in the Calgary Health Region: emergence of CTX-M-15–producing isolates. Antimicrob Agents Chemother. 2007;51:1281–6. 10.1128/AAC.01377-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mugnaioli C, Luzzaro F, De Luca F, Brigante G, Perilli M, Amicosante G, et al. CTX-M–type extended-spectrum beta-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob Agents Chemother. 2006;50:2700–6. 10.1128/AAC.00068-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branger C, Zamfir O, Geoffroy S, Laurans G, Arlet G, Thien HV, et al. Genetic background of Escherichia coli and extended-spectrum beta-lactamase type. Emerg Infect Dis. 2005;11:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the ESBL- or plasmidic ampC-type _-lactamase-carrying Escherichia coli strains (N = 35), Cambodia, 2004-2005*