Abstract
Whole-genome sequence analysis of Mycobacterium leprae has revealed a limited number of protein-coding genes, with half of the genome composed of pseudogenes and noncoding regions. We previously showed that some M. leprae pseudogenes are transcribed at high levels and that their expression levels change following infection. In order to clarify the RNA expression profile of the M. leprae genome, a tiling array in which overlapping 60-mer probes cover the entire 3.3-Mbp genome was designed. The array was hybridized with M. leprae RNA from the SHR/NCrj-rnu nude rat, and the results were compared to results from an open reading frame array and confirmed by reverse transcription-PCR. RNA expression was detected from genes, pseudogenes, and noncoding regions. The signal intensities obtained from noncoding regions were higher than those from pseudogenes. Expressed noncoding regions include the M. leprae unique repetitive sequence RLEP and other sequences without any homology to known functional noncoding RNAs. Although the biological functions of RNA transcribed from M. leprae pseudogenes and noncoding regions are not known, RNA expression analysis will provide insights into the bacteriological significance of the species. In addition, our study suggests that M. leprae will be a useful model organism for the study of the molecular mechanism underlying the creation of pseudogenes and the role of microRNAs derived from noncoding regions.
Mycobacterium leprae, the causative agent of leprosy, cannot be cultivated in vitro. Therefore, bacteriological and pathological information, such as the mechanisms of infection, parasitization, and replication, are still largely unknown. However, whole-genome sequencing has provided insight into many biological characteristics of M. leprae (5). The M. leprae genome consists of 3.3 Mbp, which is much smaller than the 4.4 Mbp of the Mycobacterium tuberculosis genome. M. leprae has 1,605 genes and 1,115 pseudogenes, while M. tuberculosis has 3,959 genes and only 6 pseudogenes. The number and ratio of pseudogenes in M. leprae are exceptionally large by comparison with the pseudogene numbers and ratios for other pathogenic and nonpathogenic bacteria and archaea (21). A feature of M. leprae pseudogenes is the massive fragmentation caused by many insertions of stop codons (26). The functional roles, if any, of these unique pseudogenes and noncoding regions are unknown. However, we have shown that some M. leprae pseudogenes are highly expressed as RNA and that their expression levels change following macrophage infection (36). In that study, a membrane-based DNA array was created utilizing a cosmid DNA library that covered >98% of the M. leprae genome. mRNAs purified from M. leprae-infected macrophages and control bacilli were enriched by cDNA subtraction and hybridized to these arrays. Southern blot analysis of the positive cosmid clones identified 12 genes that might be important for the survival and infection of M. leprae. Six of the 12 genes were pseudogenes.
Pseudogenes are described as functionally silent relatives of normal genes. Since they are usually eliminated from the genome, it was speculated that the number of pseudogenes correlates with the size of the genome (28). Most pseudogenes are thought to result from a transposon insertion or inactivation of one copy after a gene duplication event (7). Because they do not create functional proteins, they are also called “junk” genes. However, some pseudogenes are expressed and function to regulate the expression of other genes (14, 20).
About one-quarter of the M. leprae genome is composed of noncoding regions, which constitutes a much larger proportion of the genome than the noncoding regions in M. tuberculosis. Gene-regulatory short RNA fragments generated from noncoding regions have been found in many organisms (17). In those cases, precursor microRNAs are transcribed independently and processed into mature forms. In eukaryotes, most of the transcriptome, which includes thousands of microRNAs, consists of noncoding RNA (24). In addition, the abundance of small RNAs in Escherichia coli has been estimated at 1 to 2% of the number of open reading frames (ORFs) (12).
Microarrays have facilitated transcriptome analysis through the use of probes that target a large number of genes. The technique has identified unexpected gene activity in a number of areas and in some cases has served to elucidate entire microbial metabolic processes, as exemplified by caloric restriction or oxidative stress in E. coli (10, 30). Moreover, RNA expression profiling has been valuable in the analysis of pathogenic bacteria. Analyses of changes in RNA expression upon infection of host macrophages has identified genes related to oxidative stress, proliferation, and other unknown functions in Yersinia pestis (causative agent of plague) (42) and Salmonella enterica serovar Typhi (causative agent of typhoid fever) (9). DNA microarray analysis has also found genes involved in the acid stress response (2) and transcriptional hierarchy of the flagellar system (27).
Only known or predicted genes were examined in the experiments described above. Therefore, it was not possible to analyze the RNA expression of noncoding regions and potential pseudogenes that did not have the appropriate annotation. Clone-based microarrays were developed to solve this problem (29), but they were still unable to detect genome-wide RNA expression. Finally, tiling arrays have become a useful tool for the analysis of whole-genome or chromosome expression (19) and have been used to uncover several novel RNA expression patterns (15, 38). Although the genome sequence and its annotation are known, comprehensive analysis of M. leprae RNA expression has not been performed. The results of our previous study and the availability of tiling arrays prompted a detailed investigation of RNA expression throughout the M. leprae genome. In this study, tiling arrays were used to analyze comprehensive RNA expression of genes, pseudogenes, and noncoding regions in M. leprae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Footpads of hypertensive nude rats (SHR/NCrj-rnu), in which the Thai-53 strain of M. leprae was grown, were kindly provided by Y. Yogi, Leprosy Research Center, National Institute of Infectious Diseases. M. leprae was isolated as previously described (40, 41). Briefly, the skin and bones were removed from the footpad tissues. The tissues were then extensively homogenized in Hanks' balanced salt solution with 0.025% Tween 80 and centrifuged at 700 × g and 4°C for 10 min to remove tissue debris. The supernatant was treated with 0.5% trypsin at 37°C for 1 h, followed by centrifugation at 5,000 × g and 4°C for 20 min. The supernatant was discarded, and the pellet was resuspended in 10 ml Hanks' balanced salt solution with 0.025% Tween 80 and 0.25 N NaOH. A further incubation at 37°C for 15 min was followed by another centrifugation, and the pellet was resuspended in 2 ml phosphate-buffered saline. Two microliters of solution was spread on a glass slide and subjected to acid-fast staining to count the number of bacilli.
RNA extraction.
M. leprae cells (2.8 × 1011) were suspended in 2 ml of RNA Protect bacterial reagent (Qiagen, Germantown, MD), subjected to a vortex, and incubated for 10 min at room temperature. The cells were pelleted and resuspended in 2 ml of RNA Protect bacterial reagent, 0.4 ml of 1.0-mm zirconia beads (BioSpec Products, Bartlesville, OK), and 0.6 ml of lysis/binding buffer from the mirVana miRNA isolation kit (Ambion, Austin, TX). The mixture was homogenized at 3,000 rpm for 3 min using a Micro Smash homogenizer (Tomy, Tokyo, Japan) followed by four freeze-thaw cycles. RNA was then extracted according to the manufacturer's guidelines (Ambion) and treated with DNase Ι (TaKaRa, Kyoto Japan).
Preparation of labeled double-stranded DNA.
Twenty micrograms of total RNA from M. leprae was reverse transcribed using SuperScript ΙΙ (Invitrogen, Carlsbad, CA). The generated cDNA was incubated with 10 ng of RNase A (Novagen, Madison, WI) at 37°C for 10 min, phenol-chloroform extracted, and precipitated with ethanol. Cy3 labeling was performed as follows: 1 μg double-stranded cDNA was incubated for 10 min at 98°C with 1 optical-density-at-600-nm unit of Cy3-9-mer Wobble primer (TriLink Biotechnologies, San Diego, CA). The addition of 8 mmol of deoxynucleoside triphosphates and 100 U of Klenow fragment (New England Biolabs, Ipswich, MA) was followed by incubation at 37°C for 2 h. The reaction was stopped by adding 0.1 volumes of 0.5 M EDTA, and the labeled cDNA was precipitated with isopropanol.
Array design.
The tiling array was designed based on sequences obtained from the GenBank database (accession no. NC_002677) (5). Each probe was a 60-mer, and the adjacent probe was shifted by 18 nucleotides (a 42-nucleotide overlap). A total of 363,116 probes were designed for the sense and antisense strands and arranged on a glass plate with 22,000 control probes of randomly chosen sequences. Another array on which the probes were chosen from M. leprae ORFs (NimbleGen Systems, Madison, WI) was made. On this ORF array, 20 different probes were designed for each of the 1,605 ORFs. The probes were spotted onto five blocks on the glass plate, resulting in an arrangement of 160,500 probes on the ORF array.
Hybridization and analysis of tiling and ORF arrays.
Cy3-labeled samples were resuspended in 40 μl of hybridization buffer (NimbleGen Systems, Madison, WI), denatured at 95°C for 5 min, and hybridized to arrays in a MAUI hybridization system (BioMicro Systems, Salt Lake City, UT) for 18 h at 42°C. The arrays were washed using a wash buffer kit (NimbleGen Systems), dried by centrifugation, and scanned at a 5-μm resolution using the GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). NIMBLESCAN 2.3 (NimbleGen Systems) was used to obtain fluorescence intensity data from the scanned arrays.
Quantitative real-time PCR.
The cDNA used for tiling array was also subjected to real-time PCR analysis. The primers were designed using GENETYX version 7 (Genetyx Corporation, Tokyo, Japan) and are listed in Table S1 in the supplemental material. Preparation of M. leprae genomic DNA and real-time PCRs was carried out as described previously (37) with 200 nM of each primer and 0.5 ng of cDNA or 0.2 ng of genomic DNA as a control.
RESULTS
Tiling array detected highly expressed regions in genes, pseudogenes, and noncoding regions.
The 116 μg of total RNA isolated from 2.8 × 1011 M. leprae cells was treated with DNase Ι. RNA quality and quantity were evaluated with an Agilent Bioanalyzer 2100 (Agilent, Foster City, CA). The ratio of 23S rRNA to 16S rRNA was 0.83, indicating that the quality of the purified RNA was good enough to proceed with array hybridization. After hybridization and detection, the scanned row signals were normalized against the signal intensities from the control probes and converted to log2 scores with the median set at zero. The corrected intensities of all probes distributed between −2.762 and 6.282 were then calculated. When the intensities of four probes within 500 bp were higher than 60% of the maximum intensity (>3.769), the region (i.e., gene, pseudogene, or noncoding region) was considered positive. When each probe was evaluated independently, 8,658 probes (2.38%) showed >60% of the maximum intensity.
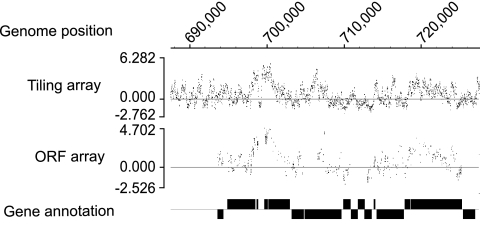
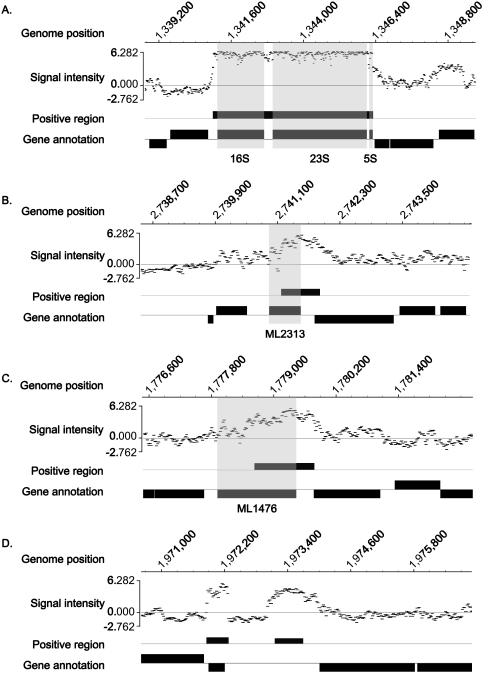
In order to confirm the specificity of the tiling array, RNA from the same sample was simultaneously hybridized with the ORF array on which multiple sequence-specific probes were designed for each gene. The positive signals detected on the ORF array were consistent with those detected on the tiling array (Fig. 1). Moreover, because the tiling array probes include ORFs in their coverage of the entire genome, it is expected that more detailed information would be obtained from them. The strongest signal was identified in the rRNA; most probes in this region showed significantly higher intensity (Fig. 2A). Other highly expressed areas were detected in the genes (Fig. 2B), pseudogenes (Fig. 2C), and noncoding regions (Fig. 2D). In this study, noncoding regions were defined as regions that are not annotated. rRNA and tRNA are usually considered noncoding RNA but are dealt with separately here since they are annotated in the database. An interesting feature of some highly expressed areas was that positive signals sometimes overlapped both gene/pseudogene and noncoding regions, as illustrated in Fig. 2B and C. The expression levels of each probe within a single ORF were not constant but rather quite variable, which might reflect a difference in melting temperature based on the GC content of each probe.
FIG. 1.
Typical array data from an approximately 40-kbp region. Data from the tiling and ORF arrays are shown with the gene annotation of Cole et al. from 2001 (5) depicted as rectangles.
FIG. 2.
Signal intensity patterns detected as highly expressed areas in the tiling array. Scanned data were normalized to log2, divided by the median, and arrayed against the corresponding M. leprae genome sequence. Positive areas were extracted and are depicted under the signal pattern of probes with gene and pseudogene annotations. (A) Genomic region of rRNA showing almost saturated signal intensity. (B) Highly expressed region of the gene for the hypothetical protein ML2313 (shaded area). (C) Highly expressed region of the ML1476 pseudogene (probable oxidoreductase alpha subunit; shaded area). (D) Highly expressed noncoding region in the genomic position from bp 1973155 to 1973700, which showed no homology to genes or other functional sequences by BLASTN search. Gene annotations are from reference 5.
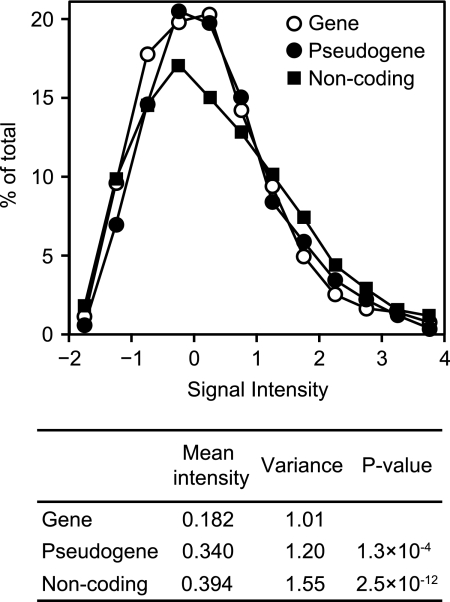
The distribution of signal intensities among the genes, pseudogenes, and noncoding regions was evaluated by calculating the average intensity of each probe within a single region and plotting the relative values (Fig. 3, upper panel). If hybridization occurred in a random fashion independent of RNA expression levels, the expectation is that all of the probes would exhibit the same distribution of signal intensities among the genes, pseudogenes, and noncoding regions. However, while positive regions were detected in similar proportions in genes, pseudogenes, and noncoding regions, with no difference in the mean lengths of the positive regions among the three groups, the array data showed stronger signal intensities in the noncoding regions (Fig. 3, right shoulder of the graph). The mean intensity in coding genes (0.182) was significantly lower than that in noncoding regions (0.394) (P = 2.5 × 10−12) and pseudogenes (0.340) (P = 1.3 × 10−4) (Fig. 3, lower panel). High RNA expression from a noncoding region (Fig. 2D) suggests that those RNAs have a biological function. However, no sequence homology was identified in these regions after intensive database searches.
FIG. 3.
Distribution of signal intensity in each region. Mean signal intensities of individual regions were calculated, and the ratio against the corresponding total number in the M. leprae genome was plotted for genes, pseudogenes, and noncoding regions. Mean signal intensities, variances, and P values from Student's t test were calculated for the entire region and are shown below the graph.
A total of 168 positive areas, some spanning more than one region, were found based on the applied criteria (>60% of the maximum level). When an expressed area overlapped two or more annotated genes or noncoding regions, they were counted separately based on each annotation (as shown in Fig. 2B and C). A noncoding region longer than 114 bp, which is the minimum length of an evaluated area, was counted as a single expressed region. As a result, 209 positives from genes, pseudogenes, and noncoding regions were classified as strong expressers. The number from each region, the mean length of the positive regions, and the mean peak signal intensities are summarized in Table 1.
TABLE 1.
Numbers of highly expressed genes, pseudogenes, and noncoding regions identified by tiled microarray analysis
| Genetic material | No. identified | % of total | Mean length (bp) | Mean peak intensitya |
|---|---|---|---|---|
| Genes | 63 | 30.1 | 637 | 4.88 |
| Pseudogenes | 78 | 37.3 | 611 | 5.11* |
| Noncoding regions | 68 | 32.5 | 634 | 5.38** |
| Total | 209 | 100 |
Mean peak intensities of pseudogenes and noncoding regions were statistically compared with the intensity of coding genes (*, P < 0.05; **, P < 0.00001 by Student's t test).
Functional classification of expressed genes and pseudogenes.
Gene expression profiles obtained from tiling array analysis were classified based on criteria that were originally determined during whole-genome sequence analysis of M. tuberculosis (4) and later applied to M. leprae (5) (Table 2). Among genes, the “cell processes” class (constituting genes with functions such as transport, secretion, and chaperone function) was highly expressed (9.8%) compared to genes overall (3.9%) (χ2 = 7.1, P = 0.008). Among the “small-molecule metabolism” class, the “amino acid biosynthesis” (4 out of 77) and “purines, pyrimidines, nucleosides, and nucleotides” (4 out of 52) subsets were highly expressed, while expression of the “biosynthesis of cofactors, prosthetic groups, and carriers” subset was not observed (0 out of 63). Similarly, in the “macromolecule metabolism” class, the “cell envelope” subset was expressed (13 out of 256), but the “degradation of macromolecules” subset was not (0 out of 43) (χ2 = 2.8, P = 0.251). Three out of 11 PE and PPE protein gene families found in the “other functions” class were expressed among the coding genes.
TABLE 2.
Numbers and percentage of expressed genes and pseudogenes based on functional classificationa
| Gene function/type | No. of expressed genes or pseudogenes/total no. of genes or pseudogenes (%)
|
|
|---|---|---|
| Genes | Pseudogenes | |
| Small-molecule metabolismb | 19/467 (4.1) | 19/334 (5.7) |
| Macromolecule metabolismc | 16/458 (3.5) | 10/163 (6.1) |
| Cell processesd | 10/102 (9.8) | 2/67 (3.0) |
| Other functionse | 6/77 (7.8) | 29/133 (21.8) |
| Conserved hypotheticals | 6/360 (1.7) | 18/416 (4.3) |
| Unknowns | 6/141 (4.3) | 0/2 (0) |
| Total | 63/1,605 (3.9) | 78/1,115 (7.0) |
Synthesis and degradation of amino acid, polyamine, nucleotide, cofactor and lipid, and energy metabolism enzymes.
Synthesis and degradation of protein, RNA, DNA, and cell envelope.
Transporter and chaperone.
Virulence, repeated sequence, and PE and PPE families.
Pseudogenes were classified based on criteria defined by the function of their counterpart genes (5) (Table 2). Pseudogene expression was significantly higher in the “other functions” class than in other classes (χ2 = 40.9, P = 1.00 × 10−7). No significance was detected when this class was excluded (χ2 = 1.7, P = 0.793). In the “other functions” class, 15 expressed pseudogenes contained parts of the LEPREP repeat sequence. Markedly expressed pseudogenes were also found in the “degradation” (5 out of 74) and “energy metabolism” (7 out of 118) subsets of the “small-molecule metabolism” class, although the expression was not statistically significant among pseudogenes (78 out of 1,115). The overall expression level of pseudogenes (7.0%) was higher than that of genes (3.9%) (χ2 = 11.3, P = 0.001). However, the “cell processes” class showed significantly higher gene expression (9.8%) than pseudogene expression (3.0%) (χ2 = 6.6, P = 0.010).
Real-time PCR confirmation of RNA expression profiles.
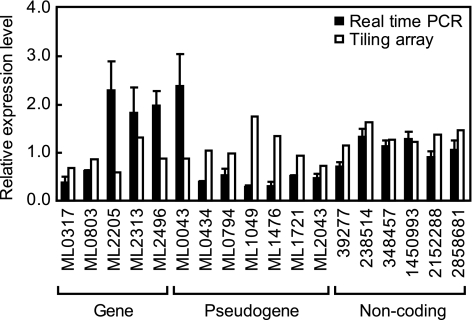
Specific primers were designed for five genes, seven pseudogenes, and six noncoding regions that were highly expressed in the tiling array analysis (see Table S1 in the supplemental material). Although M. leprae RNA was pretreated with DNase I prior to reverse transcription, the RNA was checked by PCR to exclude possible contamination by genomic DNA (data not shown).
Each primer set generated a specific reverse transcription-PCR product (data not shown). The RNA expression levels determined by real-time PCR analysis were comparable to the signal intensities from the tiling array (Fig. 4). Of interest, coding genes produced higher expression levels in real-time PCR, in contrast to the higher level of pseudogene expression detected by the tiling array.
FIG. 4.
Comparison of RNA expression between real-time PCR and tiling array. Relative RNA expression levels detected by tiling array analysis and quantitative real-time PCR were compared. Genes and pseudogenes are indicated by accession numbers. Noncoding regions are indicated by their starting position in the M. leprae genome. Data are from three independent real-time PCRs and are expressed as means ± standard errors.
DISCUSSION
We designed and performed a whole-genome tiling array analysis of M. leprae RNA expression and demonstrated that pseudogenes and noncoding regions are not silent but instead are strongly expressed. Statistical analysis indicated that RNA expression from noncoding regions was the highest in both peak (Table 1) and mean (Fig. 3) signal intensities and that RNA expression from genes (ORFs) was the lowest. The reliability of the tiling array results was confirmed in part by a comparison with an ORF array, in which multiple gene-specific probes were designed (Fig. 1). RNA expression detected by tiling array was also confirmed by quantitative real-time PCR analysis. Therefore, the tiling array was a reliable tool for the detection of specific RNA expression from M. leprae genome.
The roles of RNA derived from M. leprae noncoding regions and pseudogenes are not known, but the aberrant expression of pseudogenes has been reported in some cancers (22, 35). In addition, a nitric oxide synthase pseudogene is expressed in the central nervous system of the snail Lymnaea stagnalis, and its transcript is thought to have antisense activities (18). Pseudogenes also have some biological functions in processes such as cell growth and organogenesis (16). Computational analysis of the mouse genome showed that 10% of the mRNA fraction can be derived from pseudogenes (11). Our results suggest that pseudogenes and genes are similarly transcribed. If some pseudogenes function to regulate gene expression, it may explain why M. leprae is able to survive with only a limited number of protein-coding genes. Comprehensive analysis of small RNA revealed that small interfering RNAs are expressed from pseudogenes and regulate gene expression (37). In this study, we found that pseudogenes in the functional categories of “degradation” and “energy metabolism” in the “small-molecule metabolism” class were strongly transcribed on a frequent basis. Further functional analysis is needed to elucidate their roles and the reason behind the biased transcription between functional classes. One hypothesis is that pseudogenes are transcribed because the organism has not yet evolved so as to switch them off. The strength of the selective pressure in M. leprae to dispense with useless transcription is unclear.
It has been speculated that the massive genomic degeneration seen in M. leprae is the result of dysfunctional sigma factors (23). Up to 2% of the M. leprae genome consists of repetitive DNA sequences, potential remnants of past transposons (6). Such repetitive sequences are found in pseudogenes in the “other functions” class and in noncoding regions. Of interest, we detected high RNA expression from those regions, suggesting the existence of functional roles now and/or in the past. Mycobacterium ulcerans, a close relative of M. leprae, has a similar genome structure. M. ulcerans has 771 pseudogenes, but the proportion of pseudogenes based on genome size is about 40% of that of M. leprae (34). It was also shown that Mycobacterium marinum has 65 pseudogenes (33). These species appear to have preserved past genomic evolution and heterotrophic circumstances as they adapted.
Except for rRNA and tRNA, noncoding RNAs are classified as components of ribonucleoproteins, ribozymes, or microRNA; the rest are thought to be junk derived from transposons or splicing remnants (25). The noncoding region occupying one-quarter of the M. leprae genome was presumed to be silent. The highly expressed areas of the noncoding regions were thought to be derived from RLEP and LEPREP (6). However, a large number of other noncoding regions that are more highly expressed than genes and pseudogenes have no homology with known sequences of noncoding RNA. Consequently, these RNAs might have a hitherto unrecognized function.
Different classes of M. leprae genes exhibited different levels of RNA expression. RNA expression was relatively high from genes in the “small-molecule metabolism” class related to amino acid and nucleotide synthesis, probably because these small molecules are necessary for protein and RNA synthesis. Moreover, a low level of pseudogene expression in these classification subsets may support the idea that the genes in this class have very essential roles. Similarly, highly expressed genes in the “cell processes” class are responsible for the folding of synthesized proteins. On the other hand, genes related to DNA replication were not strongly expressed, reflecting the fact that the proliferation of M. leprae is very slow. Also, although high expression was not detected in some functional subclasses, such as the “biosynthesis of cofactors, prosthetic groups, and carriers” and “degradation of macromolecules” subclasses, these genes are expressed at a low level (data not shown). In fact, genes targeted by particular drugs are included in these subsets. Thus, RNA polymerase ΙΙΙ and folic acid synthesis genes, targeted by rifampin and dapsone, respectively (8), are not highly expressed (data not shown). These data indicate that high RNA expression does not necessarily correlate with the functional importance of the genes, such as those related to drug resistance.
High expression was detected from lipoproteins and the PE and PPE families, which is characteristic of M. leprae. Lipoproteins function in infection and survival, as exemplified in M. tuberculosis (38). The PE and PPE families are specific to Mycobacterium species and by definition contain a Pro-Glu or Pro-Pro-Glu motif near the N terminus (4). Since the PE and PPE families are associated with the early secreted antigenic target 6-kDa (ESAT-6) antigen (29), they may play an important role in virulence. Because M. leprae has fewer PE, PPE, and ESAT-6-like genes than M. tuberculosis, information on these expressed genes will facilitate further functional analysis of a PE, PPE, and ESAT-6-like protein complex.
There were some differences in the levels of RNA expression detected by tiling array and real-time PCR. The level of expression from coding genes detected by tiling array was lower than the level from these genes detected by real-time PCR, while pseudogene expression was more abundant in the tiling array analysis than in real-time PCR. This discrepancy might reflect the difference in the target length for these methods as well as the difference in the length of transcribed RNA.
The genome size of microbes, as well as the proportion of noncoding regions, is much smaller than that of eukaryotes. Therefore, RNA expression from these regions has been extensively studied. One such study resulted in the discovery of an essential protein homolog, Argonaute, which is necessary for microRNA maturation (13). RNA expression from noncoding regions was also detected from the whole-genome analyses of E. coli (39) as well as Prochlorococcus and Synechococcus spp. (3). The tiling array has facilitated far more in-depth transcriptome analysis, including noncoding regions, than previous techniques such as shotgun cloning (1). For example, a Saccharomyces cerevisiae tiling array analysis identified 98 novel noncoding RNAs (32). The present tiling array will be similarly useful for the identification of noncoding RNA in bacteria (31) and for further functional analysis. This is the first genome-wide expression profile of M. leprae genes, pseudogenes, and noncoding regions, which can used as the foundation for the screening of drug candidates and the study of host-bacillus interactions.
Supplementary Material
Acknowledgments
This work was supported by a grant-in-aid for Scientific Research on Priority Areas from The Ministry of Education, Culture, Sport, Science, and Technology of Japan (to K.S.); by a grant of the Japan Health Sciences Foundation (to T.A.); and by a grant-in-aid for Research on Emerging and Reemerging Infectious Diseases from the Ministry of Health, Labor, and Welfare of Japan (to N.I.).
We thank M. Mishima, P. D. Bang, S. Aizawa, M. Hayashi Y. Ishido, and S. Sekimura (Leprosy Research Center, National Institute of Infectious Diseases) for invaluable discussions and M. Kenmotsu and H. Kawauchi (Roche Diagnostics) for helpful assistance with the tiling array analysis.
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altuvia, S. 2007. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10257-261. [DOI] [PubMed] [Google Scholar]
- 2.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 691679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axmann, I. M., P. Kensche, J. Vogel, S. Kohl, H. Herzel, and W. R. Hess. 2005. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 6R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honoré, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 4091007-1011. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., P. Supply, and N. Honoré. 2001. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr. Rev. 72449-461. [PubMed] [Google Scholar]
- 7.D'Errico, I., G. Gadaleta, and C. Saccone. 2004. Pseudogenes in metazoa: origin and features. Brief. Funct. Genomics Proteomics 3157-167. [DOI] [PubMed] [Google Scholar]
- 8.Dhople, A. M. 2000. Search for newer antileprosy drugs. Indian J. Lepr. 725-20. [PubMed] [Google Scholar]
- 9.Faucher, S. P., S. Porwollik, C. M. Dozois, M. McClelland, and F. Daigle. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. USA 1031906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchini, A. G., and T. Egli. 2006. Global gene expression in Escherichia coli K-12 during short-term and long-term adaptation to glucose-limited continuous culture conditions. Microbiology 1522111-2127. [DOI] [PubMed] [Google Scholar]
- 11.Frith, M. C., L. G. Wilming, A. Forrest, H. Kawaji, S. L. Tan, C. Wahlestedt, V. B. Bajic, C. Kai, J. Kawai, P. Carninci, Y. Hayashizaki, T. L. Bailey, and L. Huminiecki. 2006. Pseudo-messenger RNA: phantoms of the transcriptome. PLoS Genet. 2e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58303-328. [DOI] [PubMed] [Google Scholar]
- 13.Hall, T. M. 2005. Structure and function of argonaute proteins. Structure 131403-1408. [DOI] [PubMed] [Google Scholar]
- 14.Hirotsune, S., N. Yoshida, A. Chen, L. Garrett, F. Sugiyama, S. Takahashi, K. Yagami, A. Wynshaw-Boris, and A. Yoshiki. 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 42391-96. [DOI] [PubMed] [Google Scholar]
- 15.Kampa, D., J. Cheng, P. Kapranov, M. Yamanaka, S. Brubaker, S. Cawley, J. Drenkow, A. Piccolboni, S. Bekiranov, G. Helt, H. Tammana, and T. R. Gingeras. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 14331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kandouz, M., A. Bier, G. D. Carystinos, M. A. Alaoui-Jamali, and G. Batist. 2004. Connexin43 pseudogene is expressed in tumor cells and inhibits growth. Oncogene 234763-4770. [DOI] [PubMed] [Google Scholar]
- 17.Kin, T., K. Yamada, G. Terai, H. Okida, Y. Yoshinari, Y. Ono, A. Kojima, Y. Kimura, T. Komori, and K. Asai. 2007. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 35D145-D148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korneev, S. A., J. H. Park, and M. O'Shea. 1999. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 197711-7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence, J. G., R. W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9535-540. [DOI] [PubMed] [Google Scholar]
- 20.Lin, H., A. Shabbir, M. Molnar, and T. Lee. 2007. Stem cell regulatory function mediated by expression of a novel mouse Oct4 pseudogene. Biochem. Biophys. Res. Commun. 355111-116. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y., P. M. Harrison, V. Kunin, and M. Gerstein. 2004. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 5R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, W., D. Zhou, G. Glusman, A. G. Utleg, J. T. White, P. S. Nelson, T. J. Vasicek, L. Hood, and B. Lin. 2006. KLK31P is a novel androgen regulated and transcribed pseudogene of kallikreins that is expressed at lower levels in prostate cancer cells than in normal prostate cells. Prostate 66936-944. [DOI] [PubMed] [Google Scholar]
- 23.Madan Babu, M. 2003. Did the loss of sigma factors initiate pseudogene accumulation in M. leprae? Trends Microbiol. 1159-61. [DOI] [PubMed] [Google Scholar]
- 24.Mattick, J. S. 2001. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2986-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattick, J. S., and I. V. Makunin. 2006. Non-coding RNA. Hum. Mol. Genet. 15(special no. 1)R17-R29. [DOI] [PubMed] [Google Scholar]
- 26.Nakata, N., M. Matsuoka, Y. Kashiwabara, N. Okada, and C. Sasakawa. 1997. Nucleotide sequence of the Mycobacterium leprae katG region. J. Bacteriol. 1793053-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52947-961. [DOI] [PubMed] [Google Scholar]
- 28.Ochman, H., and L. M. Davalos. 2006. The nature and dynamics of bacterial genomes. Science 3111730-1733. [DOI] [PubMed] [Google Scholar]
- 29.Okkels, L. M., and P. Andersen. 2004. Protein-protein interactions of proteins from the ESAT-6 family of Mycobacterium tuberculosis. J. Bacteriol. 1862487-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overton, T. W., L. Griffiths, M. D. Patel, J. L. Hobman, C. W. Penn, J. A. Cole, and C. Constantinidou. 2006. Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem. Soc. Trans. 34104-107. [DOI] [PubMed] [Google Scholar]
- 31.Rivas, E., R. J. Klein, T. A. Jones, and S. R. Eddy. 2001. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 111369-1373. [DOI] [PubMed] [Google Scholar]
- 32.Samanta, M. P., W. Tongprasit, H. Sethi, C. S. Chin, and V. Stolc. 2006. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc. Natl. Acad. Sci. USA 1034192-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stinear, T. P., T. Seemann, P. F. Harrison, G. A. Jenkin, J. K. Davies, P. D. Johnson, Z. Abdellah, C. Arrowsmith, T. Chillingworth, C. Churcher, K. Clarke, A. Cronin, P. Davis, I. Goodhead, N. Holroyd, K. Jagels, A. Lord, S. Moule, K. Mungall, H. Norbertczak, M. A. Quail, E. Rabbinowitsch, D. Walker, B. White, S. Whitehead, P. L. Small, R. Brosch, L. Ramakrishnan, M. A. Fischbach, J. Parkhill, and S. T. Cole. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18729-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stinear, T. P., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. L. Porter, J. Ryan, P. D. Johnson, J. K. Davies, G. A. Jenkin, P. L. Small, L. M. Jones, F. Tekaia, F. Laval, M. Daffe, J. Parkhill, and S. T. Cole. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suo, G., J. Han, X. Wang, J. Zhang, Y. Zhao, and J. Dai. 2005. Oct4 pseudogenes are transcribed in cancers. Biochem. Biophys. Res. Commun. 3371047-1051. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, K., N. Nakata, P. D. Bang, N. Ishii, and M. Makino. 2006. High-level expression of pseudogenes in Mycobacterium leprae. FEMS Microbiol. Lett. 259208-214. [DOI] [PubMed] [Google Scholar]
- 37.Tanigawa, K., K. Suzuki, K. Nakamura, T. Akama, A. Kawashima, H. Wu, M. Hayashi, S. Takahashi, S. Ikuyama, T. Ito, and N. Ishii. 2008. Expression of adipose differentiation-related protein (ADRP) and perilipin in macrophages infected with Mycobacterium leprae. FEMS Microbiol. Lett. 28972-79. [DOI] [PubMed] [Google Scholar]
- 38.Vandal, O. H., L. M. Pierini, D. Schnappinger, C. F. Nathan, and S. Ehrt. 2008. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med. 14849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 316435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yogi, Y., T. Banba, M. Kobayashi, H. Katoh, N. Jahan, M. Endoh, and H. Nomaguchi. 1999. Leprosy in hypertensive nude rats (SHR/NCrj-rnu). Int. J. Lepr. Other Mycobact. Dis. 67435-445. [PubMed] [Google Scholar]
- 41.Yogi, Y., M. Endoh, T. Banba, M. Kobayashi, H. Katoh, K. Suzuki, and H. Nomaguchi. 2002. Susceptibility to Mycobacterium leprae of congenic hypertensive nude rat (SHR/NCrj-rnu) and production of cytokine from the resident peritoneal macrophages. Jpn. J. Lepr. 7139-45. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 42.Zhou, D., Y. Han, J. Qiu, L. Qin, Z. Guo, X. Wang, Y. Song, Y. Tan, Z. Du, and R. Yang. 2006. Genome-wide transcriptional response of Yersinia pestis to stressful conditions simulating phagolysosomal environments. Microbes Infect. 82669-2678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.