Abstract
Cobalamin-independent methionine synthase (MetE) catalyzes the final step in Escherichia coli methionine biosynthesis but is inactivated under oxidative conditions, triggering a methionine deficiency. This study demonstrates that the mutation of MetE cysteine 645 to alanine completely eliminates the methionine auxotrophy imposed by diamide treatment, suggesting that modulation of MetE activity via cysteine 645 oxidation has significant physiological consequences for oxidatively stressed cells.
In addition to its well-known role as one of the amino acid building blocks of proteins, methionine also influences numerous processes in Escherichia coli, including methylation reactions, initiation of translation, and DNA replication. Thus, it is not surprising that several mechanisms exist for control of its biosynthetic pathway. In E. coli, metabolite flux is largely regulated at the transcriptional level by the MetJ and MetR systems (16). However, we have also recently discovered that E. coli appears to arrest methionine biosynthesis under oxidative stress conditions by modulating the activity of the enzyme that catalyzes the terminal step in de novo methionine biosynthesis, cobalamin-independent methionine synthase (MetE) (17).
MetE employs a zinc cofactor to catalyze the chemically difficult transfer of a methyl group from methyltetrahydrofolate to homocysteine to form methionine (16). In E. coli, this reaction may also be catalyzed by the cobalamin-dependent enzyme (MetH); however, in the absence of exogenously supplied cobalamin, MetE is the sole means of methionine biosynthesis. Yet in cells experiencing disulfide stress conditions, MetE was found to be oxidized, which was directly linked to a cellular methionine auxotrophy (17). Oxidized glutathione (GSSG) stoichiometrically and specifically oxidizes the purified protein via the formation of a glutathione adduct, resulting in enzyme inactivation. Surprisingly, glutathionylation was found to occur on cysteine 645, one of five nonessential cysteines within MetE (the two zinc-coordinated cysteines, 643 and 726, are required for activity). The crystal structure of the Thermotoga maritima homolog suggests that cysteine 645 lies at the entrance to the active site, which is positioned within a cleft between two β8α8 barrels (31). Hence, glutathionylation of cysteine 645 likely causes considerable structural perturbations at the active site; in fact, glutathionylation was found to be concomitant with a conformational change (17).
In order to further dissect the mechanism of MetE inactivation in vivo and gain insight into its physiological consequences, we focused on the role played by the redox-active cysteine, cysteine 645. Thus, experiments were performed to assess how the mutation of cysteine 645 to alanine affects E. coli experiencing disulfide stress conditions. Strains were constructed in which wild-type or mutant MetE (which is missing the redox-active cysteine) was expressed under its own promoter from a single-copy plasmid in a background lacking MetE. The promoter region and coding regions of wild-type and Cys645Ala MetE were introduced into the pCC1 cloning vector (Epicentre), which possesses the E. coli F factor single-copy origin of replication; these plasmids were then transformed into E. coli strain MTD23, which has an in-frame deletion of metE (see Fig. S1 in the supplemental material) (38).
The response of these strains to disulfide stress was evaluated by monitoring the growth of cultures in minimal medium either containing or lacking methionine. Disulfide stress was induced by the addition of diamide, which freely diffuses through the cell membrane to oxidize intracellular thiols, thereby leading to a lag in growth that is proportional to the initial concentration used (20). Following treatment with diamide, E. coli expressing wild-type MetE responded in a manner identical to that previously observed with wild-type strains of E. coli (17): upon diamide challenge, cells grown in the presence of methionine are able to resume growth significantly faster than those that must rely upon MetE, which is oxidized and presumably inactivated, for their methionine (Fig. 1). A different pattern is seen for cells expressing the Cys645Ala protein (which retains ≥90% of the wild-type MetE activity). For both strains, growth on media containing methionine is faster than in media lacking methionine before diamide addition, and these growth rates resume following the lag. However, cells containing the mutant protein resume growth at approximately the same time whether or not methionine is added to the medium, in contrast to cells containing the wild-type protein, where methionine affects the duration of the growth lag. The mutant cells, which have a MetE protein that no longer contains the redox-active cysteine and presumably cannot be inactivated, are not limited for methionine during diamide treatment. Hence, this experiment demonstrates that oxidation of cysteine 645 clearly has physiological consequences and is responsible for the transient methionine auxotrophy induced by diamide treatment.
FIG. 1.
Cysteine 645 prolongs the lag phase induced by diamide in medium lacking methionine. Exponentially growing cultures of pEM14/MTD23 (wild type [WT]) and pEM15/MTD23 (C645A) were inoculated into glucose minimal morpholinepropanesulfonic acid (MOPS) medium either containing or lacking 0.2 mM l-methionine (+ Met or − Met, respectively) (17). When the cultures reached an OD at 600 nm (OD600) of 0.2, diamide was added to a concentration of 0.9 mM. A representative trace monitoring growth by OD600 reveals the impact that the mutation of cysteine 645 has upon the resumption of growth following diamide treatment.
To further analyze the impact of cysteine 645, a high-throughput assay was devised that employs 96-well plates to measure the MIC of diamide for these two strains. Aliquots of exponentially growing starter cultures were added to various concentrations of diamide in the wells, and the optical density (OD) was measured at intervals during the subsequent incubation. The Cys645Ala mutation was found to confer resistance to diamide when cells are grown in media lacking methionine, but not when cells are grown in the presence of methionine (Fig. 2). Moreover, a comparison of the strains in disk diffusion assays elicited similar results: E. coli utilizing wild-type MetE had dramatically larger zones of inhibition than cells containing the Cys645Ala protein (data not shown). Taken together, our data provide convincing evidence that cysteine 645 serves to modulate the activity of MetE in vivo in response to disulfide stress.
FIG. 2.
Cysteine 645 confers resistance to diamide. MICs of diamide were determined for strains pEM14/MTD23 (wild type [WT]) and pEM15/MTD23 (C546A) using a 96-well plate assay. In one column of a 96-well plate, glucose minimal MOPS medium either containing or lacking 0.2 mM l-methionine (+ Met or − Met, respectively) was inoculated with exponentially growing cultures to an OD600 of 0.2 in a total volume of 1 ml. Cultures were incubated at 37°C for a single doubling and then diluted to an OD600 of 0.02 into 1 ml of the same medium containing a series of diamide concentrations at 0.1 mM intervals in the adjacent columns of the plate. Each strain was tested in duplicate and yielded identical results. Following 24 or 48 h of incubation at 37°C with shaking at 250 rpm, 50-μl aliquots were transferred to a 96-well plate, and the OD600 was measured to determine the MIC of diamide for culture growth (defined as the lowest diamide concentration where the OD600 was <0.03).
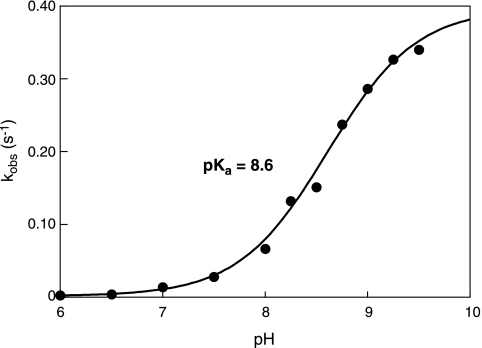
Highly reactive cysteines often have a decreased pKa, because the thiolate is the reactive species in thiol-disulfide exchange reactions. Thus, to better understand the reactivity of cysteine 645, it was important to determine its pKa. We previously demonstrated that diamide uniquely oxidizes cysteine 645 of MetE in a reaction that can be monitored by following the reduction of diamide at 350 nm using stopped-flow spectroscopy (17). Hence, the experiment was adapted to quantify the rate of diamide reduction by cysteine 645 as a function of pH (Fig. 3). Fitting these data indicates that the pKa of cysteine 645 is approximately 8.6, which is not dramatically altered compared to the pKa of free cysteine, which is around 9.1 (13). This result is in contrast to those for enzymes that specialize in thiol-disulfide exchange reactions, such as thioredoxin, glutaredoxin (Grx), protein disulfide isomerase, and DsbA, where significant reactivity is derived from increasing the concentrations of the attacking thiolate by lowering its pKa (to 7.5, <5.5, 4.5, and 3.5, respectively) (5, 19, 24, 26). However, it is clear that cysteine 645 is exquisitely sensitive to oxidation in vivo. Thus, the reactivity of cysteine 645 is likely due not simply to a decreased pKa but perhaps to its high level of accessibility.
FIG. 3.
pKa of cysteine 645. The oxidation of cysteine 645 by diamide was monitored using stopped-flow spectroscopy. Purified MetE (17) (100 μM) in 1 mM potassium phosphate buffer (pH 7.2) was rapidly mixed with 1 mM diamide in AMT buffer (50 mM acetic acid, 50 mM morpholineethanesulfonic acid [MES], 100 mM Tris) at various pHs. This allowed for the reaction to be monitored as a function of pH in a buffer that has a constant ionic strength over the pH range analyzed (9). Diamide reduction at 25°C was monitored at 325 nm, each sample was run in duplicate or triplicate, results were averaged, and the buffer was subtracted. The data were fit to two exponentials; the second phase accounted for the slow drop in absorbance observed at later times. The first high rate was taken as kobs and plotted versus the pH. The standard error associated with kobs was ≤2% of the parameter value for each pH. Data were then fit to the equation kobs = kmax/(10pKa − pH + 1) to calculate a pKa of 8.6 for cysteine 645.
There are relatively few reported examples of glutathionylation of cysteines with unaltered pKas; most notably, thioredoxin and actin both contain glutathionylation-sensitive cysteines in which susceptibility to oxidation is postulated to stem from surface accessibility (4, 39). Nonetheless, it is increasingly recognized that in vivo reactivity cannot be fully explained by a decreased pKa (3, 40). For example, in vitro GSSG glutathionylates low-pKa thiols in the protein-tyrosine phosphatases PTP1B, SHP-1, and SHP-2, but detailed kinetic studies argue against a significant effect in vivo (11). However, glutathionylation of PTP1B occurs in cells, while SHP-1 and SHP-2 are unaffected (34). Similar kinetic considerations would limit in vivo glutathionylation of MetE via simple thiol-disulfide exchange with the free glutathione pool, yet oxidation of cysteine 645 clearly occurs in stressed bacteria (17). Hence, MetE contributes to growing evidence that raises the possibility that glutathionylation could be catalyzed in vivo. Moreover, a recent study found that glutathionylation of the Arabidopsis MetE homolog requires a protein in cell extracts (8).
Although Grx is known largely for its function as a cellular reductant, it is also capable of catalyzing the reverse oxidation reaction and has been reported to promote the glutathionylation of proteins such as PTP1B, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin, and the p65 subunit of NF-κB (2, 21, 32, 35, 37). In fact, kinetic analyses indicate that Grx is quite an efficient oxidase, due to the reactivity of the enzyme-glutathione mixed disulfide (42). It has been suggested that Grx could temporarily switch roles (from a reductase to an oxidase), depending on the cellular conditions (23). Likewise, recent studies found that exposure of Grx to highly oxidizing conditions in vivo (by export to the E. coli periplasm or manipulation of yeast glutathione/GSSG levels) resulted in Grx-catalyzed protein oxidation (10, 28). Thus, there is reason to believe that the addition of diamide, which rapidly oxidizes cytoplasmic glutathione, could lead to enzyme-catalyzed oxidation of MetE. As cells attempt to restore their redox balance during an oxidative assault, the coincident modulation of MetE activity may prove advantageous by allowing methionine availability to serve as a metabolic checkpoint to prevent active growth before the stress is completely abated.
Deciphering the specificity factors that govern in vivo glutathionylation is a prevailing challenge in the field (7, 12). Therefore, MetE provides a valuable paradigm of a redox-regulated protein in which the cellular milieu must be considered, as the reactivity of cysteine 645 cannot readily be attributed to an abnormally low pKa. Yet it is possible that modulation of the pKa of cysteine 645 could still play a role in MetE glutathionylation. In particular, conformational changes can alter the local environment of a thiol, thereby having an impact on its pKa. For example, different pKa values for the reactive cysteines in the cysteine-based peroxidase AhpC and carbonic anhydrase are measured depending on the conformational state and electrostatic environment (18, 25). Similarly, glutathionylation of GAPDH does not seem to occur unless the protein is first oxidized by H2O2, and glutathionylation of 1-cysteine peroxiredoxin requires heterodimerization with glutathione S-transferase π (6, 33). Furthermore, there is precedent for structural mobility of MetE, as glutathionylation was previously found to be concomitant with a significant conformational change (17). Hence, glutathionylation could be coupled to an enzyme-induced conformational change that serves to activate cysteine 645 by modulating its pKa.
In the past few years, several studies have confirmed the sensitivity of methionine biosynthesis to oxidative conditions and circumstances likely to elicit oxidative stress (1, 29, 30, 36). MetE homologs have also been found to be susceptible to oxidation, most notably the Bacillus subtilis enzyme, which is reported to be cysteinylated rather than glutathionylated (8, 14, 15, 41). Interestingly, many of those proteins do not possess a homologous cysteine 645, which is conserved only within the Enterobacteriaceae. And indeed, oxidation of the B. subtilis MetE homolog is reported to occur on the cysteine that aligns with cysteine 726 of the E. coli enzyme, one of the essential zinc ligands (15). However, it seems that treatment of B. subtilis cultures with diamide results in a similar transient methionine auxotrophy (14). In contrast, it is clear from this study that expression of MetE lacking cysteine 645 (but containing cysteine 726) in E. coli completely eliminates the methionine limitation induced by diamide. It is possible that for E. coli, access to cysteine 726, which is buried within the cleft of the active site, is precluded for glutathione; however, in B. subtilis, which does not synthesize glutathione, oxidation of cysteine 726 via a smaller cysteine adduct can occur. Hence, the means of controlling methionine levels via oxidation of MetE may be species dependent. Such a nuanced system is reminiscent of the organic peroxide-sensing regulator OhrR, for which the mechanism of oxidation and inactivation depends upon the number of cysteine residues within the homolog (27). Thus, an understanding of the pivotal role played by cysteine 645 of E. coli MetE provides crucial insight into what may be a general mode for controlling methionine availability, so as to modulate cellular growth under stressful conditions.
Supplementary Material
Acknowledgments
We thank Robert Blumenthal (University of Toledo) for helpful advice and discussions.
This work was supported in part by National Institutes of Health grant GM29408 (to R.G.M.).
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arai, H., J. H. Roh, and S. Kaplan. 2008. Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 190286-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer, S. M., E. R. Taylor, S. E. Brown, C. C. Dahm, N. J. Costa, M. J. Runswick, and M. P. Murphy. 2004. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant defense. J. Biol. Chem. 27947939-47951. [DOI] [PubMed] [Google Scholar]
- 3.Bindoli, A., J. M. Fukuto, and H. J. Forman. 2008. Thiol chemistry in peroxidase catalysis and redox signaling. Antioxid. Redox Signal. 101549-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casagrande, S., V. Bonetto, M. Fratelli, E. Gianazza, I. Eberini, T. Massignan, M. Salmona, G. Chang, A. Holmgren, and P. Ghezzi. 2002. Glutathionylation of human thioredoxin: a possible crosstalk between the glutathione and thioredoxin systems. Proc. Natl. Acad. Sci. USA 999745-9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chivers, P. T., K. E. Prehoda, B. F. Volkman, B. M. Kim, J. L. Markley, and R. T. Raines. 1997. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry 3614985-14991. [DOI] [PubMed] [Google Scholar]
- 6.Cotgreave, I. A., R. Gerdes, I. Schuppe-Koistinen, and C. Lind. 2002. S-Glutathionylation of glyceraldehyde-3-phosphate dehydrogenase: role of thiol oxidation and catalysis by glutaredoxin. Methods Enzymol. 348175-182. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne, I., R. Rossi, G. Colombo, D. Giustarini, and A. Milzani. 2009. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 3485-96. [DOI] [PubMed] [Google Scholar]
- 8.Dixon, D. P., M. Skipsey, N. M. Grundy, and R. Edwards. 2005. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 1382233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, K. J., and J. F. Morrison. 1982. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87405-426. [DOI] [PubMed] [Google Scholar]
- 10.Eser, M., L. Masip, H. Kadokura, G. Georgiou, and J. Beckwith. 2009. Disulfide bond formation by exported glutaredoxin indicates glutathione's presence in the E. coli periplasm. Proc. Natl. Acad. Sci. USA 1061572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forman, H. J. 2007. Use and abuse of exogenous H2O2 in studies of signal transduction. Free Radic. Biol. Med. 42926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallogly, M. M., and J. J. Mieyal. 2007. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 7381-391. [DOI] [PubMed] [Google Scholar]
- 13.Harris, T. K., and G. J. Turner. 2002. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life 5385-98. [DOI] [PubMed] [Google Scholar]
- 14.Hochgräfe, F., J. Mostertz, D. Albrecht, and M. Hecker. 2005. Fluorescence thiol modification assay: oxidatively modified proteins in Bacillus subtilis. Mol. Microbiol. 58409-425. [DOI] [PubMed] [Google Scholar]
- 15.Hochgräfe, F., J. Mostertz, D. C. Pöther, D. Becher, J. D. Helmann, and M. Hecker. 2007. S-Cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J. Biol. Chem. 28225981-25985. [DOI] [PubMed] [Google Scholar]
- 16.Hondorp, E. R., and R. G. Matthews. April 2006, posting date. Chapter 3.6.1.7, Methionine. In A. Böck et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 17.Hondorp, E. R., and R. G. Matthews. 2004. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, G., and R. L. Levine. 2005. Molecular determinants of S-glutathionylation of carbonic anhydrase 3. Antioxid. Redox Signal. 7849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kortemme, T., N. J. Darby, and T. E. Creighton. 1996. Electrostatic interactions in the active site of the N-terminal thioredoxin-like domain of protein disulfide isomerase. Biochemistry 3514503-14511. [DOI] [PubMed] [Google Scholar]
- 20.Kosower, N. S., and E. M. Kosower. 1995. Diamide: an oxidant probe for thiols. Methods Enzymol. 251123-133. [DOI] [PubMed] [Google Scholar]
- 21.Lind, C., R. Gerdes, I. Schuppe-Koistinen, and I. A. Cotgreave. 1998. Studies on the mechanism of oxidative modification of human glyceraldehyde-3-phosphate dehydrogenase by glutathione: catalysis by glutaredoxin. Biochem. Biophys. Res. Commun. 247481-486. [DOI] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Mieyal, J. J., M. M. Gallogly, S. Qanungo, E. A. Sabens, and M. D. Shelton. 2008. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid. Redox Signal. 101941-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson, J. W., and T. E. Creighton. 1994. Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry 335974-5983. [DOI] [PubMed] [Google Scholar]
- 25.Nelson, K. J., D. Parsonage, A. Hall, P. A. Karplus, and L. B. Poole. 2008. Cysteine pKa values for the bacterial peroxiredoxin AhpC. Biochemistry 4712860-12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordstrand, K., F. Aslund, S. Meunier, A. Holmgren, G. Otting, and K. D. Berndt. 1999. Direct NMR observation of the Cys-14 thiol proton of reduced Escherichia coli glutaredoxin-3 supports the presence of an active site thiol-thiolate hydrogen bond. FEBS Lett. 449196-200. [DOI] [PubMed] [Google Scholar]
- 27.Oh, S. Y., J. H. Shin, and J. H. Roe. 2007. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 1896284-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Østergaard, H., C. Tachibana, and J. R. Winther. 2004. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell Biol. 166337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partridge, J. D., G. Sanguinetti, D. P. Dibden, R. E. Roberts, R. K. Poole, and J. Green. 2007. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 28211230-11237. [DOI] [PubMed] [Google Scholar]
- 30.Partridge, J. D., C. Scott, Y. Tang, R. K. Poole, and J. Green. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J. Biol. Chem. 28127806-27815. [DOI] [PubMed] [Google Scholar]
- 31.Pejchal, R., and M. L. Ludwig. 2005. Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication. PLoS Biol. 3e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qanungo, S., D. W. Starke, H. V. Pai, J. J. Mieyal, and A. L. Nieminen. 2007. Glutathione supplementation potentiates hypoxic apoptosis by S-glutathionylation of p65-NFκB. J. Biol. Chem. 28218427-18436. [DOI] [PubMed] [Google Scholar]
- 33.Ralat, L. A., Y. Manevich, A. B. Fisher, and R. F. Colman. 2006. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase π with activity changes in both enzymes. Biochemistry 45360-372. [DOI] [PubMed] [Google Scholar]
- 34.Rinna, A., M. Torres, and H. J. Forman. 2006. Stimulation of the alveolar macrophage respiratory burst by ADP causes selective glutathionylation of protein tyrosine phosphatase 1B. Free Radic. Biol. Med. 4186-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruoppolo, M., J. Lundström-Ljung, F. Talamo, P. Pucci, and G. Marino. 1997. Effect of glutaredoxin and protein disulfide isomerase on the glutathione-dependent folding of ribonuclease A. Biochemistry 3612259-12267. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, B., L. Ruiz, C. G. de los Reyes-Gavilan, and A. Margolles. 2008. Proteomics of stress response in Bifidobacterium. Front. Biosci. 136905-6919. [DOI] [PubMed] [Google Scholar]
- 37.Starke, D. W., P. B. Chock, and J. J. Mieyal. 2003. Glutathione-thiyl radical scavenging and transferase properties of human glutaredoxin (thioltransferase). Potential role in redox signal transduction. J. Biol. Chem. 27814607-14613. [DOI] [PubMed] [Google Scholar]
- 38.Thanbichler, M., B. Neuhierl, and A. Böck. 1999. S-Methylmethionine metabolism in Escherichia coli. J. Bacteriol. 181662-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, J., E. S. Boja, W. Tan, E. Tekle, H. M. Fales, S. English, J. J. Mieyal, and P. B. Chock. 2001. Reversible glutathionylation regulates actin polymerization in A431 cells. J. Biol. Chem. 27647763-47766. [DOI] [PubMed] [Google Scholar]
- 40.Winterbourn, C. C., and D. Metodiewa. 1999. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic. Biol. Med. 27322-328. [DOI] [PubMed] [Google Scholar]
- 41.Wolf, C., F. Hochgräfe, H. Kusch, D. Albrecht, M. Hecker, and S. Engelmann. 2008. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 83139-3153. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, R., J. Lundström-Ljung, A. Holmgren, and H. F. Gilbert. 2005. Catalysis of thiol/disulfide exchange. Glutaredoxin 1 and protein-disulfide isomerase use different mechanisms to enhance oxidase and reductase activities. J. Biol. Chem. 28021099-21106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.