Abstract
Regulation of β-catenin stability is essential for Wnt signal transduction during development and tumorigenesis. It is well known that serine-phosphorylation of β-catenin by the Axin–glycogen synthase kinase (GSK)–3β complex targets β-catenin for ubiquitination–degradation, and mutations at critical phosphoserine residues stabilize β-catenin and cause human cancers. How β-catenin phosphorylation results in its degradation is undefined. Here we show that phosphorylated β-catenin is specifically recognized by β-Trcp, an F-box/WD40-repeat protein that also associates with Skp1, an essential component of the ubiquitination apparatus. β-catenin harboring mutations at the critical phosphoserine residues escapes recognition by β-Trcp, thus providing a molecular explanation for why these mutations cause β-catenin accumulation that leads to cancer. Inhibition of endogenous β-Trcp function by a dominant negative mutant stabilizes β-catenin, activates Wnt/β-catenin signaling, and induces axis formation in Xenopus embryos. Therefore, β-Trcp plays a central role in recruiting phosphorylated β-catenin for degradation and in dorsoventral patterning of the Xenopus embryo.
Wnt genes encode secreted signaling molecules that play important roles in development and tumorigenesis (1, 2). Initially identified as oncogenes that cause murine mammary tumors, the wnt family of genes are now recognized as essential developmental regulators in all animals, from nematodes and Drosophila to mammals (1). Deregulation of Wnt signaling is responsible for several human malignancies (3, 4).
β-catenin has emerged as a central component in the Wnt-1 signal transduction pathway, which is required for diverse biological processes, such as segment polarity determination during Drosophila development, dorsoventral patterning in early Xenopus embryogenesis, and the homeostasis of human colorectal tissues (1–4). Stabilization of the β-catenin protein plays a pivotal role in this Wnt signaling pathway, as β-catenin is an obligatory transcriptional coactivator for the T cell factor (TCF)/lymphocyte enhancer factor family of transcription factors in the activation of Wnt-responsive genes (5–7). Deregulated stabilization of β-catenin, caused by either loss-of-function mutations in the human tumor suppresser protein APC or gain-of-function mutations of β-catenin, is associated with pathogenesis of human colorectal cancer, melanoma, and possibly other neoplasia (3, 4, 8–13). In addition, loss of β-catenin stability recently has been associated with presenilin-1 mutations in the pathogenesis of Alzheimer’s disease (14). How β-catenin stability is regulated by the Wnt signal transduction pathway is just beginning to be defined. In the absence of Wnt stimulation, β-catenin forms a multiprotein complex with several known Wnt signaling components, including APC, developmental regulator/scaffolding protein Axin or related Axil/conductin, and serine/threonine kinase GSK-3β (15–22). In this complex brought together by Axin, GSK-3β is suggested to phosphorylate β-catenin at multiple serine or threonine residues at the amino-terminal region of β-catenin (23, 24), thereby marking β-catenin for degradation by the ubiquitination–proteosome pathway (25, 26). Wnt signaling down-regulates glycogen synthase kinase (GSK)–3β activity, thereby resulting in the inhibition of β-catenin phosphorylation and increased β-catenin stability (24, 27–29). The significance of β-catenin phosphorylation in its stability is most clearly manifested in several types of human cancers (8–13). In these tumors, single amino acid mutations occur at one of the four critical phosphoserine or threonine residues (serine-33, -37, and -45 and threonine-41) in the consensus GSK-3β phosphorylation site at the β-catenin amino-terminal region. These mutations result in deregulated accumulation of β-catenin and, thereby, increased signaling through the TCF/β-catenin transcriptional complex, contributing to tumorigenesis (8–10).
In early Xenopus embryos, activation of the Wnt/β-catenin signaling pathway after fertilization results in the accumulation of β-catenin protein on the dorsal side, leading to the establishment of the dorsoventral polarity (2, 30). Ectopic activation of Wnt signaling on the ventral side by manipulating Wnt signaling components, such as by interference with GSK-3β or Axin function or by overexpression of β-catenin, causes dorsal axis duplication; conversely, depletion of β-catenin protein inhibits endogenous dorsal axis formation (2, 30). Despite the significance of regulating β-catenin degradation in Wnt signal transduction during development and tumor formation, the mechanism by which β-catenin phosphorylation leads to its degradation is unknown.
β-catenin is degraded via the ubiquitination–proteosome pathway (25, 26). Protein ubiquitination involves at least three enzymatic steps, which are catalyzed by the ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-ligase (E3) (31, 32). Substrate specificity is largely controlled by the ubiquitin-ligase, which can be either a single polypeptide or, in many cases, a protein complex (31, 32). Regulated protein ubiquitination–degradation process has been best characterized in yeast cell cycle control (32). These studies have led to the identification of a family of the so-called F-box proteins as the subunit that determines the substrate specificity of a ubiquitin–ligase complex (33–35). These yeast F-box proteins have been shown to bind to, via the F-box motif, the Skp1 protein, which in turn complexes with other components of the ubiquitination apparatus, including a ubiquitin-conjugating enzyme (34, 35). These F-box proteins also contain a WD40-repeat domain or a leucine-rich repeat domain that appears to be responsible for specific recognition of phosphorylated substrates (34, 35), thereby recruiting phosphorylated substrates to Skp1 and the ubiquitination machinery. Recently, slimb, which encodes an F-box/WD40-repeat containing protein, was identified in Drosophila as a negative regulator of both Wingless (Drosophila Wnt-1) and Hedgehog signaling (36, 37). How Slimb negatively regulates Wingless signaling, directly or indirectly (37), is unknown. Here we report that β-Trcp, the vertebrate homolog of slimb (38), plays a central role in coupling β-catenin phosphorylation–degradation and, in Xenopus, dorsal axis formation.
MATERIALS AND METHODS
Plasmids.
A detailed description of all of the plasmids constructed for this work is posted as supplementary material on the PNAS web site (www.pnas.org).
In Vitro Transcription and Translation.
Messenger RNAs were transcribed by using a MEGAscript Kit (Ambion, Austin, TX) according to kit instruction. 35S-labeled β-Trcp protein and β-Trcp mutant proteins were synthesized by using the TNT system (Promega).
Immunoprecipitation and Western Blotting.
In vitro transcribed mRNAs were injected into the animal pole of Xenopus laevis embryos at the two-cell stage. Embryo extracts were made by homogenizing stage 9 embryos in Nonidet P-40 buffer (140 mM NaCl/1% Nonidet P-40/5 mM EDTA/10 mM Tris, pH 7.5) with protease inhibitors, using 0.4 ml of buffer for 10 embryos. The extracts were immunoprecipitated with appropriate antibodies (2–4 μg) plus protein-G beads at 4°C overnight. Precipitates were washed at least three times with the same Nonidet P-40 buffer (1 ml) and were analyzed by Western blotting. Immobilon-P Membrane (Millipore) were incubated in blocking buffer (5% nonfat dry milk in PBS with 0.05% Tween-20) for 1 hour at room temperature and then were incubated with primary antibodies diluted in 1% BSA in the PBS/Tween-20 buffer for 1 hour, followed by incubation with horseradish peroxide-conjugated secondary antibodies diluted at 1: 10,000 in 1% BSA in the PBS/Tween-20 buffer for 1 hour. Protein detection was performed by using the ECL system (Amersham Pharmacia). The following antibodies were used for Western blotting (dilutions in parenthesis): anti-myc (1:1000), from Upstate Biotechnology, Lake Placid, NY (06549); or 9E10 (1:1000), from American Type Culture Collection; anti- β-catenin (1:4000), from Sigma (C2206); anti-Flag (M2) (1:1000), from Sigma (F3165); anti-Skp1 (1:5000), from Transduction Laboratories, Lexington, KY (P46020).
Glutathione S-Transferase (GST) Fusion Proteins and Phosphorylation.
Recombinant GST-β-catenin fusion proteins were purified by using glutathione agarose beads (Sigma) from bacterial extracts and were washed at least three times with a kinase reaction buffer for GSK-3β (25 mM Hepes, pH7.5/10 mM MgCl2/1 mM EDTA/0.1 mM DTT). Fusion proteins (10 μg) bound to beads were resuspended in 20 μl of 2× Kinase buffer, 10 μl of 0.8 mM ATP, 1 μl of purified recombinant GSK-3β protein (2 μg), and 9 μl of H2O, and incubation was done at 30°C for 1 hour. For visualization of phosphorylation, 2 μl of [γ-32P]ATP (5,000 Ci/mmol) plus 8 μl of H2O replaced regular ATP.
GST Pull-Down Assay.
After phosphorylation by GSK-3β plus nonradiolabeled ATP, GST-fusion proteins bound to beads were washed three times with cold PBS buffer containing 1% Triton-100 to remove GSK-3β and ATP and were incubated at 4°C with 35S-labeled β-Trcp proteins in the same buffer for 1 hour. The incubation mix was washed three times with the same buffer, and the resulting precipitates were subjected to SDS/PAGE. The gel was fixed, dried, and exposed to x-ray film to visualize 35S-labeled β-Trcp proteins.
Embryo Manipulation and in Situ Hybridization.
X. laevis embryos were raised in 0.1× MMR buffer (0.1 M NaCl/2.0 mM KCl/1 mM MgSO4/2 mM CaCl2/5 mM Hepes, pH 7.8/0.1 mM EDTA). Injections were done in 3% Ficoll and 0.1× MMR buffer. For the axis duplication assay and examination of Xnr3 expression, RNAs were injected into the ventral equatorial region at four-cell stage, with two blastomeres injected with equal amount of RNA. Whole-mount in situ hybridization was performed according to Harland (47).
RESULTS
β-Trcp Forms a Complex with β-Catenin and Axin.
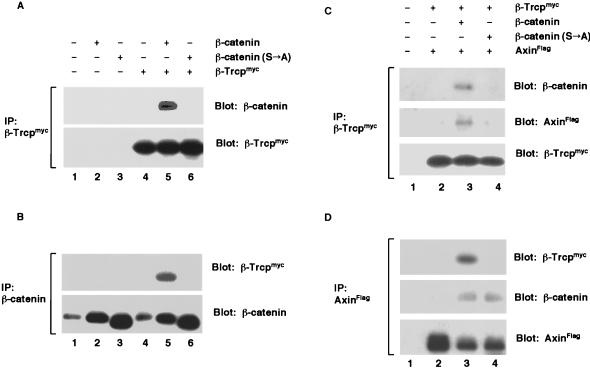
β-Trcp is a Xenopus protein that shares 80% amino acid sequence identity to the slimb gene product (36–39). Intriguingly, two major maternal β-Trcp transcripts in Xenopus oocytes are preferentially localized in the vegetal cortex that is known to contain the maternal dorsal determinant(s) (39). To investigate a potential role for β-Trcp in regulating β-catenin stability, we first examined whether the two proteins associate in vivo. In coimmunoprecipitation assays, we found that β-Trcp associated with β-catenin (Fig. 1 A and B). Importantly, the association between β-Trcp and β-catenin depended on the four serine/threonine residues at the amino terminus of β-catenin, as β-catenin (S→A), which is a mutant β-catenin with alanine substitutions of these serine/threonine residues (see Fig. 4A), completely lost the ability to associate with β-Trcp (Fig. 1 A and B).
Figure 1.
β-Trcp forms a complex with β-catenin and Axin in vivo. (A and B) Association between β-Trcp and β-catenin. (A) Immunoprecipitation of β-Trcpmyc with an anti-myc antibody. Embryo extracts were from stage 9 embryos expressing β-catenin, β-catenin (S→A), or β-Trcpmyc alone or in combination. The precipitates were examined for β-catenin (Upper) and for β-Trcpmyc (Lower). A longer exposure revealed that β-Trcpmyc also coprecipitated endogenous β-catenin (data not shown). (B) Immunoprecipitation of β-catenin using the same embryo extracts as in A. The precipitates were examined for β-Trcpmyc (Upper) and for β-catenin (Lower). Endogenous β-catenin is seen in lanes 1 and 4. RNA injected per embryo: 1 ng each for β-catenin, β-catenin (S→A), and β-Trcpmyc. (C and D) A complex formation between β-Trcp and Axin mediated by β-catenin. (C) Immunoprecipitation of β-Trcpmyc. The extracts were from stage 9 embryos expressing β-Trcpmyc plus AxinFlag in the presence or absence of β-catenin or β-catenin (S→A). The precipitates were examined for β-catenin (Top), AxinFlag (Middle), and β-Trcpmyc (Bottom). (D) Immunoprecipitation of AxinFlag using the same extracts as in C. The precipitates were examined for β-Trcpmyc (Top), β-catenin (Middle), and AxinFlag (Bottom). RNA injected per embryo: same as in A and B except 2 ng for AxinFlag.
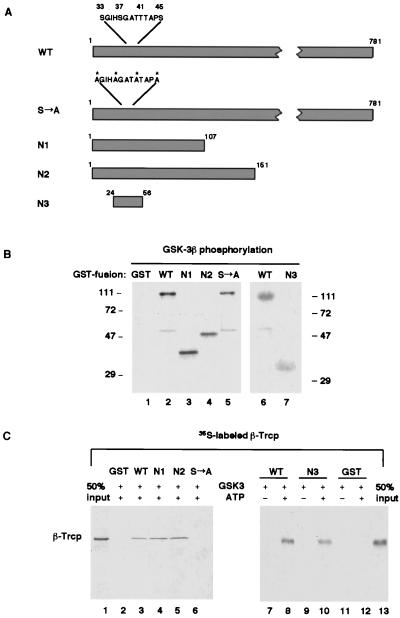
Figure 4.
Phosphorylation of the amino-terminal region of β-catenin is necessary and sufficient for recognition by β-Trcp in vitro. (A) Schematic diagram of the wild-type and mutant derivatives of β-catenin. The four critical serine/threonine residues (S33, S37, T41, and S45), alanine substitutions of these residues in the S→A mutant, and surrounding residues are highlighted. Note that the Arm-repeat region that is required for Axin or TCF binding starts from residue 131. (B) Phosphorylation of β-catenin and its mutant derivatives (as GST-fusion proteins) by purified recombinant GSK-3β. Note that GST was not a substrate for GSK-3β. (C) On phosphorylation, N1, N2, and N3, but not β-catenin (S→A) were effectively recognized by β-Trcp. Lanes 1 and 13 represent 50% input of 35S-labeled β-Trcp in each GST-pull-down assay.
We also observed β-Trcp in a complex with Axin (Fig. 1 C and D). This association was mediated by the wild-type β-catenin: It occurred only in the presence of β-catenin but not in the absence of β-catenin, nor in the presence of β-catenin (S→A) mutant (Fig. 1 C and D). The failure of β-catenin (S→A) to mediate β-Trcp-Axin complex formation was caused by the inability of β-catenin (S→A) to associate with β-Trcp because β-catenin (S→A) nonetheless interacted effectively with Axin (Fig. 1D). Therefore, β-Trcp is associated, via β-catenin, with the Axin/β-catenin complex.
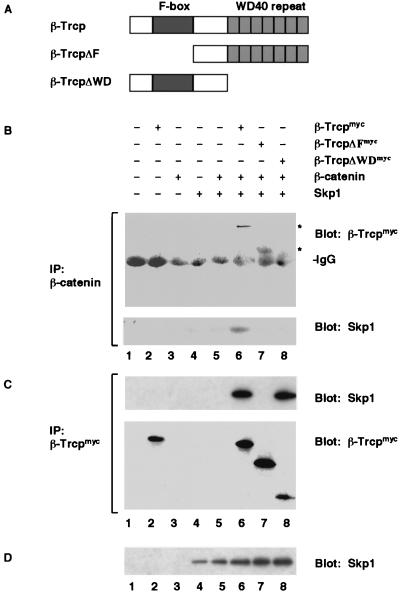
β-Trcp Binds β-Catenin via the WD40-Repeat and Binds Skp1 via the F-Box Motif.
β-Trcp protein has two recognizable motifs: the central F-box motif and the carboxyl-terminal WD40-repeat domain (Fig. 2A). We found that the WD40-repeat domain of β-Trcp was responsible for β-catenin recognition: β-catenin did not bind to β-TrcpΔWD, which had the WD40-repeat domain deleted, but bound to β-TrcpΔF, which harbored the F-box deletion (Fig. 2B). We also found that, similar to the yeast F-box proteins (33–35), β-Trcp bound to the vertebrate Skp1 protein via its F-box motif. Both the wild-type β-Trcp and β-TrcpΔWD, but not β-TrcpΔF, coprecipitated Skp1 (Fig. 2C), consistent with a similar observation made in a yeast two-hybrid assay (40). Thus, β-Trcp binds, via two distinct domains, both β-catenin and Skp1. Importantly, we found that Skp1 was coprecipitated with β-catenin only in the presence of wild-type β-Trcp but not in the absence of β-Trcp or in the presence of β-TrcpΔF or β-TrcpΔWD (Fig. 2B), demonstrating the formation of a β-catenin/β-Trcp/Skp1 ternary complex that was assembled by β-Trcp. Taken together, these results suggest that the Axin/β-catenin complex associates with β-Trcp that in turn binds to the Skp1 ubiquitination complex.
Figure 2.
β-Trcp forms a complex with both β-catenin and Skp1. (A) Schematic diagram of wild-type β-Trcp, β-TrcpΔF, and β-TrcpΔWD. (B) Immunoprecipitation of β-catenin. The precipitates were examined for β-Trcp (Upper) and Skp1 (Lower). A longer exposure revealed that endogenous β-catenin also precipitated β-Trcpmyc (data not shown). (C) Immunoprecipitation of β-Trcpmyc using the same embryo extracts as in B. The precipitates were examined for Skp1 (Upper) and β-Trcp (Lower). (D) Immunoblot of Skp1 in the same embryo extracts as in B. RNA injected per embryo: 1 ng each for β-Trcpmyc and for deletion mutants β-catenin and Skp1.
β-Trcp Specifically Recognizes β-Catenin Phosphorylated by GSK-3β in Vitro.
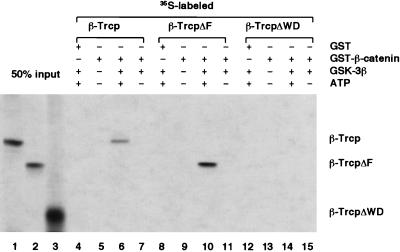
β-catenin (S→A) did not bind to β-Trcp in vivo, suggesting that β-catenin phosphorylation may be required for its recognition by β-Trcp. We purified the GST-β-catenin fusion protein and recombinant GSK-3β protein and examined the ability of β-Trcp to distinguish unphosphorylated β-catenin versus β-catenin phosphorylated by GSK-3β. Strikingly, in GST-pull-down assays, 35S-labeled β-Trcp specifically bound to GST-β-catenin phosphorylated by GSK-3β but not to GST-β-catenin or to GST-β-catenin incubated with GSK-3β in the absence of ATP (Fig. 3). GSK-3β and ATP were needed solely for phosphorylation of β-catenin, as β-Trcp showed identical binding to phosphorylated GST-β-catenin regardless of the presence of GSK-3β and ATP during binding assays (Fig. 3; data not shown). Consistent with in vivo immunoprecipitation results, the WD40-repeat domain of β-Trcp mediated binding to phosphorylated β-catenin because β-TrcpΔWD exhibited no binding to phosphorylated β-catenin whereas β-TrcpΔF exhibited strong binding (Fig. 3). Thus, β-Trcp specifically recognizes phosphorylated β-catenin via the WD40-repeat domain.
Figure 3.
β-Trcp WD40-repeat domain recognizes GSK-3β-phosphorylated β-catenin in vitro. In this GST-pull-down assay, purified GST or GST-β-catenin was examined for its ability to bind 35S-labeled β-Trcp. GST-β-catenin on beads was either untreated or were treated with purified GSK-3β in the presence or absence of ATP before incubation with β-Trcp. Note that, in this experiment, GSK-3β and ATP were removed from phosphorylated GST-β-catenin by extensively washing the GST-β-catenin on beads after phosphorylation and before incubation with β-Trcp. Lanes 1–3 represent 50% of the input 35S-labeled β-Trcp or β-Trcp mutants used in each GST-pull-down assay.
Phosphorylation of the β-Catenin Amino-Terminal Region Is Necessary and Sufficient for Recognition by β-Trcp in Vitro.
To directly examine the role of the four amino-terminal serine/threonine residues of β-catenin in β-Trcp recognition of β-catenin, GST-β-catenin (S→A) was incubated with GSK-3β and ATP. As was noticed previously (24), GST-β-catenin (S→A) could be phosphorylated by GSK-3β in vitro (Fig. 4B). However, phosphorylated GST-β-catenin (S→A) did not lead to significant binding of β-Trcp (Fig. 4C), indicating that phosphorylation of the four amino-terminal serine/threonine residues played a central role in β-Trcp recognition. Because mutations at these four serine/threonine residues are frequently detected in various human tumors that exhibit high levels of β-catenin protein (8–13), this result suggested that these oncogenic mutations cause β-catenin accumulation by preventing β-catenin from being recognized by β-Trcp and, thereby, from degradation.
To test whether the amino-terminal phosphorylation is sufficient for binding by β-Trcp, β-catenin deletion mutants that retain the amino-terminal region were purified as GST fusion proteins (Fig. 4A). These mutants, referred to as N1 (residue 1 to 107), N2 (residue 1 to 151), and N3 (residue 24 to 56, thus spanning the four serine/threonine positions) (Fig. 4A), were strongly phosphorylated by GSK-3β (Fig. 4B) and were recognized effectively by β-Trcp in a phosphorylation-dependent manner (Fig. 4C). Therefore, the 33-aa region in N3 spanning the four serine/threonine residues is necessary and sufficient, on phosphorylation, for recognition by β-Trcp.
Inhibition of β-Trcp Function Causes Activation of Wnt Signaling and Induces Axis Duplication in Xenopus Embryos.
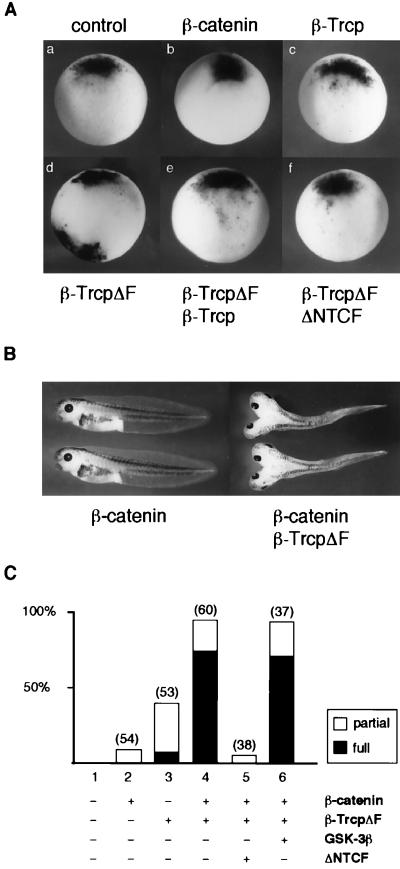
In the early Xenopus embryo, accumulation of β-catenin specifies dorsal development (2, 30). Experimental manipulations to stabilize β-catenin ventrally cause dorsal axis duplication (2, 30). Because β-TrcpΔF binds to phosphorylated β-catenin but does not bind to Skp1 that connects with the ubiquitination machinery, we reasoned that overexpression of β-TrcpΔF in the Xenopus embryo may interfere with the endogenous β-Trcp function, thereby stabilizing β-catenin. We first examined, in β-TrcpΔF-injected embryos, the expression of Xnr3 gene that is directly activated by the TCF/β-catenin transcriptional complex (41). Ventral injection of β-TrcpΔF RNA strongly induced ectopic Xnr3 expression (Fig. 5A), indicating activation of the Wnt signal transduction pathway. The ectopic Xnr3 induction was inhibited by coinjected wild-type β-Trcp RNA (Fig. 5A), suggesting that β-TrcpΔF specifically interfered with the endogenous β-Trcp function. We further examined whether Xnr3 induction by β-TrcpΔF relies on TCF/β-catenin association. Indeed, the ability of β-TrcpΔF to activate Xnr3 expression was completely blocked by ΔNTCF (Fig. 5A), a dominant-negative XTCF-3 mutant that is known to block TCF/β-catenin-dependent signaling (6).
Figure 5.
Inhibition of endogenous β-Trcp function by β-TrcpΔF induces ectopic Xnr3 expression and dorsal axis duplication. (A) Ventral injection of β-TrcpΔF RNA induced Xnr3 expression as examined by whole-mount in situ hybridization. All panels shown were ventral injection except in the control panel (a) (no injection). RNA amount injected per embryo: 1 ng for β-Trcp or β-TrcpΔF each, 100 pg for β-catenin, and 500 pg for ΔNTCF. The numbers of embryos with ectopic Xnr3 expression were 0 of 10 (number of embryos examined) in a; 0 of 8 in b; 10 of 10 for β-catenin (at 500 pg RNA per embryo; data not shown); 0 of 10 in c; 7 of 10 in d; 8 of 8 for β-TrcpΔF (at 4 ng RNA per embryo; data not shown); 0 of 10 in e; and 0 of 10 in f. (B and C) Axis duplication by β-TrcpΔF or β-catenin plus β-TrcpΔF. All shown were ventral injection except the control with no injection. Partial, partial axis duplication, defined as lacking eyes and other anterior structures in the duplicated axis. Full, complete axis duplication, defined as having eyes and other anterior structures in the duplicated axis. Number of injected embryos are indicated above each bar. RNA injected per embryo: β-catenin, 100 pg; β-TrcpΔF, 1 ng; GSK-3β, 1 ng; ΔNTCF, 500 pg.
TrcpΔF RNA injected ventrally also induced axis duplication and strongly synergized with β-catenin in axis induction (Fig. 5 B and C). Thus, ventral injection of a low level of β-catenin RNA alone did not affect normal development in most of the injected embryos (Fig. 5 B and C) and did not induce Xnr3 expression (Fig. 5A). In contrast, coinjection of β-catenin RNA together with β-TrcpΔF RNA led to a high percentage (98%) of axis duplication, including complete axis duplication (75%) (Fig. 5 B and C). This axis duplication was almost completely inhibited by coinjected ΔNTCF RNA but was not significantly affected by coinjected GSK-3β RNA (Fig. 5C). These results were consistent with the notion that β-TrcpΔF induces axis duplication via β-catenin stabilization, thus functioning epistatically downstream of GSK-3β and upstream of TCF.
Inhibition of β-Trcp Function Stabilizes β-Catenin in Vivo.
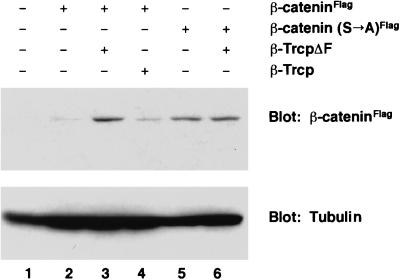
To directly determine whether inhibition of endogenous β-Trcp function stabilizes β-catenin, we compared the accumulation of wild-type β-catenin protein and the β-catenin (S→A) mutant in β-TrcpΔF-injected embryos. When the same amount of RNA for β-catenin or β-catenin (S→A) was expressed alone, only a low level of wild-type β-catenin protein was detected whereas a significantly higher level of β-catenin (S→A) was found (Fig. 6). However, after coexpression with β-TrcpΔF, β-catenin accumulated to a similar level to that of β-catenin (S→A) (Fig. 6). β-TrcpΔF did not increase the protein level of β-catenin (S→A) (Fig. 6) as expected because β-catenin (S→A) was not recognized by β-Trcp and was thus resistant to degradation. In addition, overexpression of β-Trcp did not decrease the basal level of β-catenin (Fig. 6), implying that the endogenous β-Trcp level is not rate-limiting. This is consistent with our observation that overexpression of β-Trcp dorsally does not perturb endogenous dorsal axis formation (data not shown). These results indicate that β-Trcp is involved in β-catenin stability in vivo.
Figure 6.
Inhibition of endogenous β-Trcp function by β-TrcpΔF stabilizes β-catenin in vivo. Accumulation of β-cateninFlag or β-catenin (S→A)Flag protein was examined in stage 9 embryo extracts by immunoblotting with an anti-Flag antibody. Immunoblotting of tubulin was used as a protein loading control (Lower). This experiment were performed twice with similar results. RNA injected per embryo: 50 pg for β-cateninFlag or β-catenin (S→A)Flag; 4 ng for β-Trcp or β-TrcpΔF.
DISCUSSION
We presented evidence that β-Trcp specifically recognizes β-catenin that has been phosphorylated by GSK-3β and recruits phosphorylated β-catenin into the Skp1 complex, thereby coupling β-catenin phosphorylation to the ubiquitination–proteosome pathway. Mutations at the four critical amino-terminal serine/threonine residues of β-catenin prevent β-catenin recognition by β-Trcp, thus providing insight into the mechanism by which these mutations cause β-catenin accumulation that leads to cancers. The WD40-repeat domain of β-Trcp and the amino-terminal region of β-catenin containing the phosphoserine/threonine residues are necessary and sufficient for the specific interaction between β-Trcp and β-catenin.
We also showed that inhibition of endogenous β-Trcp function in the Xenopus embryo stabilizes β-catenin, resulting in the activation of Wnt signaling and dorsal axis duplication. These results provide compelling evidence that β-Trcp regulates β-catenin stability in vivo and that β-Trcp plays an essential role in dorsoventral patterning during Xenopus embryogenesis. It is intriguing that two forms of maternal β-Trcp transcripts are specifically localized in the vegetal cortex of the Xenopus oocyte (39), which is known to contain maternal determinant(s) of dorso-ventral patterning (30). The function of β-Trcp and the localization of β-Trcp transcripts make it an excellent candidate for such a maternal determinant.
β-Trcp is the first vertebrate member of a growing family of F-box proteins (33) that function as specificity determination units of ubiquitin–ligase complexes (34, 35). Two features of the β-Trcp/β-catenin association deserve discussion. First, β-Trcp only recognizes β-catenin that has been phosphorylated by GSK-3β, thus permitting strict regulation of β-catenin degradation by phosphorylation. Second, β-Trcp is found associated with the β-catenin-Axin complex in which GSK-3β phosphorylates β-catenin. This suggests that β-catenin phosphorylation is likely to be followed immediately by β-Trcp recognition, thereby promoting rapid ubiquitination and degradation of phosphorylated β-catenin.
Our results suggest the following working model for Wnt regulation of β-catenin stability. Without Wnt signaling, β-catenin associates with the Axin/APC/GSK-3β complex in which GSK-3β phosphorylates β-catenin. This phosphorylation provides a specific docking site for the WD40-repeat domain of β-Trcp. β-Trcp, via its F-box motif, also complexes with the Skp1 protein of the ubiquitination apparatus, thus targeting phosphorylated β-catenin for ubiquitination–degradation. In the presence of Wnt signaling, GSK-3β kinase activity and thus β-catenin phosphorylation are inhibited. Because unphosphorylated β-catenin is not recognized by β-Trcp, β-catenin accumulation ensues, leading to the formation of the TCF/β-catenin transcriptional complex that activates Wnt responsive genes. In tumor cells, mutations at the critical phosphorylation sites in β-catenin prevent mutant β-catenin from being recognized by β-Trcp, resulting in accumulation of β-catenin and, ultimately, tumorigenesis. Thus, β-Trcp is an excellent candidate for a tumor suppressor protein. β-Trcp represents a central link between the Wnt signal transduction pathway and the ubiquitination–degradation machinery and plays a pivotal role in β-catenin-dependent signaling in embryonic development and in human carcinogenesis.
While this manuscript was in preparation, similar findings regarding β-Trcp function in Xenopus dorsal axis formation (42, 43) and in the degradation of β-catenin (44) and of IκB (44–46) were reported.
Supplementary Material
Acknowledgments
We thank R. Harland for Xnr3 plasmid, M. Castanon for β-Trcp cDNA, F. Wang and K. P. Lu for purified GSK-3β, O. Destree for TCF plasmids, and J. Green and M. Greenberg for comments on the manuscript. X.H. acknowledges the support from Children’s Hospital Division of Neuroscience start-up fund, the Harvard Milton Fund and Hearst Fund, Johnson and Johnson Focused Giving Program Award, a Career Development Award from the U.S. Army Breast Cancer Research Program (DAMD17-98-1-8047), a National Institutes of Health grant (RO1GM 57603), and a National Institutes of Health Mental Retardation Research Center grant. X.H. is a Pew Scholar in Biomedical Sciences.
ABBREVIATION
- GST
glutathione S-transferase
- GSK
glycogen synthase kinase
- TCF
T cell factor
References
- 1.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 2.Moon R T, Brown J D, Torres M. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Peifer M. Science. 1997;275:1752–1754. doi: 10.1126/science.275.5307.1752. [DOI] [PubMed] [Google Scholar]
- 5.Behrens J, Von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 6.Molenaar M, Van de Wetering M, Oosterwegel M, Petersonmaduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 7.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 8.Korinek V, Barker N, Morin P J, vanWichen D, deWeger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 9.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 10.Rubinfeld B, Robbins P, El Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 11.Zurawel R H, Chiappa S A, Allen C, Raffel C. Cancer Res. 1998;58:896–899. [PubMed] [Google Scholar]
- 12.Voeller H J, Truica C I, Gelmann E P. Cancer Res. 1998;58:2520–2523. [PubMed] [Google Scholar]
- 13.Miyoshi Y, Iwao K, Nagasawa Y, Aihara T, Sasaki Y, Imaoka S, Murata M, Shimano T, Nakamura Y. Cancer Res. 1998;58:2524–2527. [PubMed] [Google Scholar]
- 14.Zhang Z, Hartmann H, Do V M, Abramowski D, Sturchler-Pierrat C, Staufenbiel M, Sommer B, van de Wetering M, Clevers H, Saftig P, et al. Nature (London) 1998;395:698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]
- 15.Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek T J, Perry W L, Lee J J, Tilghman S M, Gumbiner B M, Costantini F. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;5:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto H, Kishida S, Uochi T, Ikeda S, Koyama S, Asashima M, Kikuchi A. Mol Cell Biol. 1998;18:2867–2875. doi: 10.1128/mcb.18.5.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behrens J, Jerchow B, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 20.Sakanaka C, Weiss J B, Williams L T. Proc Natl Acad Sci USA. 1998;95:3020–3023. doi: 10.1073/pnas.95.6.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh K, Krupnik V E, Sokol S Y. Curr Biol. 1998;8:591–594. doi: 10.1016/s0960-9822(98)70229-5. [DOI] [PubMed] [Google Scholar]
- 22.Hart M J, de los Santos R, Albert I N, Rubinfeld B, Polakis P. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 23.Peifer M, Sweeton D, Casey M, Wieschaus E. Development (Cambridge, UK) 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- 24.Yost C, Torres M, Miller R R, Huang E, Kimelman D, Moon R T. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 25.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orford K, Crockett C, Jensen J P, Wessman A M, Byers S W. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 27.Van Leeuwen F, Harryman Samos C, Nusse R. Nature (London) 1994;368:342–344. doi: 10.1038/368342a0. [DOI] [PubMed] [Google Scholar]
- 28.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook D, Fry M J, Hughes K, Sumathipala R, Woodgett J R, Dale T C. EMBO J. 1996;15:4526–4536. [PMC free article] [PubMed] [Google Scholar]
- 30.Heasman J. Development (Cambridge, UK) 1997;124:4179–4191. doi: 10.1242/dev.124.21.4179. [DOI] [PubMed] [Google Scholar]
- 31.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 32.Hoyt M A. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 33.Bai C, Sen P, Hofmann K, Ma L, Eoebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 34.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 35.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 36.Jiang J, Struhl G. Nature (London) 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 37.Theodosiou N A, Zhang S, Wang W Y, Xu T. Development (Cambridge, UK) 1998;125:3411–3416. doi: 10.1242/dev.125.17.3411. [DOI] [PubMed] [Google Scholar]
- 38.Spevak W, Keiper B D, Stratowa C, Castanon M J. Mol Cell Biol. 1993;13:4953–4966. doi: 10.1128/mcb.13.8.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson J W, Alarcon V, Elinson R. Dev Genet. 1996;19:190–198. doi: 10.1002/(SICI)1520-6408(1996)19:3<190::AID-DVG2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Margottin F, Bour S P, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 41.McKendry R, Hsu S C, Harland R M, Grosschedl R. Dev Biol. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- 42.Marikawa Y, Elinson R P. Mech Dev. 1998;77:75–80. doi: 10.1016/s0925-4773(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 43.Lagna G, Carnevali F, Marchioni M, Hemmati-Brivanlou A. Mech Dev. 1999;80:101–106. doi: 10.1016/s0925-4773(98)00208-1. [DOI] [PubMed] [Google Scholar]
- 44.Winston J T, Strack P, Beer-Romero P, Chu C Y, Elledge S J, Harper J W. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning A M, Andersen J S, Mann M, Mercurio F, Ben-Neriah Y. Nature (London) 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 46.Spencer E, Jiang J, Chen Z J. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harland R M. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.