Abstract
Height is a classic polygenic trait, reflecting the combined influence of multiple as-yet-undiscovered genetic factors. We carried out a meta-analysis of genome-wide association study data of height from 15,821 individuals at 2.2 million SNPs, and followed up the strongest findings in >10,000 subjects. Ten newly identified and two previously reported loci were strongly associated with variation in height (P values from 4 × 10-7 to 8 × 10-22). Together, these 12 loci account for ~2% of the population variation in height. Individuals with ≤8 height-increasing alleles and ≥16 height-increasing alleles differ in height by ~3.5 cm. The newly identified loci, along with several additional loci with strongly suggestive associations, encompass both strong biological candidates and unexpected genes, and highlight several pathways (let-7 targets, chromatin remodeling proteins and Hedgehog signaling) as important regulators of human stature. These results expand the picture of the biological regulation of human height and of the genetic architecture of this classical complex trait.
The advent of genome-wide association studies1, made possible by knowledge gained from the HapMap Consortium2 and recent advances in genome-wide genotyping technologies and analytic methods, have had a dramatic impact on the field of human genetics. Recent genome-wide studies have led to the identification of common genetic variants reproducibly associated with complex human diseases3. Genome-wide association (GWA) studies have also been used successfully to identify genetic variation associated with quantitative traits, such as lipid levels4 and body mass index5,6. These discoveries, through the identification of previously unknown and often unanticipated genes, have opened an exciting period in the study of human complex traits and common diseases.
The small effect sizes that have characterized most of the variants recently identified present a challenge to the study of polygenic diseases and traits, as large sample sizes have generally been required to identify associated common variants. It is not yet known whether increasing sample size further will accelerate the pace of discovery, and to what extent multiple loci with modest effect will reveal previously unsuspected biological pathways. To begin to answer these important questions, we used adult height as a model phenotype. Adult height is a complex trait with high heritability (h2 ~0.8-0.9 within individual populations)7,8. Furthermore, height is accurately measured and relatively stable over a large part of the lifespan9, and data is available for very large numbers of individuals. Thus, the study of height is an ideal opportunity to dissect the architecture of a highly polygenic trait in humans. In addition, because height is associated with several common human diseases (for example, cancers)10, loci associated with height may be pleiotropic, influencing the risk or severity of other diseases11.
Using data from GWA studies, we and our colleagues identified the first two common variants to be robustly associated with adult height variation: a SNP in the 3′ UTR of the HMGA2 gene12 and a SNP at the GDF5-UQCC locus11. The overall variation in height explained by these two polymorphisms is small (0.3-0.7% of the total variance), suggesting that most common variants that influence height will have a small effect. The modest effects observed also highlight the importance of using large datasets to identify ‘true’ stature variants: the HMGA2 SNP was first found in a combined analysis of the DGI and Wellcome Trust Case Control Consortium UKT2D datasets (n = 4,921 individuals), and the GDF5-UQCC finding was identified initially in an analysis of the SardiNIA and FUSION results (n = 6,669 individuals). As these findings are likely to be among the upper range of effect sizes for common variants associated with height, we considered the likely possibility that progressively larger sample sizes would be required to identify additional height loci.
Encouraged by these earlier successes, we proceeded to carry out a larger meta-analysis of six GWA datasets, including height association results for 15,821 individuals at ~2.2 million SNPs, to find additional loci associated with height. Here we report the identification and validation of ten newly identified associations between common SNPs and height variation (each with P < 5 × 10-7), and an additional four associations with strongly suggestive evidence (each with P < 5 × 10-6). We also confirm the two previously reported associations (HMGA2 and GDF5-UQCC). The newly identified loci associated with height implicate several biological pathways or gene sets—including targets of the let-7 microRNA, chromatin remodeling proteins and Hedgehog signaling—as important regulators of human stature. Finally, we examine the interaction with gender, test for epistatic interactions between loci, and estimate the explanatory power of each locus individually and in combination. These results broaden our understanding of the biological regulation of human growth and set the stage for further genetic analysis of this classical complex trait.
RESULTS
Identification of loci associated with height
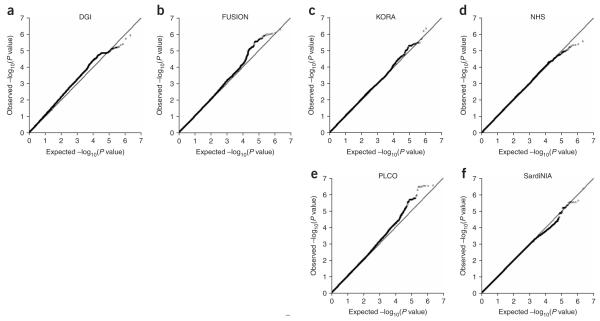
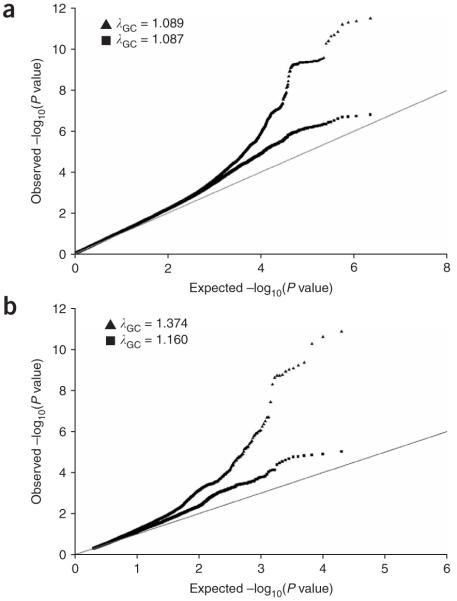
We carried out a meta-analysis of GWA data for height that included 15,821 individuals from six studies: two type 2 diabetes case-control datasets (DGI4, n = 2,978; FUSION13, n = 2,371), two nested cancer case-control datasets (NHS14, n = 2,286; PLCO15, n = 2,244) and two datasets from population-based cohorts (KORA16, n = 1,644; SardiNIA17, n = 4,305)(Supplementary Table 1 online). All participants were of European ancestry. Because genome-wide genotyping in these studies was done on different platforms (Affymetrix 500K for DGI, KORA, and SardiNIA, Illumina 317K for FUSION and Illumina 550K for NHS and PLCO), we imputed genotypes for all polymorphic markers in the HapMap Phase II CEU reference panel in each GWA scan using the program MACH (Y. Li and G.R.A., unpublished data), thereby generating compatible datasets of 2,260,683 autosomal SNPs. Adult height was tested for association with these SNPs in each study under an additive genetic model, and association results were combined by meta-analysis using a weighted Z-score method (Methods). Whereas the distribution of test statistics for each individual GWA study was consistent with the expectation under the null hypothesis (Fig. 1), the quantile-quantile plot of the meta-analysis results clearly showed a large excess of low P values at the right tail of the distribution, despite minimal evidence of overall systematic bias (λGC = 1.089; Fig. 2a). This result suggests that true associations with height variation, which were not discernable from the background in the individual studies, were brought to light in the combined analysis. Indeed, the second- and third-strongest association signals in these height meta-analysis results—HMGA2 rs1042725 (P = 2.6 × 10-11) and GDF5-UQCC rs6060369 (P = 1.9 × 10-10)—have recently been shown to be robustly associated with stature in humans not selected for tall or short stature11,12. These findings validate our meta-analytic approach and suggest that other loci associated with height are represented at or near the top of our results.
Figure 1.
Quantile-quantile plot of ~2.2 million SNPs for each of the six genome-wide association scans meta-analyzed. (a) DGI (n = 2,978). (b) FUSION (n = 2,371). (c) KORA (n = 1,644). (d) NHS (n = 2,286). (e) PLCO (n = 2,244). (f) SardiNIA (n = 4,305). Each black circle represents an observed statistic (defined as the -log10(P)) versus the corresponding expected statistic. The gray line corresponds to the null distribution.
Figure 2.
Quantile-quantile plots supporting the presence of additional loci associated with height. (a) Plot of 2,260,683 SNPs from the meta-analysis of the six GWA scans (n = 15,821). Each black triangle represents an observed statistic (defined as the -log10(P)) versus the corresponding expected statistic. The black squares correspond to the distribution of test statistics after removing markers correlated to the 12 height signals with P < 5 ×10-7 described in Table 1. The gray line corresponds to the null distribution. (b) Association results from Weedon et al.18 provide independent evidence that the top 10,000 SNPs from our meta-analysis exceed the null expectations (gray line). We selected the top 10,000 SNPs from our meta-analysis before (black triangles) and after (black squares) removing SNPs marking the known height loci (identified by us and in the accompanying manuscript) and retrieved the corresponding P values from Weedon et al. We show one-tailed P values because we corrected for the direction of effects, if needed. Inflation factors are unusually high because the analysis is restricted to the top 10,000 SNPs, which are likely enriched for SNPs that are truly associated with height variation.
To distinguish true variants associated with height from other SNPs that could have achieved low P values by chance or confounding effects, and also to collect direct genotype data for associations based on imputed genotypes, we set up a two-stage follow-up strategy (Supplementary Fig. 1 online). In the first stage, we selected 78 SNPs representative of the top association signals (ranked by P values, and taking into account linkage disequilibrium (LD) to minimize redundancy); we then genotyped these 78 SNPs in a panel of European Americans (n = 2,189) ascertained from the near-ends of the normal height distribution (short, 5-10th percentile; tall, 90-95th percentile) (Supplementary Table 2 online). This panel has been used previously to replicate very strongly the HMGA2 rs1042725 association12. To decrease the number of false positives taken into the second stage and retain true associations with height, we selected for further study 29 SNPs that had an odds ratio in the European American panel that was consistent with the direction of the effect observed in the meta-analysis (Methods). We genotyped these SNPs in four large validation (replication) panels: all 29 SNPs were genotyped in the population-based FINRISK97 cohort (n = 7,803), and a subset was genotyped in the population-based KORA S4 (n = 4,130) and PPP (n = 3,402) cohorts and the type 2 diabetes case-control FUSION stage 2 panel (n = 2,466) (Supplementary Tables 3-6 online). Because of logistic and technical issues, not all 29 SNPs could be genotyped in all four DNA panels, thus leading to a loss in power for some of these SNPs in our follow-up strategy. Nevertheless, these combined efforts led to the identification of 12 SNPs with combined P < 5 × 10-7 (using evidence from the meta-analysis and the validation panels except the European American panel, because of its specific ascertainment), a level of significance strongly suggestive of true association (Table 1; detailed association results and LD plots are given in Supplementary Table 7 and Supplementary Fig. 2 online, respectively)3. Of the three loci with P values between 5 × 10-7 and 5 × 10-8, two were confirmed in the accompanying manuscript18 (SH3GL3-ADAMTSL3 and CDK6), and one had strong independent evidence of association in the European American tall-short (USHT) panel (CHCHD7-RDHE2, P = 9 × 10-6; Table 1). This indicates that loci with P values in this range are at least enriched for loci with valid associations with height. In addition, four loci showed overall suggestive evidence (combined P < 5 × 10-6) of association with adult height (Table 1). Although SNP rs2730245 at the WDR60 locus has a combined P = 3 × 10-7, we chose to include this marker in our list of suggestive associations because, unlike the 12 loci in the top section of Table 1, this signal had a follow-up P > 0.05.
Table 1. Summary association results for 29 SNPs genotyped in the follow-up panels.
| Chromosome (position) | SNP | MAF (allele)a | Meta-analysis P beta (s.e.m.)b | Meta-analysis heterogeneity I2c (%) | Follow-up P beta (s.e.m.)b | Follow-up heterogeneity I2c (%) | Combined P beta (s.e.m.)d | USHT P OR (95% CI)e | Nearby genesf |
|---|---|---|---|---|---|---|---|---|---|
| Validated associations (combined P < 5 × 10-7) | |||||||||
| 3 (142588268) | rs724016 | 0.48 (G) | 5.0 × 10-12 0.461 (0.070) |
0 | 2.5 × 10-11 0.307 (0.051) |
71 | 8.3 × 10-22 0.365 (0.038) |
7.8 × 10-8 1.4 (1.2-1.6) |
ZBTB38 |
| 12 (64644614) | rs1042725 | 0.49 (T) | 2.6 × 10-11 -0.403 (0.070) |
8 | 1.7 × 10-10 -0.493 (0.077) |
0 | 2.7 × 10-20 -0.484 (0.051) |
4.2 × 10-6 0.8 (0.7-0.9) |
HMGA2 |
| 6 (142745570) | rs4896582 | 0.27 (A) | 3.2 × 10-8 -0.397 (0.077) |
0 | 6.3 × 10-12 -0.365 (0.058) |
24 | 2.4 × 10-18 -0.378 (0.051) |
2.2 × 10-5 0.8 (0.7-0.9) |
GPR126 |
| 6 (26341366) | rs10946808 | 0.28 (G) | 3.3 × 10-8 -0.448 (0.083) |
40 | 1.9 × 10-10 -0.314 (0.058) |
63 | 3.8 × 10-17 -0.358 (0.045) |
3.4 × 10-8 0.7 (0.7-0.8) |
HIST1H1D |
| 20 (33370575) | rs6060369 | 0.36 (C) | 1.9 × 10-10 0.454 (0.077) |
0 | 1.3 × 10-7 0.410 (0.077) |
0 | 1.4 × 10-16 0.435 (0.051) |
0.012 1.2 (1.0-1.3) |
GDF5-UQCC |
| 4 (146007626) | rs1492820 | 0.48 (G) | 3.6 × 10-8 -0.435 (0.077) |
0 | 3.9 × 10-5 -0.218 (0.051) |
0 | 1.2 × 10-11 -0.288 (0.038) |
0.002 0.8 (0.7-0.9) |
HHIP |
| 14 (91529711) | rs8007661 | 0.30 (T) | 8.9 × 10-8 -0.486 (0.090) |
0 | 0.0015 -0.326 (0.102) |
n.a. | 5.5 × 10-10 -0.416 (0.064) |
0.64 1.0 (0.9-1.1) |
TRIP11-ATXN3 |
| 6 (105514355) | rs314277 | 0.13 (A) | 5.9 × 10-9 0.608 (0.109) |
0 | 0.035 0.230 (0.102) |
0 | 1.1 × 10-8 0.410 (0.077) |
0.26 1.1 (0.9-1.3) |
LIN28B |
| 19 (2121954) | rs12986413 | 0.45 (T) | 2.0 × 10-5 0.333 (0.077) |
23 | 3.7 × 10-4 0.275 (0.077) |
60 | 2.9 × 10-8 0.307 (0.051) |
0.20 1.1 (1.0-1.2) |
DOT1L |
| 15 (82077496) | rs2562784 | 0.17 (G) | 2.9 × 10-5 0.307 (0.083) |
3 | 5.5 × 10-4 0.397 (0.115) |
n.a. | 6.4 × 10-8 0.339 (0.064) |
0.28 1.1 (1.0-1.2) |
SH3GL3-ADAMTSL3 |
| 8 (57318152) | rs9650315 | 0.13 (T) | 9.6 × 10-6 -0.474 (0.102) |
46 | 0.010 -0.352 (0.134) |
26 | 3.8 × 10-7 -0.429 (0.083) |
9.0 × 10-6 0.7 (0.6-0.8) |
CHCHD7-RDHE2 |
| 7 (91901556) | rs2040494 | 0.50 (C) | 4.8 × 10-5 -0.288 (0.077) |
29 | 0.0020 -0.237 (0.077) |
0 | 3.8 × 10-7 -0.262 (0.051) |
0.18 0.9 (0.8-1.0) |
CDK6 |
| Suggestive associations (5 × 10-6 > combined P > 5 × 10-7) | |||||||||
| 7 (158224265) | rs2730245 | 0.33 (G) | 2.6 × 10-7 0.410 (0.083) |
4 | 0.082 0.179 (0.102) |
23 | 3.0 × 10-7 0.320 (0.064) |
0.99 1.0 (0.9-1.1) |
WDR60 |
| 9 (130493638) | rs7466269 | 0.33 (G) | 1.2 × 10-5 -0.339 (0.077) |
0 | 0.011 -0.198 (0.077) |
0 | 7.5 × 10-7 -0.269 (0.058) |
0.48 1.0 (0.9-1.1) |
FUBP3 |
| 9 (116214350) | rs7869550 | 0.24 (G) | 9.5 × 10-6 -0.429 (0.090) |
0 | 0.18 -0.218 (0.090) |
2 | 1.2 × 10-6 -0.326 (0.064) |
0.58 1.0 (0.8-1.1) |
PAPPA |
| 17 (51785154) | rs12449568 | 0.47 (C) | 4.7 × 10-6 0.333 (0.077) |
19 | 0.047 0.153 (0.077) |
35 | 2.4 × 10-6 0.250 (0.051) |
0.49 1.0 (0.9-1.2) |
ANKFN1 |
| Others (combined P > 1×10-5) | |||||||||
| 16 (615681) | rs763014 | 0.43 (C) | 2.5 × 10-5 0.307 (0.077) |
41 | 0.033 0.166 (0.077) |
59 | 5.1 × 10-6 0.237 (0.058) |
0.05 1.1 (1.0-1.3) |
RAB40C |
| 14 (36069800) | rs17104630 | 0.04 (G) | 1.4 × 10-5 -0.621 (0.141) |
0 | 0.062 -0.250 (0.128) |
2 | 8.1 × 10-6 -0.422 (0.096) |
0.02 0.8 (0.6-1.0) |
NKX2-1 |
| 8 (41596148) | rs11786297 | 0.05 (G) | 4.9 × 10-5 0.544 (0.128) |
20 | 0.10 0.320 (0.186) |
52 | 3.9 × 10-5 0.474 (0.102) |
0.39 1.1 (0.9-1.4) |
AGPAT6 |
| 14 (91477446) | rs3783937 | 0.28 (T) | 5.5 × 10-5 -0.326 (0.083) |
35 | 0.15 -0.128 (0.090) |
0 | 8.2 × 10-5 -0.237 (0.064) |
0.02 0.9 (0.8-1.0) |
FBLN5 |
| 16 (2225358) | rs26840 | 0.43 (T) | 1.6 × 10-4 0.326 (0.077) |
0 | 0.29 -0.128 (0.186) |
46 | 1.5 × 10-4 0.269 (0.064) |
0.02 1.2 (1.0-1.3) |
E4F1 |
| 5 (4573856) | rs1450822 | 0.48 (A) | 4.1 × 10-4 0.282 (0.077) |
10 | 0.43 0.141 (0.186) |
6 | 3.3 × 10-4 0.262 (0.070) |
0.21 1.1 (1.0-1.2) |
|
| 8 (129118629) | rs13249999 | 0.05 (T) | 2.8 × 10-4 -0.768 (0.198) |
18 | 0.30 -0.205 (0.198) |
n.a. | 3.6 × 10-4 -0.480 (0.141) |
0.59 0.9 (0.8-1.2) |
|
| 20 (5034939) | rs6116651 | 0.10 (T) | 4.8 × 10-5 -0.531 (0.122) |
36 | 0.46 -0.122 (0.147) |
0 | 4.7 × 10-4 -0.371 (0.096) |
0.004 0.8 (0.6-0.9) |
C20orf30 |
| 4 (4394990) | rs2916448 | 0.13 (C) | 3.9 × 10-7 -0.531 (0.102) |
0 | 0.67 0.070 (0.160) |
23 | 5.5 × 10-4 -0.358 (0.090) |
0.25 0.9 (0.8-1.1) |
LYAR |
| 4 (82511461) | rs1662845 | 0.35 (T) | 5.7 × 10-5 0.326 (0.083) |
0 | 0.60 0.038 (0.077) |
77 | 9.1 × 10-4 0.179 (0.058) |
8.5 × 10-6 1.3 (1.2-1.5) |
PRKG2 |
| 15 (77391837) | rs11858942 | 0.37 (G) | 8.2 × 10-6 0.339 (0.077) |
0 | 0.86 -0.013 (0.083) |
0 | 0.0015 0.173 (0.058) |
0.54 1.0 (0.9-1.2) |
TMED3 |
| 7 (50439131) | rs12540874 | 0.29 (G) | 7.3 × 10-5 -0.288 (0.077) |
0 | 0.91 0.006 (0.083) |
0 | 0.0027 -0.160 (0.058) |
0.98 1.0 (0.9-1.1) |
GRB10 |
| 18 (48613000) | rs12958987 | 0.23 (T) | 8.8 × 10-6 -0.339 (0.083) |
0 | 0.16 0.154 (0.109) |
n.a. | 0.0045 -0.160 (0.064) |
0.69 1.0 (0.9-1.1) |
DCC |
The minor allele frequency (MAF) and corresponding allele are given for the HapMap European American CEU population, positive strand, Build NCBI 35.
Effect size (beta) and standard error (s.e.m.) are expressed in centimeters. The direction of the effect is given for the CEU minor allele. The meta-analytic P value is calculated using a weighted Z-score method. Effect sizes are calculated using the inverse variance method (fixed effect).
Values in bold have a Cochran’s Q heterogeneity test P value < 0.05.
The combined P value is calculated by combining the P values from the GWA meta-analysis and the follow-up panels using a weighted Z score method (results from the European American (USHT) tall-short panel are excluded from the combined analysis because of its specific ascertainment). The combined effect size is calculated using the inverse variance method (fixed effect), and by excluding results from the related component of DGI and the tall-short USHT panel.
The odds ratio corresponds to the increased odds of being in the group of tall individuals for each additional minor allele.
Nearby genes are defined as the closest genes to (200-kb window), and/or genes in LD with, the SNP. n.a., not applicable for SNPs genotyped in only one follow-up panel.
Characterization of signals associated with height
Population stratification is a possible strong confounder for association studies of height and other human phenotypes19. We took several steps to ensure that the association signals for height confirmed in our study were not due to population stratification. Association tests in the NHS and PLCO datasets were corrected for residual population structure using principal component methods20; a similar analysis on the unrelated component of DGI did not change association results for the SNPs reported in Table 1 (Supplementary Table 8 online). An analysis of the FINRISK97 replication panels stratified for geographic origins within Finland did not alter the strength of the associations with height reported in Table 1 (Supplementary Table 9 online). Finally, using 279 variants known to correlate with the major axes of ancestry in European-derived populations21, we calculated a small inflation factor (λ) of 1.09 for our height meta-analysis results, suggesting that no substantial stratification is shown even by markers selected for this purpose. Taken together, these results indicate that the genetic associations with height found in our study are unlikely to be due to population stratification.
For the 16 SNPs identified with combined P < 5 ×10-6 (Table 1), there was, in the FINRISK97 panel, no evidence of departure from an additive genetic model (P > 0.05 for likelihood ratio test of additive versus unconstrained two degree-of-freedom test), no significant difference in effect size between males and females (P > 0.05), and no strong epistatic interactions between loci (the most significant interaction was observed between SNP rs1492820 and SNP rs10946808 (P = 0.001), which is not significant after correcting for the 120 pairwise tests of interaction done).
Explanatory power of loci associated with height
Each of the common height SNPs reported here explains a small fraction of the residual phenotypic variation in height (0.1-0.8%; Supplementary Table 7). When analyzed together, the additive effects of the 12 SNPs with combined P < 5 × 10-7 only contribute 2.0% of the height variation in the FINRISK97 panel, far from the estimated ~80-90% attributable to genetic variation. To assess the cumulative predictive value of this initial set of variants, we created a ‘height score’ by counting the number of height-increasing alleles (12 SNPs; height score 0-24) in each participant with complete genotype for these 12 SNPs in the FINRISK97 panel (n = 7,566). On average, males and females with ≤8 ‘tall’ alleles (4.7% of FINRISK97) are 3.5 cm and 3.6 cm shorter than males and females with ≥16 ‘tall’ alleles (7.1% of FINRISK97), respectively (Fig. 3). Individual effect sizes range from 0.3 cm per T allele for CDK6 rs2040494 to 0.5 cm per C allele for HMGA2 rs1042725 (Table 1).
Figure 3.
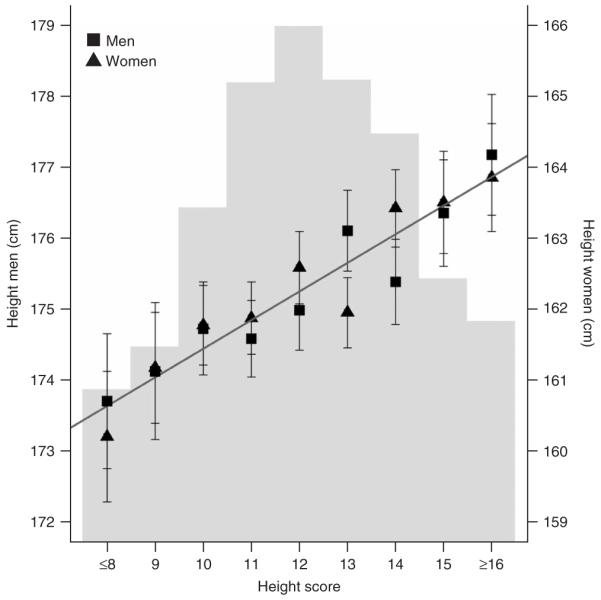
Analysis of combined effects. For each participant in the FINRISK97 panel with complete genotype at the 12 SNPs with P < 5 × 10-7 in Table 1 (n = 7,566), we counted the number of height-increasing alleles to create a height score. Individuals with ≤8 or ≥16 ‘tall’ alleles were grouped. For each height score group, in men and women separately, the mean ± 95% confidence interval is plotted. The axis for men is on the left and the axis for women is on the right (same scale). The regression line in dark gray indicates that, for both men and women, each additional ‘tall’ allele increases height by 0.4 cm. The light gray histogram in the background represents the relative fraction of individuals in each height score group (height score ≤8: 4.7%; height score 9: 6.2%; height score 10: 11.1%; height score 11: 15.5%; height score 12: 17.5%; height score 13: 15.6%; height score 14: 13.7%; height score 15: 8.6%; height score ≥16: 7.1%).
DISCUSSION
Some of the SNPs reported here fall in or near strong candidate height genes, such as the recently described associations with HMGA2 (ref. 12) and GDF5-UQCC11, whereas others identify previously unsuspected loci. Together, these associations highlight biological pathways that are important in regulating human growth.
Hedgehog-interacting protein (HHIP; rs1492820) is a transcriptional target and an antagonist of Hedgehog signaling; it binds with high affinity to the three mammalian Hedgehog proteins22. The mouse homolog, Hhip, is expressed in the perichondrium, including regions flanking Indian hedgehog (Ihh) expression in the appendicular and axial skeleton. Ectopic overexpression of Hhip in mouse cartilage causes severe skeletal defects, including short-limbed dwarfism, a feature reminiscent of the phenotype observed in Ihh null mice22,23.
We identified several associated SNPs in or near genes related to chromatin structure. In addition to HMGA2, which encodes a chromatin-binding protein, we found associations with a SNP (rs10946808) in a histone cluster on chromosome 6, a SNP (rs12986413) in the histone methyltransferase DOT1L gene and a SNP (rs724016) in an intron of the methyl-DNA-binding transcriptional repressor gene ZBTB38. It is currently unclear how genetic variation at these loci modulate height, but there is a precedent for a connection between regulation of chromatin structure and stature: Sotos syndrome (MIM117550), characterized by extreme tall stature, is caused by mutations and deletions in the histone methyltransferase gene NSD1. It would be interesting to test whether the height variants at HMGA2, the chromosome 6 histone cluster, DOT1L and ZBTB38 modify clinical outcome in Sotos syndrome, or whether severe mutations in these genes, particularly DOT1L, could cause a Sotos syndrome-like phenotype.
That the variant rs1042725, strongly associated with adult and childhood height12, falls in the 3′ UTR of HMGA2 is notable in part because HMGA2 is the human gene with the greatest number of validated let-7 microRNA binding sites24,25. In fact, rs1042725 is 13 base pairs away from a let-7 site, suggesting a possible mechanism of action whereby the SNP alters microRNA binding and therefore expression of HMGA2. When we examined our list of 12 height loci, we were somewhat surprised to find three additional previously described targets of let-7: the cell cycle regulator CDK6 (ref. 26), the histone methyltransferase DOT1L27 and the gene LIN28B28. PAPPA, a locus with a combined P = 1.2 × 10-6 in our study, also contains a predicted let-7 binding site27. Thus, genes that influence height seem to be enriched for validated or potential let-7 targets: 5 of the 16 (31%) confirmed or suggestive loci associated with height have let-7 binding sites, compared with 2% of the genes in the human genome (Fisher’s exact test P = 3 × 10-5). Because microRNAs can co-regulate genes involved in the same biological process, it will be interesting to test whether the other targets of let-7, or let-7 itself, are regulators of adult height.
There were also noteworthy candidate genes among the variants that showed strong but as yet less conclusive levels of significance for association with height in our meta-analysis of GWA studies and replication cohorts. A SNP 28 kb upstream of PRKG2 (rs1662845), which encodes the cGMP-dependent protein kinase II (cGKII), showed strong association with height in our meta-analysis of GWA scans (P = 5.7 × 10-5), and in the same direction in the European American height panel (P = 8.5 × 10-6) and the FUSION stage 2 sample (P = 0.001), but not in the FINRISK97 (P = 0.93, opposite direction) and PPP (P = 0.16, opposite direction) panels. a This locus is very strong candidate for a role in height variation. First, Prkg2-/- mice developed dwarfism that is caused by a severe defect in endochondral ossification at the growth plates29. Second, the naturally occurring Komeda miniature rat Ishikawa mutant, which has general longitudinal growth retardation, results from a deletion in the rat gene encoding cGKII30. Therefore, in rodents, it is clear that cGKII has a role in skeletal growth, acting as a molecular switch between chondrocyte proliferation and differentiation. We predict that larger replication studies will demonstrate that common genetic variation at the PRKG2 locus does contribute to height variation in humans, but it seems possible that there will also be heterogeneity among studies.
Several newly identified loci associated with height are located near genes with less immediately apparent connections to stature, including the G protein-coupled receptor gene GPR126, a locus that encompasses the thyroid hormone receptor interactor TRIP11 and the ataxin ATXN3 genes, a locus with the Huntingtin-interacting gene SH3GL3 and the glycoprotein metalloprotease gene ADAMTSL3 (the later often mutated in colon cancer31), a locus with gene CHCHD7, frequently fused to the PLAG1 oncogene in salivary gland adenomas32, and the epidermal retinal dehydrogenase 2 gene RDHE2. Because of LD (Supplementary Fig. 2), it is possible that the causal alleles at these loci are not located in these genes; fine-mapping in larger cohorts or in populations of different ancestry may be required to pinpoint the relevant gene and functional variant(s). Alternatively, these genes may themselves influence height, and further work will be needed to elucidate the relevant pathways and mechanisms.
We note that the accompanying manuscript by Weedon et al.18 identifies association with height for several of the loci reported in our study (ZBTB38, HMGA2, GDF5-UQCC, HHIP, SH3GL3-ADAMTSL3, CDK6), and reports, as we do, a suggestive association for a SNP at the FUBP3 locus (P = 7.5 × 10-7 in our study; P = 2.0 × 10-5 in Weedon et al.). FUBP3, a gene implicated in c-myc regulation, is therefore likely to represent an additional locus associated with height.
The variants associated with height that we validated do not have strong enough effects to generate detectable linkage signals33. Three of our loci lie under previously reported height linkage peaks8: ZBTB38, lod score 2.03; TRIP11-ATXN3, lod score 2.01; and CDK6, lod score 2.26. However, because 17.6% of the genome overlaps with a height linkage peak with lod score >2, the number of such co-localizations is not greater than expected by chance (3 observed versus 2.12 expected). It remains possible that some genes harbor both common and rare variants that influence height, so some overlaps may yet emerge between associated and linked loci that have a real genetic basis. Furthermore, regions of linkage may indicate the locations of rare variants or other types of genetic variation that are not well captured by our current association methods.
As expected, the estimated effect sizes in the GWA meta-analysis were generally larger than the effect sizes observed in replication samples, because of the well-known ‘winner’s curse’ phenomenon34. Perhaps less well appreciated is that the magnitude of the winner’s curse effect depends on the underlying distribution of effect sizes: the greater the number of variants with small effects, the more likely it is that one or more of these variants will approach genome-wide significance even in a study that is not well powered to detect these very modest effects35. Such variants will then prove difficult to convincingly replicate, unless very large replication cohorts are used. Thus, it is possible that even some of the initial associations that we failed to replicate will eventually be validated.
Given the modest effect sizes observed for the validated variants associated with height (Table 1; average = 0.4 cm per additional allele), it is not surprising that the quantile-quantile plots for the individual GWA studies are essentially indistinguishable from the null expectation (Fig. 1). Indeed, we calculate that a study of 3,000 unrelated individuals has 1% power to detect a variant (minor allele frequency 10%) that increases height by 0.4 cm at a statistical threshold of P = 1 × 10-5. In comparison, a study of 16,000 individuals has 72% power to identify the same variant (in fact, there is a slight loss in power when using meta-analytic methods to combine results). Our discovery of valid associations by combining individual studies with nearly null P-value distributions highlights the importance of using large datasets to find common variants with small effects. When we remove the 12 validated height variants (and nearby correlated SNPs) from the meta-analysis results, the number of low P values still exceeds the null expectation (Fig. 2a, filled squares). Furthermore, the 10,000 SNPs with the best P values also showed excess evidence of association in an independent meta-analysis18, even when all loci known to be associated with height were excluded (Fig. 2b). These results indicate that there are other associations with common alleles yet to be discovered, but that our meta-analysis is not sufficiently powered to identify these associations because the effect sizes are small.
Our results have several implications. First, they outline a role for multiple genes and biological pathways that were previously not known to regulate height, substantiating the ability of unbiased genetic approaches to yield new biological insights. The identification of these genes not only expands our knowledge of human growth but also promotes these genes as candidates for as yet unexplained syndromes of severe tall or short stature. Second, these findings convincingly confirm the polygenic nature of height, a classic complex trait, and demonstrate that, at least for this trait, increasingly large GWA studies can uncover increasing numbers of associated loci. Third, each variant makes only a small contribution to phenotypic variation (although determining the total contribution of each of the loci reported here requires much more comprehensive resequencing and genotyping); thus, either many hundreds of common variants influence complex traits such as height and/or other genetic contributors (for example, gene-gene or gene-environment interactions, rare variants with large effects, or uncaptured genomic features such as structural polymorphisms) will play a significant role. In particular, because the quality-control criteria used in the GWA studies analyzed here would have removed SNPs affected by copy number polymorphisms, we cannot conclude anything regarding the role of these variants on adult height. With the development of new platforms and improved analytical tools applicable to large cohorts, it is likely that the role of common structural variants on human complex traits such as adult height will soon be elucidated. Finally, if height is indeed a good model for other complex traits, these results suggest that large meta-analyses of GWA studies will provide insights not only into human growth but also into the underlying biological mechanisms of common disease.
METHODS
Description of genome-wide association study samples
The individuals analyzed by the Diabetes Genetics Initiative (DGI) have been described elsewhere4. In total, there were 1,464 type 2 diabetes cases and 1,467 matched controls of European ancestry from Finland and Sweden. The Finland-United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) GWA study included 1,161 Finnish type 2 diabetes (T2D) cases, 1,174 normal glucose tolerant (NGT) controls, and 122 offspring of case-control pairs13. Cases and controls were matched as previously described, taking into account age, sex and birth province within Finland. The KORA genome-wide association study samples were recruited from the KORA S3 survey, which is a population-based sample from the general population living in the region of Augsburg, Southern Germany. The 1,644 study participants had a German passport and were of European origin16. The Nurses’ Health Study (NHS) GWA scan included DNA from 2,286 registered nurses of European ancestry: 1,145 postmenopausal women with invasive breast cancer and 1,142 matched controls14. The Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO) GWA scan included 1,172 non-Hispanic prostate cancer cases of European ancestry and 1,105 matched controls15. The SardiNIA GWAS examined a total of 4,305 related individuals participating in a longitudinal study of aging-related quantitative traits in the Ogliastra region of Sardinia, Italy17. More details can be found in the Supplementary Methods online.
Description of follow-up samples
The European American (n = 2,189) sample is a tall-short study with subjects ranking in the 5-10th percentiles in adult height (short women, 152-155 cm; short men, 164-168 cm) and in the 90-95th percentiles in adult height (tall women, 170-175 cm; tall men, 185-191 cm)19. All individuals were self-described ‘white’ or ‘of European descent’. All subjects were born in the United States, and all of their grandparents were born in either the United States or Europe. Using the Genetic Power Calculator36 and assuming a purely additive genetic effect across the whole phenotypic distribution, we calculated that the study design of the European American tall-short panel provides 33.0, 79.0 and 98.6% power to detect variants that explain ≥0.1, 0.25 and 0.5% of the phenotypic variation in height, respectively (at an α-threshold of 0.01), and power was greater to meet our less stringent screening criteria of any odds ratio in the same direction than the effect observed in the meta-analysis. FINRISK 1997 is one of the population-based risk factor surveys carried out by the National Public Health Institute of Finland every five years37, and was approved by the Ethical Committee of the National Public Health Institute (decision number 38/96). The sample was drawn from the National Population Register for five geographical areas. The FUSION stage 2 study includes a series of cases and controls matched to take into account age, sex, and birth province within Finland13. The KORA S4 samples were recruited from Augsburg, Southern Germany, and do not overlap with the KORA S3 population. Prevalence, Prediction and Prevention of Diabetes (PPP) in the Botnia study is a population-based study started in 2004 to study the epidemiology of type 2 diabetes. More details can be found in the Supplementary Methods. These studies were approved by the appropriate ethical review boards.
Genotype imputation
Because the GWA scans used different genotyping platforms, we imputed genotypes for all polymorphic HapMap SNPs in each scan, using a hidden Markov model as implemented in MACH (Y. Li and G.R.A., unpublished data). This approach allowed us to evaluate association at the same SNPs in all scans. The imputation method combines genotype data from each sample with the HapMap CEU samples (July 2006 phased haplotype release) and then infers the unobserved genotypes probabilistically. The inference relies on the identification of stretches of haplotype shared between study samples and individuals in HapMap CEU reference panel. For each SNP in each individual, imputation results are summarized as an ‘allele dosage’ defined as the expected number of copies of the minor allele at that SNP (a fractional value between 0.0 and 2.0). As previously described, r2 between each imputed genotype and the true underlying genotype is estimated and serves as a quality-control metric (rsq_hat in Supplementary Table 7). We chose an estimated r2 >0.3 as a threshold to flag and discard low-quality imputed SNPs (ref. 13 and Y. Li and G.R.A., unpublished data).
Association analyses
For all studies, except for the European American height panel, we converted height to Z score, taking into account sex, age and disease status when appropriate. For the DGI, KORA S3, NHS, PLCO, FINRISK97, KORA S4 and PPP cohorts, we carried out association testing using a regression framework implemented in PLINK38 for genotyped markers, and in MACH2QTL (Y. Li and G.R.A., unpublished data), which takes into account dosage information (0.0-2.0), for imputed SNPs. For DGI, we used a genomic control method to correct for the presence of related individuals. Association testing in the FUSION and SardiNIA datasets was done for both genotyped and imputed SNPs using a method that allows for relatedness, estimating regression coefficients in the context of a variance components model39. Statistical analysis for the European American tall-short panel was done using a Cochran-Mantel-Haenszel (CMH) test38 stratified by the European region of origin of the grandparents.
Meta-analysis
Association results presented in this manuscript take into account the posterior probability on each imputed genotype. To combine results, we used a weighted Z-score method:
zw is the weighted Z score from which the meta-analytic two-tailed P value is calculated, zi is the Z score from study i (calculated as the cumulative normal probability density for the corresponding one-tailed P value, adjusted if needed by subtracting the P value from one when the direction of the effect is reversed), Ni is the sample size of study i and Ntot is the total sample size. In total, we combined association results at 2,260,683 autosomal SNPs in 15,821 individuals (DGI, n = 2,978; FUSION, n = 2,371; KORA, n = 1,644; NHS, n = 2,286; PLCO, n = 2,244; SardiNIA, n = 4,305). The I2 statistic40 and Cochran’s Q test were used to assess heterogeneity.
Genotyping and quality control
Genotyping of the initial GWA studies is described elsewhere4,11,13-15, except for KORA, which is described in the Supplementary Methods. Genotyping in the European American height panel and the replication panels FINRISK97, PPP, FUSION stage 2 and KORA S4 was done using the Sequenom MassARRAY iPLEX platform. From the meta-analysis, we selected 78 SNPs (three iPLEX pools; six SNPs failed) for genotyping in the European American tall-short panel (n = 2,189): SNPs were first ranked using their meta-analytic P values, and then binned using the LD pattern from the HapMap Phase II CEU population (SNPs with r2 >0.5 to a SNP with a lower P value were binned together with that SNP). When a locus had more than one high-ranking bin, we only genotyped the most significant SNP such that only one SNP per locus or gene was genotyped. Because of the specific design of this DNA panel (near-ends of the height distribution) but also because effect sizes on height are small (and were inflated by the winner’s curse in the meta-analysis), we promoted for genotyping in larger follow-up cohorts markers that had an odds ratio consistent with the direction of the effect observed in the meta-analysis (without considering the magnitude of the European American CMH P values). For the FINRISK97 panel, 29 SNPs were attempted and two failed. For the PPP panel, 23 SNPs were attempted and two failed. For the FUSION stage 2 panel, 27 SNPs were attempted and one failed. For the KORA S4 panel, five SNPs were attempted and one failed. For all passing SNPs, the genotyping success rate was >96% and the consensus error rate, estimated from replicates, was <0.1%.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of our laboratories for helpful discussion, and gratefully acknowledge all of the participants in the studies. Contributing members for the DGI, FUSION, KORA and SardiNIA GWA scans are listed in the Supplementary Note. The authors acknowledge C. Chen of Bioinformed Consulting Services Inc. for expert programming, and L. Qi for his assistance. The authors thank C. Berg and P. Prorok, Division of Cancer Prevention, National Cancer Institute, the Screening Center investigators and staff of the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, T. Riley and staff, Information Management Services, Inc., and B. O’Brien and staff, Westat, Inc. KORA gratefully acknowledges the contribution of T. Meitinger and all other members of the GSF genotyping staff in generating the SNP dataset. We thank the Mayor and the administration in Lanusei for providing and furnishing the clinic site; and the mayors of Ilbono, Arzana and Elini, the head of the local Public Health Unit ASL4. Support for this work was provided by the following: US National Institutes of Health grants 5P01CA087969 and CA49449 (S.E. Hankinson), 5UO1CA098233 (D.J.H.), DK62370 (M.B.), DK72193 (K.L.M.), HG02651 and HL084729 (G.R.A.); Novartis Institutes for BioMedical Research (D. Altshuler); March of Dimes grant 6-FY04-61 (J.N.H.); EU Projects GenomEUtwin grant QLG2-CT-2002-01254 and the Center of Excellence in Complex Disease Genetics of the Academy of Finland (L.P.); the Sigrid Juselius Foundation (L.C.G., V.S. and PPP); the Finnish Diabetes Research Foundation and the Folkhaälsan Research Foundation and Clinical Research Institute HUCH (L.C.G.); this research was supported (in part) by the intramural Research Program of the NIH, National Institute on Aging; the PLCO research was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS; KORA/MONICA Augsburg studies were financed by the GSF-National Research Center for Environment and Health, Munich/Neuherberg, Germany and supported by grants from the German Federal Ministry of Education and Research (BMBF); part of this work by KORA was supported by the German National Genome Research Network (NGFN), the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ, and a subcontract of the 5 R01 DK 075787 by the NIH/NIDDK to the GSF-National Research Center for Environment and Health (to J.N.H.).
Accession numbers. Genbank Entrez Gene: data for validated loci associated with height are available with accession codes 253461 (ZBTB38), 8091 (HMGA2), 57211 (GPR126) 3007 (HIST1H1D), 8200 (GDF5), 55245 (UQCC), 64399 (HHIP), 9321 (TRIP11), 4287 (ATXN3), 389421 (LIN28B), 84444 (DOT1L), 6457 (SH3GL3), 57188 (ADAMTSL3), 79145 (CHCHD7), 195814 (RDHE2), 1021 (CDK6).
Footnotes
URLs. DGI GWA study, http://www.broad.mit.edu/diabetes/; Markov Chain Haplotyping Package, http://www.sph.umich.edu/csg/abecasis/MaCH.
References
- 1.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 2.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 5.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- 8.Perola M, et al. Combined genome scans for body stature in 6,602 European twins: evidence for common Caucasian loci. PLoS Genet. 2007;3:e97. doi: 10.1371/journal.pgen.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathias RA, et al. Comparison of year-of-exam- and age-matched estimates of heritability in the Framingham Heart Study data. BMC Genet. 2003;4(Suppl 1):S36. doi: 10.1186/1471-2156-4-S1-S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunnell D, et al. Height, leg length, and cancer risk: a systematic review. Epidemiol. Rev. 2001;23:313–342. doi: 10.1093/oxfordjournals.epirev.a000809. [DOI] [PubMed] [Google Scholar]
- 11.Sanna S, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weedon MN, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 16.Wichmann HE, Gieger C, Illig T. KORA-gen-resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl. 1):S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 17.Pilia G, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008 Apr 6; doi: 10.1038/ng.121. advance online publication, doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell CD, et al. Demonstrating stratification in a European-American population. Nat. Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 20.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 21.Price AL, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 23.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 27.Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 28.Guo Y, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Pfeifer A, et al. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science. 1996;274:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- 30.Chikuda H, et al. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004;18:2418–2429. doi: 10.1101/gad.1224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 32.Asp J, Persson F, Kost-Alimova M, Stenman G. CHCHD7-PLAG1 and TCEA1-PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromosom. Cancer. 2006;45:820–828. doi: 10.1002/gcc.20346. [DOI] [PubMed] [Google Scholar]
- 33.Hirschhorn JN, et al. Genomewide linkage analysis of stature in multiple populations reveals several regions with evidence of linkage to adult height. Am. J. Hum. Genet. 2001;69:106–116. doi: 10.1086/321287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 35.Zollner S, Pritchard JK. Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am. J. Hum. Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 37.Vartiainen E, et al. Cardiovascular risk factor changes in Finland, 1972-1997. Int. J. Epidemiol. 2000;29:49–56. doi: 10.1093/ije/29.1.49. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br. Med. J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.