Abstract
Competence for genetic transformation in Streptococcus pneumoniae is a transient physiological state whose development is coordinated by a peptide pheromone (CSP) and its receptor, which activates transcription of two downstream genes, comX and comW, and 15 other “early” genes. ComX, a transient alternative sigma factor, drives transcription of “late” genes, many of which are essential for transformation. In vivo, ComW both stabilizes ComX against proteolysis by the ClpE-ClpP protease and stimulates its activity. Interestingly, stabilization of ComX by deletion of the gene encoding the ClpP protease did not extend the period of competence. We considered the hypothesis that the rapid decay of competence arises from a rapid loss of ComW and thus of its ComX stimulating activity, so that ComX might persist but lose its transcriptional activity. Western analysis revealed that ComW is indeed a transient protein, which is also stabilized by deletion of the gene encoding the ClpP protease. However, stabilizing both ComX and ComW did not prolong either ComX activity or the period of transformation, indicating that termination of the transcriptional activity of ComX is not dependent on proteolysis of ComW.
Streptococcus pneumoniae (pneumococcus) is an important human pathogen with an inherent ability to develop competence for genetic transformation. Development of this temporary state, in which cells can both kill noncompetent neighbors (9) and take up extracellular DNA, depends on a cell-to-cell communication mechanism mediated by a two-component signal transduction system (reviewed in reference 8) (Fig. 1). As could be expected in view of its capacity to turn part of a population into killer cells, this mechanism is tightly regulated at multiple levels. The protein ComA, which belongs to the ABC transporter family of proteins, cleaves ComC and together with ComB secretes the cleavage product, CSP (competence stimulating peptide) (15, 16). As CSP accumulates, it is thought to act through its receptor, the histidine kinase ComD, to autophosphorylate and activate the response regulator ComE (13, 25). Synthetic CSP has the same effect and is useful for controlling development of competence during laboratory culture. Activated ComE acts as a transcription factor, recognizing a direct repeat sequence found upstream of comAB and comCDE, upregulating their transcription, and amplifying the amount of CSP produced and secreted (25, 36). Similar direct repeats also occur upstream of two identical copies of comX and upstream of comW (1, 26, 27, 36). The genes comAB, comCDE, comX, comW, and nine others compose the class known as early genes. Expression of these genes continues for only a few minutes, creating, by unknown mechanisms, a short window of expression followed by a long refractory period in which recently competent cells are unresponsive to CSP. ComX (σX) is an alternative sigma factor which, together with RNA polymerase (RNAP), is responsible for the transcription of a second class of genes, termed late genes, which code for proteins required for genetic transformation and other functions that are only beginning to be understood (27). ComW is also required for development of competence, but its role is not so well understood. Together, ComX and ComW are the only direct output of the cell-to-cell signaling circuit known to be required for transcription of the late genes and for full development of competence for genetic transformation (21, 22).
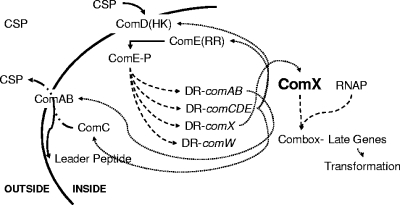
FIG. 1.
Roles of early gene products in a model of the regulation of competence development. CSP, secreted by ComAB, accumulates outside of the cell and interacts with ComD, which autophosphorylates and phosphorylates the response regulator ComE. ComE-P acts as a transcription factor recognizing a direct repeat sequence (DR) upstream of comAB, comCDE, comX, and comW. ComX, together with RNAP, recognizes a promoter sequence, the combox, to transcribe late genes, including genes that encode effector proteins of transformation. ···▸, translation; ---▸, transcriptional regulation; —·▸, transport; ➞, interactions and movement of proteins.
Transcription of the late genes, which total approximately 60, is also tightly regulated, since many are transcribed essentially only during the period of competence that follows coordinated induction of early genes. Late genes are upregulated simultaneously, and termination of their transcription is also synchronized, occurring only a few minutes after induction (27). In attempts to understand how this tight transcriptional regulation is accomplished, attention has been focused on regulation of the activity of ComX. ComX is a transient protein that first appears 5 min after addition of CSP, reaches a maximum at 15 min, and disappears thereafter with a half-life of about 5 min (20). The transient appearance of the ComX protein in vivo (23) suggests that the half-life of ComX may be controlled by one or more proteases.
It has been suggested that the cessation of late gene transcription follows from loss of the ComX sigma factor (8, 10, 23). However, close comparison of the temporal pattern of the level of the ComX protein (which reaches a maximum at 20 min) with that of late gene mRNA levels (which reach a maximum at 12 min), made under substantially similar conditions, suggests that activity of ComX ceases well before its amount declines significantly (23, 26, 27). To clarify the cause of shutoff of late gene expression, it would be useful to be able to manipulate proteolysis directly. ATP-dependent proteases control the half-lives of numerous regulatory proteins, including the stationary-phase sigma factor σS in Escherichia coli (37), the heat shock sigma factor σ32 in E. coli (33), the sporulation sigma factor σH in Bacillus subtilis (19), and the competence transcription factor ComK in B. subtilis (35). Among the four classes of ATP-dependent proteases known in bacteria, the pneumococcal genome contains single clpP and ftsH genes but lacks lon and hslUV genes (14, 34). clpP mutations can derepress competence development in pneumococcus (7, 28) and σX accumulation in group A streptococcus (24), indicating a direct or indirect negative regulatory impact on ComX activity. Since Clp proteolysis requires assembly of a multimeric complex including the serine peptidase ClpP and an ATPase subunit of the AAA+ family that mediates the specificity of substrate selection (31), we examined the stability of the ComX protein in vivo in a comprehensive set of clp mutants in order to identify a ComX-specific protease. Here we identify specific AAA+ subunits involved in proteolysis of both ComW and ComX and show that both transformation and transcription of late genes are strictly downregulated during escape from competence by a mechanism distinct from proteolysis.
MATERIALS AND METHODS
Strains, plasmids, and culture media.
The strains used are listed in Table 1. Strain CP1361 has a clpX deletion, even though clpX has been reported to be essential in S. pneumoniae (29). Careful examination of this strain revealed a second, unlinked mutation (designated slx) suppressing the lethality of the clpX deletion (data not shown). Plasmids used were pCKS01 (32) and pR410 (2). S. pneumoniae was grown in complete CAT medium (17), while E. coli was grown in LB medium (3). Antibiotics were used at the following concentrations: for S. pneumoniae, novobiocin, 2.5 μg/ml; erythromycin (Em), 0.25 μg/ml; tetracycline (Tc), 0.25 μg/ml; kanamycin (Kan), 200 μg/ml; and chloramphenicol (Cm), 2.5 μg/ml; and for E. coli, ampicillin, 100 μg/ml. Synthetic CSP-1 was obtained from Chiron Mimotopes (Raleigh, NC).
TABLE 1.
Strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| S. pneumoniae | ||
| CP1250 | Rx derivative; low β-galactosidase activity; hex malM511 str-1 bgl-1; Hex− Mal−-Smr Bga− | 25 |
| CP1343 | CP1250, but ΔclpL::PcEm; Emr | This study |
| CP1344 | CP1250, but ΔclpC::PcTet; Tcr | This study |
| CP1359 | CP1250, but ΔclpP::PcTet; Tcr | This study |
| CP1360 | CP1250, but ΔclpE::PcKan; Kanr | This study |
| CP1361 | CP1250, but ΔclpX::PcEm slx; Emr | This study |
| CP1376 | CP1250, but ΔcomW::KANT; Kanr | 32 |
| CP1500 | Rx derivative; hex novo-r byra-r str-r1 ery-rG ery-r2; Novr Emr Strr | 6 |
| CP1851 | CP1250, but ΔclpE::PcEm; Emr | 32 |
| CP1890 | CPM7, but ΔclpP::PcTet; Tcr Cmr | CPM7 × CP1359a |
| CPM7 | CP1250, but ssbB::pEVP3::ssbB+; SsbB+ Cmr | 18 |
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Invitrogen |
Recipient × donor; see Materials and Methods.
Construction of mutant strains used in this study.
To create strain CP1343 (ΔclpL::PcEm), a fragment upstream of clpL and containing part of clpL was amplified from CP1500 using the primers PL49 and PL50, a downstream fragment was amplified from CP1500 using PL51 and PL52, and a third fragment containing the PcEm marker was amplified from amplicon aMSL1 (18) using PL46 and PL47 (primers are listed in Table 2). After digestion by BamHI and/or EcoRI, the three fragments were purified, ligated, and used directly as a donor for transforming strain CP1250 with selection for Emr. (This was done by growing a 10-ml culture of strain CP1250 in complete CAT medium plus 10 mM HCl to an optical density at 550 nm [OD550] of 0.06 and inducing development of competence with CaCl2 (to 0.5 mM), bovine serum albumin (to 0.002%), and CSP-1 (to 250 ng/ml). After 5 min, 1 ml of the induced cells was incubated with 100 ng of donor DNA for 70 min. The other clp mutant strains were created similarly. For CP1344 (ΔclpC::PcTet), the upstream fragment was amplified using PL56 and PL57, the downstream fragment was amplified using PL58 and PL59, and the insert with the PcTet marker was amplified from the amplicon aMSL4 (18) using PL46 and PL48. These fragments were digested by XbaI and/or HindIII. For CP1359 (ΔclpP::PcTet), the upstream fragment was amplified using PL66 and PL65, the downstream fragment was amplified using PL63 and PL64, and the insert with the PcTet marker was amplified from the amplicon aMSL4 using PL46 and PL48. These fragments were digested by ApaI and/or KpnI. For CP1360 (ΔclpE::PcKan), the upstream fragment was amplified using PL68 and PL67 and the downstream fragment using PL69 and PL70, and the insert with a PcKan marker was amplified from the plasmid pR410 using PL41 and PL42. The three fragments were digested by BamHI and/or ApaI before ligation. For CP1361 (ΔclpX::PcEm), the upstream fragment was amplified using PL72 and PL71, the downstream fragment using PL73 and PL74, and the insert with the PcEm marker from aMSL1 using PL46 and PL47. The three fragments were digested by XbaI and/or HindIII. Each mutation was confirmed by sequencing of a fragment amplified using the outermost primers for each of the constructs. To create strain CP1890 (ssbB::lacZ clpP::PcTet), CP1359 DNA was used to transform strain CPM7 to Tcr as described above. Colonies from selective plates were characterized further, using PCR to confirm the mutation. In parallel, the picked colonies were also used to inoculate 4 ml of complete CAT medium plus the appropriate drug. These cultures were grown to an OD550 of 0.1 and stored as a 15%-glycerol stock at −80°C. One verified clone was retained as CP1890.
TABLE 2.
Primers used in this study
| Primer name | Gene or marker | Primer sequence (5′-3′)a |
|---|---|---|
| PL41 | PcKan | TGGGGATCCGTTTGATTTT |
| PL42 | PcKan | ATGGGCCCTATGGACAGTTGCGGATGTA |
| PL46 | PcTet, PcEm | GGTCTAGAGGATCCGGGTACCGGGCCCAAAATTTGTTTGATTTGT |
| PL47 | PcEm | GGGGTACCGAAGCTTGGGATCCGGAATTCAGTCGGCAGCGACTCATAGAATTA |
| PL48 | PcTet | GGGGTACCGAAGCTTGGATCCCCCAAAGTTGATCCCTTAACG |
| PL49 | mraY | CCGTATGACGCCTGTACATCAC |
| PL50 | clpL | CGTCCGATAACAGGATCCAACT |
| PL51 | clpL | TCTTCCGTCCAGAATTCCTCAA |
| PL52 | luxS | CGGCCTTCACACTATCGAGCAC |
| PL56 | spr2002 | GTGTAGCCTTACTCCGGACCTACCT |
| PL57 | clpC | ACAAGTGCCACGACTCTAGATAACG |
| PL58 | clpC | ccgaagcttAGGCAGCACACTTAAGATTG |
| PL59 | spr1998 | TCTTGTCCATCCTTAGCCATAATGA |
| PL63 | clpP | TACAGGTGGTGGTACCCAACAAACT |
| PL64 | livJ | GCAGGTCCTACGACTGCTGATACTT |
| PL65 | clpP | atgggcccTTTGTTGGTCAAATGACTGAAGATA |
| PL66 | comEB | TCAACCAGTACCGAATGGACGACTA |
| PL67 | clpE | cgcggatccTGGTGTAAAGATGAATTGTTGAGTC |
| PL68 | glnP | AGTCTCCGAAACATAAGCACCACTA |
| PL69 | clpE | atgggcccTCTAGCCGAAGATCTCAAGTCTCAT |
| PL70 | clpE | AAAGGGCAAGCTGTCAAGAAATTAC |
| PL71 | clpX | gctcTAGACATGATTTCCTTCCATTCTATACTG |
| PL72 | dpr | CGTGTTCTTGTTATCTACCGTTACT |
| PL73 | clpX | cccaagctttcATGTTTGAGGTGCCGAGTCAGGA |
| PL74 | aldR | TTACATCACGAGGAAGACGAGCTAC |
Lowercase, 5′ extension; underscore, restriction site.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described previously (30) using the Bio-Rad Mini-Protean II gel apparatus. Each gel was a 15-well, 1.5-mm-thick discontinuous gel composed of a 5% stacking gel and a 15% resolving gel prepared according to the manufacturer's recommendations. Protein samples were prepared by mixing with one volume of loading buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 0.2% bromophenol blue, 20% glycerol, and 200 mM dithiothreitol) and heating at 95°C for 10 min. The gels were run in 25 mM Tris, 250 mM glycine, 0.1% SDS, pH 8.0, at 65 V until the dye reached the resolving gel and then at 120 V until the dye reached the bottom of the gel.
Purification of His6-ComW.
An overnight culture of E. coli strain BL21(DE3)(pCKS01) in LB broth plus 100 μg/ml ampicillin was diluted 1:50 in 1 liter LB plus 100 μg/ml ampicillin in a 4-liter flask. The culture was grown at 37°C with shaking at 200 rpm to an OD600 of 0.6. After addition of isopropyl-β-d-thiogalactopyranoside to 0.5 mM and aeration at 37°C for four additional hours, the cells were chilled and harvested (7,000 × g, 20 min, 4°C). The weighed wet cell pellet was stored at −80°C, thawed on ice, and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl, 0.1 mM phenylmethylsulfonylfluoride) at 5 ml per g of cell pellet. After sonication (24 10-s pulses) on ice, the lysate was centrifuged at 10,000 × g for 20 min at 4°C. The pellet was resuspended in 15 ml wash buffer (2% Triton X-100 [vol/vol], 3 M urea, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0], 100 mM NaCl) per g of cell pellet and stirred for 1 h at room temperature. The washed inclusion bodies were repelleted (10,000 × g, 20 min, 4°C), and the wash was repeated. After a further wash in 15 ml lysis buffer per g of cell pellet, the inclusion bodies were dissolved in solubilization buffer (6 M guanidine HCl, 100 mM NaH2PO4, 10 mM Tris, 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, pH 8.0) at 15 ml per g of cell pellet by stirring for 1 h at room temperature. After centrifugation at 10,000 × g for 20 min at 4°C, the supernatant was loaded at 0.25 ml/min onto a nickel affinity column (nickel-nitrilotriacetic acid agarose; Qiagen) with a bed volume of 1 ml per g of cells. After washing with 10 column volumes of column wash buffer (50 mM NaH2PO4, 300 mM NaCl, 6 M urea, 30 mM imidazole, pH 8.0), bound protein was eluted with 4 column volumes of elution buffer (column wash buffer with 300 mM imidazole). Twenty-four fractions were collected and analyzed by SDS-PAGE as described above. The fractions with ComW were pooled and dialyzed against two changes of phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), 1 liter each, one for 4 h and one overnight. The protein began to precipitate after 10 min. The resulting protein suspension, at a concentration of 3 mg/ml and >95% pure, was used for antibody production in rabbit (Orbigen, Inc., San Diego, CA). A total of 2 mg of ComW was obtained per g of cells (wet weight).
Characterization of antibody to ComW.
To characterize antibody to ComW, sera were tested for activity using an aliquot of the ComW protein preparation used for antibody production. The preimmune serum did not react with the protein preparation, while serum from the second bleed (used throughout this study) reacted strongly with His6-ComW and four other minor components remaining in the inclusion body extract (Fig. 2). When pneumococcal cell extracts deficient for comW were used to test the antibody, the antibody reacted with two slowly migrating components, which are apparently highly antigenic and shared by S. pneumoniae and E. coli. However, when the antibody was tested against wild-type pneumococcal cell extracts, a third band appeared, corresponding to a molecular weight of 10,000, the same molecular weight as ComW, but only if the cells had been exposed to CSP for 10 or 15 min.
FIG. 2.
Characterization of antibody to ComW. Western blot analysis of anti-ComW antiserum against pneumococcal cell extracts or His-tagged ComW purified from E. coli cells. Samples are from CP1250 or CP1376 cultures induced to competence with CSP and then sampled after incubation for 0, 5, 10, and 15 min. His-tagged ComW was purified as described in Materials and Methods. Also shown is a lane from SDS-PAGE stained with Bio-Safe Coomassie (Bio-Rad) showing the His-tagged ComW preparation (His6-ComW), used as an antigen for raising ComW antibody.
Western analysis.
For Western analysis, SDS-PAGE gels were run as described above, washed three times for 5 min with 100 ml deionized water, and equilibrated for 15 min in transfer buffer (25 mM Tris, 192 mM glycine, 10% [vol/vol] methanol, pH 8.0). The proteins were transblotted from the gel to a polyvinylidene difluoride membrane (Immobilon Psq; Millipore) in transfer buffer for 2 h at 36 V at 4°C. The membrane was then blocked overnight at 4°C in TBS-T (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 1% [vol/vol] Tween 20) with 5% nonfat dry milk. The membrane was incubated for 90 min at room temperature with primary antibody specific for ComW or ComX, diluted 1:3,000 in TBS-T with 1% nonfat dry milk. After three washes with 100 ml TBS-T and then incubation for 1 h at room temperature with secondary antibody (anti-rabbit immunoglobulin G conjugated to horseradish peroxidase; Amersham) diluted 1:20,000 in TBS-T with 1% nonfat dry milk and then three washes with 100 ml TBS-T, the position of secondary antibody on the membrane was detected using an ECL substrate (ECL Plus; Amersham) and either Hyblot CL film (Denville Scientific) or the Alpha Imager charge-coupled-device (CCD) camera (Alpha Innotech). Typical exposure times were 1 to 5 min for the film and 5 to 15 min for the CCD camera. Quantification was done by spot densitometry analysis using AlphaEaseFC (Alpha Innotech).
Culture growth and analysis of response to CSP.
Cultures of pneumococcus were started by inoculating complete CAT medium plus 10 mM HCl with a 1/100 volume of a frozen stock of cells (OD550 = 0.1). At the first signs of turbidity during growth at 37°C, 10 ml was transferred to an 18-mm-by-150-mm tube for monitoring optical density. When the culture reached an OD550 of about 0.06, it was induced to competence with CaCl2 (to 0.5 mM), bovine serum albumin (to 0.002%), and CSP (to 250 ng/ml). Samples were taken periodically from the culture for various analyses as described in subsequent sections.
For the Western blot analyses represented in Fig. 3, samples were processed as described in reference 23. For the Western blot analyses represented in Fig. 4 and 5, 1.8-ml samples were withdrawn from the culture, chilled rapidly on dry ice without freezing, and then kept at 4°C until harvest by centrifugation (10,000 × g, 2 min, 4°C). After each cell pellet was resuspended with 35 μl loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 100 mM dithiothreitol) and heated at 95°C for 10 min, 15 μl of the lysate was loaded into one lane of an SDS-PAGE gel.
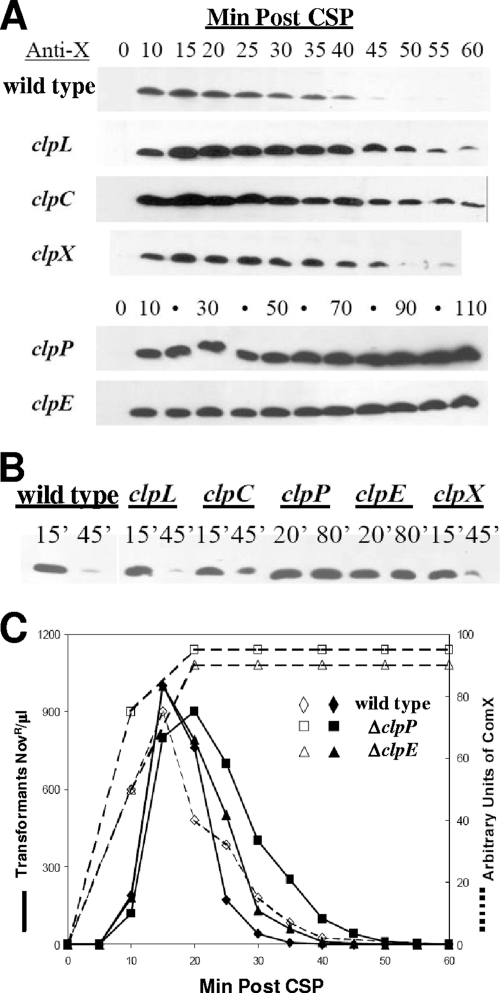
FIG. 3.
Transience or stability of ComX in Clp mutants. (A) Cell lysates from wild-type CP1250 and clp mutants, CP1343 (ΔclpL), CP1344 (ΔclpC), CP1359 (ΔclpP), CP1360 (ΔclpE), and CP1361 (ΔclpX), were analyzed using Western blotting with anti-ComX antibody. Ten-milliliter samples of each mutant culture were harvested at 5- or 10-min intervals after CSP treatment (Min Post CSP) at 37°C. (B) Comparison of ComX peak levels and decay. The 15-min and 45-min samples of each strain above (but 20 min and 80 min for ΔclpP and ΔclpE mutants) were adjusted to equalize total protein amounts, analyzed on a single gel, and probed with anti-ComX antibody simultaneously. (C) Kinetics of response to CSP. CP1250 (wild type), CP1359 (ΔclpP), and CP1360 (ΔclpE) were induced by CSP at 37°C. One-milliliter samples, withdrawn at 5-min intervals after induction, were exposed to 1 μg novo-r DNA for 5 min at 37°C, treated with 20 ng/ml DNase I for 55 min at 37°C, and plated for Novobiocin-resistant transformants (solid lines). Meanwhile, 10-ml culture samples were collected and prepared for Western analysis using anti-ComX antibody. ComX protein levels in Western blots were quantified using the Alpha Imager camera and adjusted to arbitrary units (dashed lines).
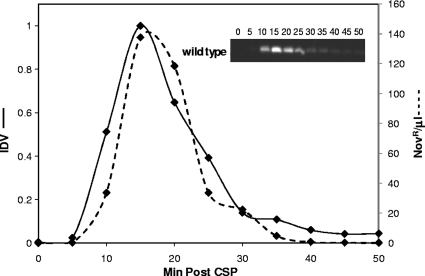
FIG. 4.
Transience of ComW. Western analysis of the relative levels of ComW (solid lines) in strain CP1250 (wild type) at different times during exposure to CSP (Min Post CSP) as described in Materials and Methods. Transformation kinetics of the culture is also plotted (dashed lines) as described in Materials and Methods. The inset represents an immunoblot scanned directly using the CCD camera. IDV, relative integrated density value, normalized to a maximum value of 1.
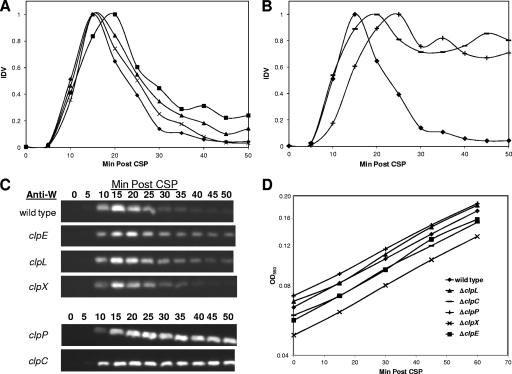
FIG. 5.
Transience or stability of ComW in Clp mutants. (A) Western analysis of relative amounts of ComW in strains CP1250 (⧫; wild type), CP1851 (▪; ΔclpE), CP1343 (▴; ΔclpL), and CP1361 (×; ΔclpX). (B) Western analysis showing relative amounts of ComW in strains CP1250 (⧫; wild type), CP1359 (+; ΔclpP), and CP1344 (−, ΔclpC). (C) Representative immunoblot images, scanned directly using an Alpha Imager camera. (D) Growth curves of CP1250 (wild type), CP1851 (ΔclpE), CP1343 (ΔclpL), CP1361 (ΔclpX), CP1359 (ΔclpP), and CP1344 (ΔclpC). IDV, relative integrated density value, determined as for Fig. 4; Min Post CSP, minutes from start of CSP exposure.
For measurement of β-galactosidase (β-Gal) activity, a 1.8-ml sample of liquid culture was chilled on ice. From this sample, three 0.4-ml portions were transferred to tubes containing 100 μl 5× Z buffer (300 mM Na2HPO4, 200 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, 0.5% Triton X-100) and kept on ice. After 75 min, all samples were heated at 37°C for 10 min to lyse the cells and then kept at room temperature. One hundred fifty microliters of each lysate was added to 50 μl of o-nitrophenyl-β-d-galactopyranoside solution (4 mg/ml o-nitrophenyl-β-d-galactopyranoside, 60 mM Na2HPO4, 40 mM NaH2PO4) in a 96-well microplate. The plate was incubated at 37°C, and absorbance at 420 nm was read automatically every 10 min for 90 min. The average slope of the three samples was used to calculate LacZ activity, reported in Miller units as described previously (30).
To measure transformation, three 1-ml samples were withdrawn from the culture and mixed with 25 ng of CP1500 chromosomal DNA for continued incubation. After 3.5 min, 0.1 ml of the mixture was added to 1.6 ml of CAT with 10 μg/ml DNase I, incubated at 37°C for 70 min, mixed with 1.5 ml of molten CAT agar, and poured onto plates containing 3 ml of CAT agar. After 3 ml of CAT agar was poured over the cell layer, a fourth 3-ml layer of CAT agar with 10 μg/ml novobiocin was poured over the hardened third layer. Novobiocin-resistant colonies were counted after overnight incubation at 37°C. Transformation is reported as the average for the three samples.
RESULTS
Degradation of the ComX protein depends largely on a ClpEP protease complex.
To define the cause of the instability of ComX with more specificity, individual deletions of the clpC, clpE, clpL, and clpX ATPase subunit genes and the clpP serine protease subunit gene were constructed by gene replacement mutagenesis in S. pneumoniae CP1250. All mutants were obtained using standard transformation at 37°C, except that 30°C was necessary to recover the clpP and clpX mutants, as noted by Robertson et al. (28). Structures of the deletion mutations were verified by PCR of both novel joints and verified by sequencing whole fragments containing the heterologous inserts and the homologous flanking DNA. To visualize the temporal pattern of ComX accumulation and loss in these protease mutants, cell lysates were prepared from each mutant at 5-min or 10-min intervals after CSP induction and subjected to Western blotting analyses with an anti-ComX antibody (Fig. 3A). The ComX protein appeared by 10 min in all mutants and was maximal at 15 or 20 min, as in the wild-type background. It then decayed in the wild type and in the clpL, clpC, and clpX mutants but remained at a high level in the clpP and clpE mutants for about 2 h, indicating that ClpP and ClpE both participate in ComX degradation. To compare ComX levels in the different mutants more directly, we applied equivalent total protein amounts from samples with peak levels of ComX on a single gel and probed it with anti-ComX antibody (Fig. 3B). ComX persisted robustly only in the clpP and clpE mutants, where it remained at the same level as the maximum found in the wild type for more than 80 min. ComX protein levels in the clpP and clpE mutants are also compared to the relative amount of ComX in the wild type in Fig. 3C. Since deletions of clpL and clpX had little or no effect on ComX accumulation or loss and since the loss of clpC stabilized ComX much less than did the loss of ClpE or ClpP, ClpP and ClpE appear to form the major Clp protease responsible for the degradation of ComX after expression of early genes ceases and cells escape from the competent state.
Since it has been proposed that a shutoff of competence reflects the disappearance of ComX (8, 10, 23), we asked whether the persistent high level of ComX that can be achieved in a clpP mutant would cause an extended high level of competence. The transformation kinetics for the clpP (CP1359) and clpE (CP1360) mutants after induction by synthetic CSP were compared directly to the wild-type pattern (Fig. 3C). Surprisingly, competence in both clp mutants had nearly the same temporal pattern as in the wild type (CP1250), with no more than a slight delay in exit from competence. In the other clp mutants, the patterns of induction and shutoff of transformability were also substantially the same as in the wild type (data not shown). Since the prolonged life of ComX was not accompanied by an extended period of competence, we conclude that ComX activity is also regulated by some other mechanisms in addition to the degradation of the protein itself. This might be explained by the requirement for ComW in transformation, since it appears to be strictly necessary for the activity of ComX (32).
ComW is a transient competence-specific protein.
Since ComW appears to be a key regulator of ComX activity, we asked whether it is unstable and whether its loss could explain the rapid shutoff of competence independent of the persistence of ComX. Transcription of comW is strongly upregulated in response to CSP (27), but the amounts and fates of its putative protein product were unknown. To allow study of the behavior of this protein, we obtained a polyclonal rabbit anti-ComW antiserum (see Methods) (Fig. 2). Using this serum in Western blots, we determined the temporal pattern of the appearance of the ComW protein by analyzing cell extracts from cultures treated with CSP to induce competence development. The Western blots revealed that ComW was indeed unstable, appearing briefly in cells developing competence (Fig. 4). ComW was first detectable 5 min after addition of CSP, reached a maximum approximately 100-fold above the background level at 15 min, and nearly disappeared by 30 min, consistent with a half-life of about 5 min. This pattern is very similar to that displayed by ComX (Fig. 3) (23), and both are similar to the temporal pattern of transformation, consistent with the idea that ComW might be controlling ComX activity in the clpP background.
ComW is transient in clpE, clpX, and clpL mutants but stable in clpP and clpC mutants.
To examine whether the instability of ComW was also dependent on a specific Clp protease, the temporal pattern of ComW accumulation was determined in each of the available clp mutants as described above for ComX. Unlike ComX, ComW appeared transiently in a clpE mutant. It was also transient in the clpX and clpL mutants (Fig. 5A and C), but like ComX, ComW became stable in a clpP mutant. Finally, it was also stable in a clpC mutant (Fig. 5B and C), suggesting that the protease ClpP and the AAA+ specificity factor ClpC together are responsible for the degradation of ComW.
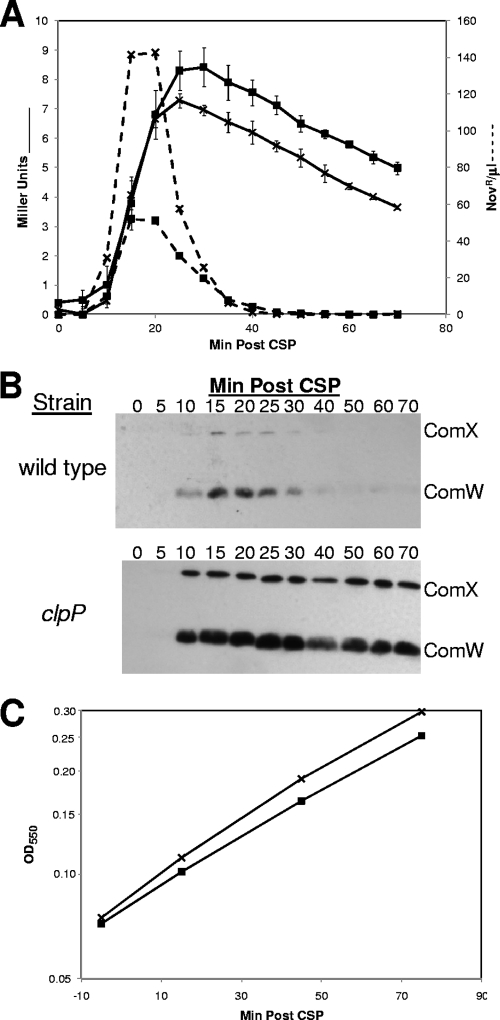
Transcription of late genes terminates despite persistently high levels of ComX and ComW in a stabilizing protease-deficient background.
As mentioned above, in clpP mutants, competence follows wild-type kinetics, with a rapid decline after 15 to 20 min, despite the stability of ComX in this background (Fig. 3C). Because ComW is required for activity of ComX (32), the rapid termination of transformation in a clpP mutant could be explained if the level of ComW drops rapidly in this background, leading to a rapid loss of ComX activity and a consequent shutoff of expression of late genes. However, the stability of ComW in a clpP mutant, revealed in Fig. 5, implies that neither the termination of competence nor the loss of late gene expression could be due simply to the loss of ComW. We wondered whether competence might be decoupled from late gene transcription so that competence was shut off despite continued transcription of late genes during the prolonged presence of both ComX and ComW in the clpP background. To define more precisely the regulatory consequences of abrogating the proteolysis of ComX and ComW, we examined specifically whether the transcription of late genes is prolonged when ComX and ComW are maintained at maximal levels in a clpP background by use of a transcriptional fusion of lacZ with the promoter of the late gene, ssbB. As expected, the strains showed similar temporal patterns of transformability but differed in temporal patterns of the amounts of ComX and ComW. Specifically, both proteins were stable in the clpP mutant but transient in the wild type (Fig. 6B). The amounts of β-Gal produced by the two strains differed slightly, as might be explained by the higher levels of ComX and ComW in the clpP background. However, the temporal patterns of β-Gal activity following CSP treatment were the same in both wild-type and clpP mutant backgrounds (Fig. 6A): in each case, LacZ activity rose sharply as cells developed competence, reached a maximum at about 22 min, and stabilized thereafter. Thus, it is clear that late gene expression terminated in the clpP mutant at about the same time as in the wild-type strain. Therefore, we conclude that late gene expression is brought to an end by a mechanism different from proteolysis of ComX and ComW.
FIG. 6.
Transient expression of the late gene ssbB in clpP and wild-type backgrounds. (A) β-Gal activity (Miller units) (-) along with transformation kinetics (- - -) in strains CPM7 (×; wild type) and CP1890 (▪; ΔclpP). (B) Western analysis of samples taken in parallel and probed with antisera specific to both ComX and ComW using the ECL substrate and imaged on film (see Materials and Methods). (C) Growth curves of cultures used in panel A: CPM7 (×; wild type) and CP1890 (▪; Δclp).
DISCUSSION
While genetic studies had indicated a role for ComW in reducing proteolysis of ComX and microarray data show that comW expression responds dramatically to competence pheromone in the temporal pattern termed “early,” nothing was known prior to this study about the presence of the ComW protein itself. Here we report that ComW accumulated rapidly in response to CSP, first appearing after 5 min of exposure to CSP, consistent with the existing microarray data (27), and reaching its maximal level 10 min later. Interestingly, the ComW temporal pattern is similar to that of the key regulator of competence, ComX. Both proteins were transient, accumulating and disappearing in parallel in wild-type cells. Since the transience of ComX depended on both clpP and clpE, we infer that ClpE-ClpP forms a proteolytic machine responsible for ridding competent cells of ComX. Since ComW became stable in clpP and clpC mutants, we conclude that ComW is subject to proteolysis by a ClpP protease as well but directed by a different AAA+ specificity subunit.
ComW appears to have two distinct roles in establishing the competent state. Deletion of comW reduces ComX levels 10-fold, but this reduction is reversed completely by deletion of clpP or clpE, showing that ComW is needed for accumulation of normal levels of ComX only in the presence of a functional ClpE-ClpP protease and suggesting that it acts to antagonize this protease. However, this antagonism is not absolute, since postcompetence stabilization of ComW in clpC mutants did not completely prevent the decay of ComX. In addition to protecting ComX from proteolysis, ComW has a second role in induction of competence, since it is required for a high level of transcription of late genes even when ComX is produced at a high level due to protection by deletion of clpE or clpP (32). It is unknown how ComW works to complete its two roles. However, here we have shown that the temporal maxima of ComX and ComW coincide at 15 min post-CSP induction, affording ample opportunity for the two proteins to interact directly. Two regulatory systems in E. coli illustrate how these functions could be carried out. The protein Crl, which stimulates activity of and has been thought to assist the alternative sigma factor, σs, in binding to RNAP to form the holoenzyme, interacts directly with σs and RNAP (4, 12). ComW may interact directly with ComX to promote its transcriptional activity in a similar way. This could also be consistent with a role for ComW in reducing proteolysis of ComX, perhaps by obscuring the signal by which ComX is recognized by ClpE. Another protein with a protective function like that of ComW is IraP. In E. coli, IraP interacts with the ClpX-ClpP adaptor RssB to protect σs from proteolysis (5). Similarly, ComW may protect ComX by interacting with an (unknown) proteolytic adaptor for ComX or by interacting directly with ClpE to reduce its activity. In E. coli, two different proteins with two different functions accomplish what ComW seems able to do by itself. It is unknown whether this reflects interaction with a single partner or two different ones.
A separate regulatory question is raised by the observation that cells escape from the competent state as rapidly in a clpP mutant, where ComX and ComW are stable, as in a wild-type strain. Although it has been thought that competence and late gene transcription might terminate due simply to the disappearance of ComX, it now appears that this cannot be the only mechanism by which late gene transcription terminates. Since ComX and ComW are the only two CSP-induced early proteins required for late gene transcription and therefore competence, it could have been predicted that their persistence in a clpP mutant would cause late gene transcription to continue for a longer period than in the wild type, giving rise to an extended period of expression of late genes and of competence. We tested this prediction by monitoring the temporal pattern of transcription of the late gene ssbB in both wild-type and clpP mutant backgrounds. Since the patterns in the two strains were nearly identical, we conclude that transcription of this late gene is not significantly prolonged by the absence of ClpP. While other late genes may in principle behave differently, this result strongly suggests that soon after competence induction, the transcription of late genes becomes subject to strict regulation that is independent of the amounts of ComX and ComW in the cell and that brings about an orderly exit from the competent state.
Regulators critical to initiating the switch to the competent state thus appear to be different from those critical for escape from that state. While the mechanism of the former is understood to involve transcriptional upregulation and evasion of proteolysis, the mechanism of the latter regulation remains to be discovered. Certainly regulation of the activity of ComX or ComW is likely to be a part of the mechanism. In addition to proteolysis, ComX or ComW may be modified or sequestered so that transcription is blocked despite their continued physical presence, or their activity may be indirectly shut off by downregulating combox promoters. Or if late mRNA is in fact still made, it may be destabilized or blocked from translation by protein or RNA products of late genes. Apparently, either at 40 min there is a factor missing that was present at 15 min to allow ComX (or ComW) activity or a new factor is present at 40 min that was missing at 15 min and that blocks ComX activity. Since ComW is a factor that appears by 15 min, it may be its activity that is directly regulated, indirectly reducing the activity of ComX. Since the principal elements that are known to change during this window of time are early and late mRNAs and their protein products, these are therefore the primary candidates for such a factor.
Finally, the decay of transformability as late gene expression ceases will require additional explanation. It may be explained by accumulation of an inhibitor or by instability of one or more late gene effectors, such as CoiA (11), which is unstable and plays an important part in DNA integration.
Acknowledgments
This material is based upon work supported in part by the National Science Foundation under grant no. MCB 0543187.
We thank an anonymous reviewer for especially insightful comments during preparation of this report and Haiying Li for assistance in strain construction.
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 2975-83. [DOI] [PubMed] [Google Scholar]
- 2.Berge, M., M. Moscoso, M. Prudhomme, B. Martin, and J. P. Claverys. 2002. Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45411-421. [DOI] [PubMed] [Google Scholar]
- 3.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bougdour, A., C. Lelong, and J. Geiselmann. 2004. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 27919540-19550. [DOI] [PubMed] [Google Scholar]
- 5.Bougdour, A., S. Wickner, and S. Gottesman. 2006. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev. 20884-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cato, A., Jr., and W. R. Guild. 1968. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double cross-over model. J. Mol. Biol. 37157-178. [DOI] [PubMed] [Google Scholar]
- 7.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 1837295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claverys, J. P., and L. S. Havarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7d1798-d1814. [DOI] [PubMed] [Google Scholar]
- 9.Claverys, J. P., B. Martin, and L. S. Havarstein. 2007. Competence-induced fratricide in streptococci. Mol. Microbiol. 641423-1433. [DOI] [PubMed] [Google Scholar]
- 10.Claverys, J. P., M. Prudhomme, and B. Martin. 2006. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu. Rev. Microbiol. 60451-475. [DOI] [PubMed] [Google Scholar]
- 11.Desai, B. V., and D. A. Morrison. 2006. An unstable competence-induced protein, CoiA, promotes processing of donor DNA after uptake during genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 1885177-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaal, T., M. J. Mandel, T. J. Silhavy, and R. L. Gourse. 2006. Crl facilitates RNA polymerase holoenzyme formation. J. Bacteriol. 1887966-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21863-869. [DOI] [PubMed] [Google Scholar]
- 14.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 1835709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui, F. M., and D. A. Morrison. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hui, F. M., L. Zhou, and D. A. Morrison. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 15325-31. [DOI] [PubMed] [Google Scholar]
- 17.Lee, M. S., B. A. Dougherty, A. C. Madeo, and D. A. Morrison. 1999. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl. Environ. Microbiol. 651883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 1815004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, J., W. M. Cosby, and P. Zuber. 1999. Role of lon and ClpX in the post-translational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol. Microbiol. 33415-428. [DOI] [PubMed] [Google Scholar]
- 20.Luo, P. 2003. Genetic transformation in Streptococcus Pneumoniae: regulation by ComX an alternative sigma factor. Ph.D. thesis. University of Illinois—Chicago.
- 21.Luo, P., H. Li, and D. A. Morrison. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50623-633. [DOI] [PubMed] [Google Scholar]
- 22.Luo, P., H. Li, and D. A. Morrison. 2004. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 54172-183. [DOI] [PubMed] [Google Scholar]
- 23.Luo, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2003. Expression of the secondary sigma factor σX in Streptococcus pyogenes is restricted at two levels. J. Bacteriol. 1854291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestova, E. V., L. S. Havarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21853-862. [DOI] [PubMed] [Google Scholar]
- 26.Peterson, S., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 1826192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 511051-1070. [DOI] [PubMed] [Google Scholar]
- 28.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 1843508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson, G. T., W. L. Ng, R. Gilmour, and M. E. Winkler. 2003. Essentiality of clpX, but not clpP, clpL, clpC, or clpE, in Streptococcus pneumoniae R6. J. Bacteriol. 1852961-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Schirmer, E. C., J. R. Glover, M. A. Singer, and S. Lindquist. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21289-296. [PubMed] [Google Scholar]
- 32.Sung, C. K., and D. A. Morrison. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 1873052-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatsuta, T., T. Tomoyasu, B. Bukau, M. Kitagawa, H. Mori, K. Karata, and T. Ogura. 1998. Heat shock regulation in the ftsH null mutant of Escherichia coli: dissection of stability and activity control mechanisms of sigma32 in vivo. Mol. Microbiol. 30583-593. [DOI] [PubMed] [Google Scholar]
- 34.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 35.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 176730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ween, O., P. Gaustad, and L. S. Havarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33817-827. [DOI] [PubMed] [Google Scholar]
- 37.Zhou, Y., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 1801154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]