Abstract
We have developed an online catalog of SNP-trait associations from published genome-wide association studies for use in investigating genomic characteristics of trait/disease-associated SNPs (TASs). Reported TASs were common [median risk allele frequency 36%, interquartile range (IQR) 21%−53%] and were associated with modest effect sizes [median odds ratio (OR) 1.33, IQR 1.20–1.61]. Among 20 genomic annotation sets, reported TASs were significantly overrepresented only in nonsynonymous sites [OR = 3.9 (2.2−7.0), p = 3.5 × 10−7] and 5kb-promoter regions [OR = 2.3 (1.5−3.6), p = 3 × 10−4] compared to SNPs randomly selected from genotyping arrays. Although 88% of TASs were intronic (45%) or intergenic (43%), TASs were not overrepresented in introns and were significantly depleted in intergenic regions [OR = 0.44 (0.34−0.58), p = 2.0 × 10−9]. Only slightly more TASs than expected by chance were predicted to be in regions under positive selection [OR = 1.3 (0.8−2.1), p = 0.2]. This new online resource, together with bioinformatic predictions of the underlying functionality at trait/disease-associated loci, is well-suited to guide future investigations of the role of common variants in complex disease etiology.
Keywords: catalog, evolution, GWAS, polymorphism, disorders
In the past 3 years, genome-wide association studies (GWAS) assaying hundreds of thousands of SNPs in thousands of individuals have reproducibly identified hundreds of associations of common genetic variants with over 80 diseases and traits (http://www.genome.gov/gwastudies). These studies have progressed from assaying fewer than 100,000 SNPs to more than one million, and sample sizes have increased dramatically as the search for variants that explain more of the disease/trait heritability has intensified (1). Important insights from these studies thus far include generally small effect sizes (odds ratios often <1.5), putative risk loci in or near genes not previously suspected of being involved in the etiology of a particular disease/trait, associated loci in common among diseases not previously thought to share etiologic pathways, and associations in many chromosomal regions currently annotated as gene poor (1).
The rapid increase in the number of GWAS provides an unprecedented opportunity to examine the potential impact of common genetic variants on complex diseases by systematically cataloging and summarizing key characteristics of the observed associations and the trait/disease associated SNPs (TASs) underlying them. Although some of these aspects have been examined on a smaller scale for individual diseases such as type 2 diabetes (2), inflammatory bowel disease (3), and cancer (4), a comprehensive genome-wide analysis across all GWAS published to date has not been conducted.
Identifying published GWAS can be challenging. For example, a simple PubMed search using the words “genome wide association studies” produced over 2,000 citations through December 2008, most of which are not actual GWAS. With this in mind, we developed the manually curated National Human Genome Research Institute (NHGRI) Catalog of Published Genome-Wide Association Studies (http://www.genome.gov/gwastudies), an online, regularly updated database of SNP-trait associations extracted from published GWAS. Here, we (i) describe the features of this resource and the methods we have used to produce it, (ii) provide and examine key descriptive characteristics of reported TASs such as estimated risk allele frequencies and odds ratios, (iii) examine the underlying functionality of reported risk loci by mapping them to genomic annotation sets and assessing overrepresentation via Monte Carlo simulations and (iv) investigate the relationship between recent human evolution and human disease phenotypes. There are several challenges in conducting these analyses within the present context wherein the actual functional variant is often unknown. Specifically, for a given TAS, the causative variant may be: (i) the TAS itself, (ii) a known common SNP in strong linkage disequilibrium (LD) with the TAS, (iii) an unknown common SNP or rare single nucleotide variant tagged by a haplotype on which the TAS occurs, or (iv) a linked copy number variant. Due to the limited annotation for categories (iii) and (iv), our analyses focus on the role of reported TASs and their LD partners (TASPs) only. Further, the power of analyses to detect overrepresentation of causative variants in specific functional categories may be weakened by the need to consider all of the TASPs within the LD block tagged by a TAS (we will refer to this as a TAS block). To circumvent this issue we use a strategy that calculates the overrepresentation of unique TAS blocks in specific categories (i.e., for a particular category, a TAS block is counted once if one or more unique member TASPs map to the category) rather than individual TASPs (Methods).
Results
Descriptive and Association Data.
We examined associations from a total of 151 (of 237) published GWAS (through December, 2008) reporting at least one TAS at p < 5 × 10−8 (supporting information (SI) Table S1). Including replication sample sizes reported in 130 (86%) of these 151 studies, the median total sample size for initial + replication studies was 7,858 participants (range, 146−91,749). From the 151 studies, we extracted information on 531 SNP-trait associations (limiting to one TAS per gene region and trait, as described in SI Text).
Unsurprisingly, since the GWAS method is primarily powered for common alleles, risk allele frequencies were well above 5% (median risk allele frequency 36%, interquartile range, IQR, 21%−53%) in the populations analyzed as well as in the HapMap populations (CEU: 37%, 21−54%; YRI: 33%, 13−65%; combined JPT+CHB 32%, 13−58%; Fig. S1).
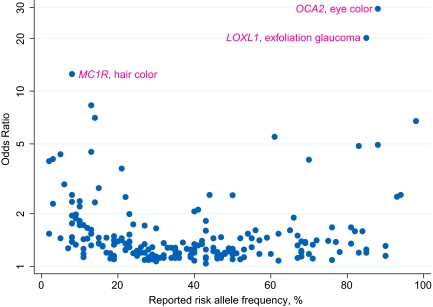
The 531 reported SNP-trait associations represented 465 unique TASs; 43% (n = 199) of which were located in an intergenic region, 45% (n = 208) were intronic, 9% (n = 41) were nonsynonymous, 2% (n = 10) were in a 5′ or 3′ untranslated region, and 2% (n = 7) were synonymous, according to the University of California Santa Cruz Genome Browser (5). Discrete traits were the focus of 227 (43%) of the 531 SNP-trait associations, which had associated odds ratios (ORs) ranging from 1.04 to 29.4 (median 1.33, IQR 1.20–1.61; Fig. S2). Among the discrete traits, the range of ORs was similar between nonsynonymous and other TASs; however, the right tail of the OR distribution for nonsynonymous TASs was slightly skewed toward higher valves. The highest ORs were reported for pigmentation traits [Fig. 1; MC1R and hair color (6) and OCA2 and eye color (6)]. SNP-trait associations were also distributed widely across diseases of high population prevalence, including heart disease, obesity, diabetes, and cancer (Table S2). Trait prevalence was not associated with the magnitude of ORs and risk allele frequencies, which were similar between the 10 most prevalent traits and all others combined (median ORs 1.26 and 1.29, respectively; median risk allele frequencies, 40% and 35%, respectively).
Fig. 1.
Published odds ratios for discrete traits by reported risk allele frequencies. Labeled SNP-trait associations are those with the highest ORs. Note that the y axis is on the log scale.
Among genes or regions harboring TASs that were reported in multiple studies of discrete traits, 18 were associated with seemingly distinct traits that may suggest clues toward common etiologic pathways (Table 1). Several TASs were located in previously characterized candidate genes, such as APOE, HLA, KCNJ11, PPARG, and CARD15, and were detected through GWAS at comparable effect sizes and stronger levels of statistical significance (Table S3). In these instances, GWAS-identified SNPs served as reasonable positive controls for known disease-associated genetic variants.
Table 1.
Reported TASs associated with two or more distinct traits
| Chromosomal region | Rs number(s) | Attributed genes | Associated traits reported in catalog |
|---|---|---|---|
| 1p13.2 | rs2476601, rs6679677 | PTPN22 | Crohn's disease, type 1 diabetes, rheumatoid arthritis |
| 1q23.2 | rs2251746, rs2494250 | FCER1A | Serum IgE levels, select biomarker traits (MCP1) |
| 2p15 | rs1186868, rs1427407 | BCL11A | Fetal hemoglobin, F-cell distribution |
| 2p23.3 | rs780094 | GCKR | CRP, lipids, waist circumference |
| 6p21.33 | rs3131379, rs3117582 | HLA / MHC region | Systemic lupus erythematosus, lung cancer, psoriasis, inflammatory bowel disease, ulcerative colitis, celiac disease, rheumatoid arthritis, juvenile idiopathic arthritis, multiple sclerosis, type 1 diabetes |
| 6p22.3 | rs6908425, rs7756992, rs7754840, rs10946398, rs6931514 | CDKAL1 | Crohn's disease, type 2 diabetes |
| 6p25.3 | rs1540771, rs12203592, rs872071 | IRF4 | Freckles, hair color, chronic lymphocytic leukemia |
| 6q23.3 | rs5029939, rs10499194 | TNFAIP3 | Systemic lupus erythematosus, rheumatoid arthritis |
| 7p15.1 | rs1635852, rs864745 | JAZF1 | Height, type 2 diabetes* |
| 8q24.21 | rs6983267 | Intergenic | Prostate or colorectal cancer, breast cancer |
| 9p21.3 | rs10811661, rs1333040, rs10811661, rs10757278, rs1333049 | CDKN2A, CDKN2B | Type 2 diabetes, intracranial aneurysm, myocardial infarction |
| 9q34.2 | rs505922, rs507666, rs657152 | ABO | Protein quantitative trait loci (TNF-α), soluble ICAM-1, plasma levels of liver enzymes (alkaline phosphatase) |
| 12q24 | rs1169313, rs7310409, rs1169310, rs2650000 | HNF1A | Plasma levels of liver enzyme (GGT), C-reactive protein, LDL cholesterol |
| 16q12.2 | rs8050136, rs9930506, rs6499640, rs9939609, rs1121980 | FTO | Type 2 diabetes, body mass index or weight |
| 17q12 | rs7216389, rs2872507 | ORMDL3 | Asthma, Crohn's disease |
| 17q12 | rs4430796 | TCF2 | Prostate cancer, type 2 diabetes |
| 18p11.21 | rs2542151 | PTPN2 | Type 1 diabetes, Crohn's disease |
| 19q13.32 | rs4420638 | APOE, APOC1, APOC4 | Alzheimer's disease, lipids |
* The well known association of JAZF1 with prostate cancer was reported with a p value of 2 × 10−6 (18), which did not meet the threshold of 5 × 10−8 for this analysis.
Functional Analysis.
To assess the underlying functionality at the trait/disease-associated genetic loci, we systematically mapped all TASPs (reported index TASs with an association p value < 5.0 × 10−8 and all HapMap phase II CEU SNPs in LD [r2 > 0.9]) to 20 nonmutually exclusive genomic annotation sets (Table S4). For each annotation set, we did the following. For every unique TAS block, we determined whether any TASPs mapped to the annotation set. If none mapped, we did not count the block. However, if one or more TASPs mapped, then we counted 1 per block. To compute the odds of a TAS block mapping to the annotation set, we divided the number of unique TAS blocks that were counted in the annotation set (n) by the number of TAS blocks that were not counted (N−n). To evaluate whether any annotation set was significantly enriched or depleted for TAS blocks, we compared the observed odds with the expected odds calculated from 100 control datasets comprised of randomly selected SNPs and their LD partners. Importantly, the mapping and counting strategies were consistent across both the test and the control datasets to ensure a fair comparison. Further, the generation of the control datasets took into account the representation biases on the genotyping arrays that were used to identify the TASs (SI Text).
For 9 annotation sets (nonsynonymous sites, 1kb promoters, 5kb promoters, most conserved sequences (MCSs), 3′ UTRs, microRNA target sites, Introns, CpG islands and experimentally validated regulatory regions from ORegAnno), the 95% confidence interval (CI) of the OR excluded 1.0 and the enrichment p values were <0.05 (Fig. S3), indicating that these categories may be significantly enriched for TAS blocks. Nonsynonymous sites had the strongest signal for enrichment (OR = 3.9 [2.2−7.0], p = 3.5 × 10−7). After restricting the analysis to only those nonsynonymous SNPs predicted by PolyPhen (7) to be potentially deleterious (which reduces the sample size by approximately 65%), TAS blocks were even more strongly enriched (OR = 5.2 [1.8–15.3], p = 0.001). Thirty nonsynonymous TASPs that are predicted to be potentially deleterious [by PolyPhen and an unpublished method, CDPred (P. Cherukuri and J. Mullikin, personal communication)] were identified as attractive candidates for functional follow-up (Table 2).
Table 2.
Predicted deleterious non-synonymous TASPs with p < 5 × 10−8. Sixteen nonsynonymous trait/disease associated SNPs and their strong linkage disequilibrium partners (TASPs defined here as r2 > 0.9), are predicted to be deleterious according to both PolyPhen (PP) and CDPred (CDP), 7 only by PP and 7 only by CDP
| Non-synonymous TASP | Amino acid change | Index TAS from GWAS | Affected gene | Reported gene | Trait/disease | Program |
|---|---|---|---|---|---|---|
| rs1046934 | Q59H | rs2274432 | TSEN15 | GLT25S2 | Height | PP & CDP |
| rs2305479 | G282R | rs7216389 | GSDMB | ORMDL3 | Asthma | PP & CDP |
| rs2305480 | P289S | rs2872507 | GSDMB | ORMDL3 | Crohn's disease | PP & CDP |
| rs3197999 | R689C | rs3197999 | MST1 | MST1 | Crohn's disease | PP & CDP |
| rs2476601 | R620W | rs2476601 | PTPN22 | PTPN22 | Rheumatoid arthritis | PP & CDP |
| rs2395029 | V112G | rs2395029 | HLA-C | HLA-C | Psoriasis | PP & CDP |
| rs4149056 | V174A | rs4149056 | SLCO1B1 | SLCO1B1 | Myopathy | PP & CDP |
| rs7578597 | T1187A | rs7578597 | THADA | THADA | T2D | PP & CDP |
| rs229527 | G21V | rs229541 | C1QTNF6 | C1QTNF6 | T1D | PP & CDP |
| rs11820589 | P148L | rs12272004 | BUD13 | APOA | LDL/TG | PP & CDP |
| rs676210 | P2739L | rs673548 | APOB | APOB | Metabolic traits | PP & CDP |
| rs1799990 | M129V | rs1799990 | PRNP | PRNP | Creutzfeldt-Jacob disease | PP & CDP |
| rs6756629 | R50C | rs6756629 | ABCG5 | ABCG5 | LDL | PP & CDP |
| rs1800562 | C282Y | rs1800562 | HFE | HFE | Serum markers of iron status | PP & CDP |
| rs328 | Nonsense | rs10503669 | LPL | LPL | HDL/TG | PP & CDP |
| rs601338 | Nonsense | rs492602 | FUT2 | FUT2 | Vitamin B12 | PP & CDP |
| rs2274432 | G19D | rs2274432 | TSEN15 | GLT25D2 | Height | PP only |
| rs11591147 | R46L | rs11591147 | PCSK9 | PCSK9 | LDL | PP only |
| rs11887534 | D19H | rs11887534 | ABCG8 | ABCG8 | Gallstones | PP only |
| rs1799969 | G241R | rs1799969 | ICAM1 | ICAM1 | Soluble ICAM1 | PP only |
| rs17467284 | R911L | rs11175593 | Predicted | LRRK2 / MUC19 | Crohn's disease | PP only |
| rs1260326 | P446L | rs1260326 | GCKR | GCKR | TG | PP only |
| rs1064608 | P290A | rs10838738 | MTCH2 | MTCH2 | BMI | PP only |
| rs1042602 | S192Y | rs1042602 | TYR | TYR | Freckles | CDP only |
| rs6046 | R413Q | rs561241 | Factor VII | Factor VII | Factor VII level | CDP only |
| rs16969968 | D398N | rs8034191 | CHRNA5 | CHRNA5 | Lung cancer | CDP only |
| rs16890979 | V282I | rs16890979 | SLC2A9 | SLC2A9 | Serum uric acid | CDP only |
| rs2231142 | Q141K | rs2231142 | ABCG2 | ABCG2 | Serum uric acid | CDP only |
| rs3761472 | D110G | rs2281135 | SAMM50 | SAMM50 | Plasma level of liver enzymes | CDP only |
| rs1169288 | I27L | rs2650000 | HNF1A | HNF1A | LDL | CDP only |
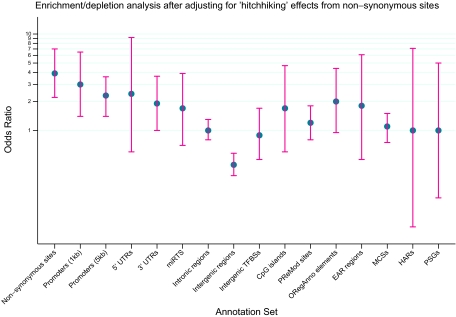
To examine the possibility that signals in other annotation sets might not represent bona fide TAS block enrichment, but rather a “hitchhiking” effect whereby TASPs closely linked with nonsynonymous SNPs map to nearby annotation sets and artificially increase their ORs, we removed all TASPs having r2 > 0.6 with any nonsynonymous HapMap CEU SNP and repeated the test (Fig. 2). Only 2 categories retained a clear signal for enrichment — 1-kb promoters (OR = 3.0 [1.4 − 6.5], p = 0.005) and 5-kb promoters (OR = 2.3 [1.5 − 3.6], p = 0.0003). After Bonferroni correction for 20 comparisons, only the enrichment signal from 5-kb promoters remained significant, although this may reflect greater power relative to 1-kb promoters due to a larger number of mapped TASPs. Although no other category had even an uncorrected enrichment p value < 0.05, a few showed nonsignificant trends toward enrichment, such as the ORegAnno elements (OR = 2.0 [0.95 − 4.4], p = 0.09). We note here that the number of TAS blocks mapping to 1kb promoter regions was similar to or smaller than several other categories (Table S4), so power is unlikely to explain the strong enrichment signal in promoter regions and lack thereof elsewhere.
Fig. 2.
Odds ratios for TAS block enrichment/depletion analysis after adjusting for “hitchhiking” effects from nonsynonymous sites. Four annotation sets (Splice sites, Validated enhancers, EvoFold elements, and noncoding RNAs) are not represented here because no TAS blocks mapped to these annotation sets. The blue circle represents the point estimate of the odds ratio (OR) and the red lines represent the 95% CI. Possible “hitchhiking” effects from nonsynonymous sites are reduced by discarding any TASP/control SNP in r2 > 0.6 with a nonsynonymous SNP. For an explanation of the annotation sets on the x axis, we refer the reader to Table S4. Note that the y axis is on the log scale. Nonsynonymous OR computation is not adjusted for “hitchhiking” effects.
Using previous predictions of high confidence transcription factor binding sites in human 1-kb promoter regions, 4 TASPs were predicted to have strong allele-specific TF binding affinities (Table S5). These predictions may lead to compelling hypotheses for trait/disease etiology, as in the case of the protective allele [G] of SNP rs1077834, which is predicted to abolish a partially conserved binding site for HNF4α 696 base pairs upstream of the LIPC transcription start site. HNF4α is an essential hepatic transcriptional activator that has been associated with metabolic pathways (8) and has been shown to activate LIPC, an important enzyme in lipid metabolism (9). One could thus hypothesize that the loss of HNF4α binding in the presence of the protective allele may lower LIPC expression, leading to increased plasma HDL levels.
Intergenic regions, despite harboring the largest fraction of TAS blocks, were significantly depleted for TAS blocks (OR = 0.44 [0.34 − 0.58], p = 2.0 × 10−9). This is consistent with the assumption that intergenic regions, although containing important regulatory sequences, have the smallest ratio of functional to total DNA. Intronic regions and several putative functional categories within intergenic regions (such as predicted intergenic transcription factor binding sites, experimentally supported enhancer regions, noncoding RNAs, and regions of conserved RNA secondary structures) did not show evidence for enrichment or depletion (Fig. 2). However, a definitive interpretation of this result is hindered by the current lack of extensive experimental annotation and noisy computational predictions of functional elements within intergenic regions.
Although conservation across species is a popular proxy for important functionality, enrichment analysis in the mammalian Most Conserved Sequences (MCSs) revealed no TAS block enrichment signal (OR = 1.07 [0.75 − 1.5], p = 0.79). However, ORegAnno sites—experimentally supported regulatory elements of which many are nonconserved—did show a trend toward TAS block enrichment (Fig. 2; OR = 2.0 [0.95−4.4], p = 0.09). This affirms the need for more experimental investigation into the architecture of noncoding regulatory elements (such as enhancers and microRNAs) to decrease the reliance on conservation and guide more integrative computational prediction methodologies.
To ensure the robustness of our results, we repeated the analyses using different r2 thresholds for defining LD partners (r2 = 1.0 and r2 > 0.8) and the results were essentially unchanged (data not shown). Although even lower r2 thresholds are reasonable for capturing more of the possible causative variants, they will also likely pick up a greater number of irrelevant variants that will only obscure the interpretation. When we repeated the analysis using a threshold of r2 > 0.6, results for 4 categories changed substantially−CpG islands and ORegAnno elements displayed a stronger enrichment signal and introns and MCSs revealed a stronger signal for depletion.
Evolutionary Analysis.
To assess evolutionary characteristics of TASs, we used the integrated haplotype score (iHS), which is a measure of recent positive selection designed to detect extended haplotypes due to selective sweeps (10). The proportion of TASs (not the full set of TASPs as we wanted to select only one SNP for each TAS block) with iHS above the 90th percentile (1.635) among HapMap Phase II CEU SNPs was slightly higher relative to background expectation (OR = 1.3 [0.8−2.1]), but only at p = 0.2. At least 50 TASs were predicted to be under positive selection (Table S6). Several risk alleles associated with melanin synthesis, immune response, and cancer were identified as having undergone recent moderate (iHS = 1.635–2.0) or strong (iHS > 2.0) positive selection (Table S6), consistent with previously established links between these traits/diseases and positive selection (11–13). Of particular interest are positively selected alleles that confer risk for obesity and related metabolic disorders (Table S6), because these are consistent with the long-standing thrifty gene hypothesis (14). Despite the popularity of this hypothesis, wherein genetic variants conferring resistance to starvation in hunter-gatherer populations were positively selected, a convincing “thrifty gene” has yet to be identified. The risk allele of rs1121980 in the fat mass and obesity associated gene (FTO), which is involved in fat storage, has undergone recent positive selection by iHS analysis and is an attractive candidate for a thrifty gene.
Discussion
The availability of an online, curated, user-friendly, and downloadable catalog of GWAS and SNP-trait associations has facilitated a multifaceted analysis of published GWAS results. The more than 150 studies and 500 SNP-trait associations reviewed here demonstrate that, in general, reported TASs tend to be common (>5% minor allele frequency), are associated with modest effect sizes, and are not highly differentiated across populations. In addition, TAS blocks are significantly enriched in nonsynonymous sites (especially for potentially deleterious sites) and in promoter regions and are depleted in intergenic regions. Despite this enrichment, 43% of reported TASs were intergenic and 45% were intronic, suggesting a greater than anticipated role for noncoding SNPs in common diseases. The mere presence of a predicted deleterious nonsynonymous associated variant does not discount the possibility that a nearby SNP in LD may (i) be the true causative variant, (ii) confer an independent molecular mechanism for the phenotype, or (iii) epistatically interact with the nonsynonymous variant to cause disease. Despite the recent interest in the role of microRNA (miRNA) targeting dysfunction in human disease (15, 16), we found no evidence that predicted miRNA target sites in the 3′ UTR were significantly enriched for TAS blocks (Fig. 2). Finally, predictions based on the integrated haplotype score (iHS) may provide important clues about the evolutionary history and underlying molecular mechanisms of certain TASPs.
Several limitations of the underlying catalog data should be noted. We extracted all eligible associations from published articles and SI Text, but the number and quality of reported SNP associations is dependent upon the preferences of the individual author and journal. Also, the studies within the catalog generally test only those SNPs that are detectable via commonly used genotyping platforms in participants who tend to be from European-descent populations. The GWAS data are likely to be subject to varying degrees of upward bias in effect size estimates (the “winner's curse” phenomenon), particularly to the extent that estimates from the GWAS discovery population, who may be less representative of the general population, influence those reported in our catalog. Nonetheless, in several instances in which known candidate SNPs have been previously identified, GWAS of the same trait tended to confirm these findings with similar effect sizes and stronger levels of statistical significance. Finally, TASs reported in published GWAS suffer from “lead TAS bias”; generally 1 or 2 TASs out of a cluster are selected from the initial study, often based on likely functional significance such as a conserved nonsynonymous site, for association analysis in the replication sample. To minimize the effect of this bias, we analyzed TAS blocks, which include the lead SNPs and their known LD partners based on HapMap phase II data. However, the true impact of the bias is difficult to quantify and it may still exert a slight effect on the enrichment/depletion signals especially for categories such as nonsynonymous sites.
An important question is to what extent GWAS have identified genetic variants likely to be of clinical or public health importance, particularly for developing preventive or therapeutic interventions. Answering this question must await better functional characterization of TASs or the true causative variants they may be tagging, evidence of effective interventions, and identification of potential modifiers of SNP-trait associations (1). However, the current study contributes empiric bounds on the expectations for the effect sizes and allele frequencies of TASs that can be identified from GWAS. It also highlights the distribution of promising SNP-trait associations across a wide variety of traits of substantial public health interest, such as obesity, hypertension, coronary artery disease, and cancer. Our results may guide future studies by highlighting genetic variants that are of particular interest from a descriptive, association, evolutionary, or functional perspective (such as predictions of TASP-mediated allele-specific transcription factor binding sites) and suggesting hypotheses for future study. Our description of GWAS-identified variants builds upon the important work previously targeted toward candidate genes, adding to a more complete picture of the contribution of common genetic variation to common diseases. It is clear, however, that the proportion of heritability explained by common variation for most common diseases to date is modest at best (17). As the power of the GWAS approach increases with access to more samples, and as the types of methods to test for genetic associations expand to include copy number variants and rarer alleles, more associations will likely be identified and timely analyses similar to those presented here will continue to update our knowledge of the influence of genomic structure and function on complex diseases.
Methods
Catalog Development and Curation.
Published GWAS were identified primarily through 2 sources: (i) weekly PubMed searches and (ii) daily NIH-distributed compilations of news and media reports. We also periodically consulted an online database of published genomic epidemiology literature (http://www.cdc.gov/hugenet). All studies assaying at least 100,000 SNPs in any subset of participants, typically referred to as the “initial scan,” were included in the catalog, regardless of the number of SNPs that ultimately passed quality control and were used in analysis. Several study level and SNP-trait association level variables were extracted (SI Text). The somewhat liberal statistical threshold of p < 1.0 × 10−5 was chosen to allow examination of borderline associations and to accommodate scans of various sizes while maintaining a consistent approach. Filters on the catalog allow thresholding at various levels of p values or odds ratios. Additional detail about the catalog curation is available in the SI Text.
Descriptive and Association Analyses.
We restricted these analyses to 151 of the 237 papers reporting a total of 531 SNP-trait associations with p < 5 × 10−8 through December 31, 2008, and corresponding to 465 unique SNPs. A complete list of references and extracted catalog data for these analyses are available from the authors upon request. Statistical analyses were done in STATA Intercooled 8.0. Further details on the descriptive and association analyses are available in the SI Text.
Functional and Evolutionary Analyses.
The 20 annotation sets were defined and obtained as described in Table S4. Each annotation set was assayed for enrichment or depletion as previously described (Results). Fisher's exact test p values were computed using the “Text∷NSP∷Measures∷2D∷Fisher” Perl module. Additional details of the functional and evolutionary analyses are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Judy Wyatt and Kent Klemm (NHGRI) for their work in developing the catalog into an online resource and Narisu Narisu, Lori Bonnycastle, and Samir Kelada (NHGRI) for several helpful discussions and insightful suggestions for the manuscript. We also would like to thank Praveen Cherukuri in the J. Mullikin Lab (NHGRI) for kindly sharing the program CDPred and the 3 reviewers for their expertise and insightful suggestions that greatly improved the manuscript. This work was supported in part by the intramural programs of the National Human Genome Research Institute and the National Library of Medicine.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903103106/DCSupplemental.
References
- 1.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy MI, Hirschhorn JN. Genome-wide association studies: Potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 4.Eeles RA, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 5.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 7.Ramensky V, Bork P, Sunyaev S. Human nonsynonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rufibach LE, Duncan SA, Battle M, Deeb SS. Transcriptional regulation of the human hepatic lipase (LIPC) gene promoter. J Lipid Res. 2006;47:1463–1477. doi: 10.1194/jlr.M600082-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summers K, Crespi B. Molecular evolution of the prostate cancer susceptibility locus RNASEL: Evidence for positive selection. Infect Genet Evol. 2008;8:297–301. doi: 10.1016/j.meegid.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Sabeti PC, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 2007;71:354–369. doi: 10.1111/j.1469-1809.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- 14.Neel JV. Diabetes mellitus: A “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 15.Georges M, Coppieters W, Charlier C. Polymorphic miRNA-mediated gene regulation: Contribution to phenotypic variation and disease. Curr Opin Genet Dev. 2007;17:166–176. doi: 10.1016/j.gde.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era–concepts and misconceptions. Nat Rev Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 18.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.