Abstract
Pathogenic streptococci and enterococci primarily rely on the conserved secretory (Sec) pathway for the translocation and secretion of virulence factors out of the cell. Since many secreted virulence factors in gram-positive organisms are subsequently attached to the bacterial cell surface via sortase enzymes, we sought to investigate the spatial relationship between secretion and cell wall attachment in Enterococcus faecalis. We discovered that sortase A (SrtA) and sortase C (SrtC) are colocalized with SecA at single foci in the enterococcus. The SrtA-processed substrate aggregation substance accumulated in single foci when SrtA was deleted, implying a single site of secretion for these proteins. Furthermore, in the absence of the pilus-polymerizing SrtC, pilin subunits also accumulate in single foci. Proteins that localized to single foci in E. faecalis were found to share a positively charged domain flanking a transmembrane helix. Mutation or deletion of this domain in SrtC abolished both its retention at single foci and its function in efficient pilus assembly. We conclude that this positively charged domain can act as a localization retention signal for the focal compartmentalization of membrane proteins.
Understanding the transport and processing of proteins in their journey from the cytosol to the extracellular milieu has driven significant advances in elucidating the molecular interactions between an organism and its environment. These interactions are particularly important at the host-pathogen interface, where bacterial adhesins, toxins, and other virulence factors interact with host tissues (31). In gram-negative organisms, transit from the cytosol to the extracellular environment occurs by several mechanisms that either bypass the periplasm or use it as an organelle to process and fold proteins destined for secretion (46). Gram-positive organisms lack a membrane-bound periplasm but nevertheless secrete many virulence factors that require posttranslational modification (21). It has been proposed that the space between the cell membrane and cell wall provides a protected environment for folding and processing of secreted proteins in gram-positive bacteria (23, 24, 36, 52). Once translocated across the membrane, many virulence factors, such as the Streptococcus pyogenes SpeB protease, are secreted into the extracellular milieu (4), while adhesins are retained at the bacterial surface, where they mediate attachment to host tissues. A large subset of adhesins characterized as virulence factors in gram-positive organisms, such as S. pyogenes M protein and Staphylococcus aureus protein A, are covalently linked to the cell wall by the presence of a cell wall sorting (CWS) signal (1, 8, 41). The CWS signal is comprised of a C-terminal LPXTG motif, a transmembrane domain, and a positively charged tail (41). Proteins containing this CWS signal are recognized by a sortase enzyme, which cleaves the CWS motif between the threonine-glycine bond. Subsequent transpeptidation links the protein to a lipid II intermediate prior to its incorporation into the cell wall (26, 47). The protein-lipid II complex is processed by penicillin binding proteins, which results in the incorporation of the CWS protein into the mature cell wall (48). Despite the great deal known about the biochemical and mechanistic aspects of cell wall synthesis and sorting, details of the spatio-temporal coordination of cell wall synthesis, sorting, and secretion are unclear. Nevertheless, a close association linking these separate processes appears to be critical, because CWS proteins become properly exposed on the surface of the bacteria only after their sortase-mediated incorporation into the cell wall (25).
Enterococcus faecalis commonly causes urinary tract infections, endocarditis, intra-abdominal infections, and bacteremia, and it relies on CWS proteins, including Esp, aggregation substance (AS), and pili, to cause disease (18, 27, 39, 42). While these studies demonstrate the importance of cell wall proteins in E. faecalis pathogenesis, the basic mechanisms by which these proteins are localized to the cell surface or secreted remains unclear. We show here that secretion, protein trafficking, and cell wall processing are colocalized at single foci in E. faecalis through the presence of a positively charged retention domain within the localized protein itself, indicating that these processes are compartmentalized into an organelle.
MATERIALS AND METHODS
Bacterial strains and culture.
Strains used in this study are listed in Table 1. Escherichia coli MC1061 (50) was grown in Luria-Bertani broth or agar at 37°C and used to propagate plasmids. E. faecalis strains were inoculated 1:1,000 and grown statically in brain heart infusion (BHI) broth or agar at 37°C for 15 to 18 h for all assays unless otherwise noted. Antibiotics were added at the following concentrations for E. coli: chloramphenicol, 20 mg/liter; kanamycin, 50 mg/liter; and erythromycin (Erm), 750 mg/liter. For E. faecalis strains, the antibiotics were added as follows: chloramphenicol, 20 mg/liter; erythromycin, 25 mg/liter; fusidic acid, 25 mg/liter, kanamycin, 500 mg/liter; rifampin (rifampicin), 25 mg/liter; streptomycin, 500 mg/liter; tetracycline, 15 mg/liter.
TABLE 1.
Strains and plasmids used in this study
| Species and strain or plasmid (antibiotic)a | Description | Reference |
|---|---|---|
| E. faecalis | ||
| OG1SS/pCF10 (Str, Tet) | prgB+srtA+srtC+ | 12 |
| OG1SS/pCF10 ΔSrtA (Str, Tet) | prgB+srtA srtC+ | This study |
| OG1SS/pCF10 ΔSrtC (Str, Tet) | prgB+srtA+srtC | This study |
| OG1RF (Rif, Fus) | prgB srtA+srtC | 10 |
| OG1X (Str) | prgB srtA+srtC | 12 |
| OG1X ΔSrtA (Str) | prgB srtA srtC+ | This study |
| OG1X ΔSrtC (Str) | prgB srtA+srtC | This study |
| OG1X ΔSrtA ΔSrtC (Str) | prgB srtA srtC | This study |
| E. coli MC1061 | 50 | |
| Plasmids | ||
| pABG5 (Kan, Cm) | Shuttle vector | 16 |
| pJRS233 (Erm) | Temperature-sensitive plasmid for generation of deletions | 33 |
| pAL1 (Kan) | Derivative of pABG5 | This study |
| pAL1::SrtA (Kan) | lacking Cmr | This study |
| pAL1::SrtA-HA (Kan) | cassette | This study |
| pAL1::SrtA-HA, native srtA promoter (Kan) | This study | |
| pAL1::SrtC-HA (Kan) | This study | |
| pAL1::SrtC-HA(+/−)tail (Kan) | This study | |
| pAL1::SrtC-HA(−)tail (Kan) | This study | |
| pAL1::SrtC-HAΔtail (Kan) | This study |
Str, streptomycin; Tet, tetracycline; Kan, kanamycin; Erm, erythromycin; Rif, rifampin; Fus, fusidic acid. See Materials and Methods for antibiotic concentrations.
Genetic manipulations.
Genes targeted for mutation were identified based on the annotated complete genome of E. faecalis V583 (32); all references to genomic loci are based on this annotation (GenBank accession number AE016830). In-frame deletions of srtA (EF3056) and srtC (EF1094) were created according to previously described methods (38). SrtB, a third sortase present in a subset of strains (18, 32), was not investigated. To construct in-frame deletions of srtA (EF3056) and srtC (EF1094), regions approximately 800 bp upstream and downstream of the genes were amplified from OG1RF using primer pairs EF3056e-f3/EF3056 sew-r or EF1094e-f3/EF1094 sew-r for upstream regions and EF3056e-r3/EF3056 sew-f or EF1094e-r3/EF1094 sew-r for downstream regions (Table 2). These products were sewn together and amplified using EF3056e-f3/EF3056e-r3 or EF1094e-f3/EF1094e-r3. These PCR products were then cloned into the pCR2.1 vector (Invitrogen) according to the manufacturer's protocol and sequenced. Correct clones were subsequently subcloned into pJRS233, a temperature sensitive gram-positive plasmid, using the flanking XbaI sites to generate deletion constructs pJRS233-ΔEF3056 and pJRS233-ΔEF1094 (33). Deletion constructs were then transformed into OG1SS/pCF10 or OG1X (12) by electroporation and the transformants selected at 30°C on Erm. Chromosomal integrants were selected by growth at 42°C in the presence of Erm. Selection for excision of the integrated plasmid by homologous recombination was accomplished by growing the bacteria at 30°C in the absence of Erm. Loss of the EF3056 or EF1094 loci in Erm-sensitive bacteria was demonstrated by PCR using primer pair EF3056e-f1/EF3056e-r1 or EF1094e-f1/EF1094e-r1.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′ → 3′) |
|---|---|
| EF3056e-f3 | GCTCTAGAAGTAATTCAGGACTTAATTTTTCGACAGGCGT |
| EF3056e-r3 | GCTCTAGAGAAGGAGTGAAACGTTGTGGTAACTGTG |
| EF3056 sew-f | GTGGTAAACTTGATACATTAAAAGGAGGGAAAATTGGGTGGCTTAATAAAAAAATCCTTGAAAAGTTTTGACACTTTTC |
| EF3056 sew-r | GAAAAGTGTCAAAACTTTTCAAGGATTTTTTTATTAAGCCACCCAATTTTCCCTCCTTTTAATGTATCAAGTTTACCAC |
| EF1094e-f3 | GCTCTAGAATAACGTGACTTCTGGAGAGTATGCTTATGC |
| EF1094e-r3 | GCTCTAGACGCGGAACAAAGGAGCTAAAAACTTAATAGAATA |
| EF1094 sew-f | CTGTCCCAGGCTCTCATGCTTTATTTTTAAGGAGGAAGCATAGTATGACGAAAAGGCTAAACATACTAAAAAAAAGAG |
| EF1094 sew-r | CTCTTTTTTTTAGTATGTTTAGCCTTTTCGTCATACTATGCTTCCTCCTTAAAAATAAAGCATGAGAGCCTGGGACAG |
| EF1094e-f1 | GGAATTCCGAAGTGGCGCAGTCTTGCTAC |
| EF1094e-r1 | GGAATTCCCAGTCGATTGATTGGCCCTTTGTCG |
| EF3056e-f1 | GGAATTCCCCCTCTACTAGCCTCCTTACCATTTTAC |
| EF3056e-r1 | GGAATTCCGTTGATAATGAGTCTGCCGCTAGTGTATG |
| EF1094i-f2 | CCGGAATTCAAGGAGGAAGCAATGAAGTCAAAAAAGAAACGTCGTATCATTG |
| EF1094i-r5 | AAAACTGCAGCTAAGCATAATCTGGAACATCATATGGATAAGCATAATCTGGAACATCATATGGATACTTTGGTTTTCTGGTCGTCTTTTTCCGTCGC |
| EF1094 tail delete-r | AAAACTGCAGCTAAGCATAATCTGGAACATCATATGGATAAGCATAATCTGGAACATCATATGGATAGTACCAGATAATGAAGCCGCTAATAATTAACGCACAGGC |
| EF1094(+/−)tail | AACTGCAGCTAAGCATAATCTGGAACATCATATGGATAAGCATAATCTGGAACATCATATGGATACTTTGGTTCTCTGGTCGTCTCTTTCTCTCGATCGTACCAGATAATGAAGCCGCTAATAATTAACGCACAG |
| EF1094(−) tail | AACTGCAGCTAAGCATAATCTGGAACATCATATGGATAAGCATAATCTGGAACATCATATGGATACTCTGGTTCTTCGGTCGTCTCTTCCTCTTCATCGTACCAGATAATGAAGCCGCTAATAATTAACGCACAG |
| EF3056i-f2 | CCGGAATTCAAGGAGGGAAAATATGCGCCCAAAAGAGAAAAAAAGAGG |
| EF3056i-r3 | AAAACTGCAGTTAAGCCACCCAATCGGCTAAAGTTTTTTGCTCCAATTGG |
| EF3056e-f5 | CGGGATCCCCGCGTTGCTGGTTACATCTATATGTAC |
| EF3056i-r7 | AAAACTGCAGTTAAGCATAATCTGGAACATCATATGGATAAGCATAATCTGGAACATCATATGGATAAGCCACCCAATCGGCTAAAGTTTTTTGCTCC |
Complementation constructs of sortase, sortase mutants, and hemagglutinin (HA)-tagged sortases were made in pAL1, a derivative of pABG5 that has the Cmr determinant inactivated by NcoI and StuI digestion, blunting, and religation (16). Wild-type srtA expressed from the rofA promoter present in the plasmid was cloned into pAL1 by amplifying the srtA coding sequence from OG1RF with EF3056i-f2/EF3056i-r3, digesting with EcoRI and PstI, and ligating into pAL1. A gene encoding a C-terminal HA-tagged SrtA expressed under the control of its native promoter was constructed by amplifying this region from OG1RF (10) using the primer pair EF3056e-f5/EF3056i-r7. This construct was then cloned into pAL1 using the BamHI and PstI restriction sites, removing the rofA promoter. Tail mutants of SrtC containing a C-terminal HA2 tag were generated in one step using primers EF1094i-r5, EF1094 tail delete-r, EF1094 (+/−) tail, or EF1094 (−) tail and EF1094i-f2 and cloned into pAL1 using EcoRI and PstI restriction sites. All constructs were confirmed by sequencing and transformed into E. faecalis. The expression and stability of complementation constructs was verified by anti-HA immunoblotting of whole-cell E. faecalis lysates.
Clumping response protocol.
The bacterial clumping protocol was adapted from previous studies (11) with the following modifications. Overnight starter cultures were diluted to an optical density (OD) at 600 nm of ∼0.06 in 5 ml BHI supplemented with 0.25 ml sterile supernatant from OG1X. These cultures were then grown with shaking for 2 to 2.5 h at 37°C before visualization and quantitative OD measurement. These bacteria were also used for localization of AS.
Electron microscopy.
Immunolocalization was performed as described previously for S. pyogenes focal protein localization with the following modifications (35, 36). Bacteria were fixed in 4% paraformaldehyde-0.5% glutaraldehyde in 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]-0.5 mM MgCl2 (pH 7.2) for 1 h at 4°C. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 M sucrose-20% polyvinyl pyrrolidone in PIPES-MgCl2 at 4°C. Samples were trimmed, frozen in liquid nitrogen, and sectioned with a Leica Ultracut UCT cryo-ultramicrotome (Leica Microsystems Inc., Bannockburn, IL). Seventy-nanometer sections were blocked with 0.01 M glycine-5% fetal bovine serum-5% normal goat serum for 30 min and subsequently incubated with polyclonal anti-HA (Sigma), anti-SecA, anti-Asc10, or affinity-purified anti-EbpA or anti-EbpC primary antibody overnight at 4°C. Sections were then washed in blocking buffer and probed with 18-nm colloidal gold-conjugated anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories, Inc., West Grove PA) for 1 h at room temperature. Sections were washed in PIPES buffer followed by an extensive water rinse and stained with 1% uranyl acetate-1.6% methylcellulose. Samples were viewed with a JEOL 1200EX transmission electron microscope (JEOL USA Inc., Peabody, MA). Parallel controls with the primary antibody omitted were consistently negative at the concentration of colloidal gold-conjugated secondary antibodies used in these studies.
Negative-stain immunogold electron microscopy experiments for labeling of pili were carried out as described previously (27) with the following modifications. Bacterial strains were grown statically overnight in BHI, diluted 1:1,000 in tryptic soy broth containing 0.25% glucose (TSBG), and again grown overnight (∼16 h) statically at 37°C. All mutant strains exhibited growth curves in TSBG that were similar to those of their wild-type controls (data not shown). The bacteria were then pelleted, washed in phosphate-buffered saline (PBS), and resuspended in PBS containing 5% calf serum. The cells were adsorbed to grids and incubated with affinity-purified rabbit anti-EbpA or anti-EbpC for 1 h. The grids were then washed with PBS and incubated with goat anti-rabbit IgG conjugated to 10-nm-diameter colloidal gold particles for 30 min. The grids were again washed with PBS, fixed with 1% glutaraldehyde for 20 min, and stained with 0.1% uranyl acetate for 30 s. After three subsequent washes with PBS, the grids were examined with a JEOL 1200 transmission electron microscope as described above. Parallel controls using species-matched preimmune serum were consistently negative.
Quantification of SecA and sortase foci.
Quantitative analysis of the frequency and region of localization was done from electron micrographs of ∼600 representative bacteria per strain. A focus of localization was defined as a focus containing ≥3 gold particles clustered together. The frequency of focal formation was determined as the total number of bacterial cells containing foci divided by the total number of cells counted. Focal localization was determined by dividing each focus-containing bacterium into three regions from youngest visible septum to pole. Cells in which septa were not visible were not included in this analysis. Fisher's exact test of significance was performed to compare the association between the numbers of foci in each region. Quantification of SrtC focal reduction in mutant strains was assessed by comparing the number of foci to the total number of bacterial cells containing ≥3 gold particle anywhere on the cell. Significance was measured by Fisher's exact test.
Theoretical mathematical predictions for SrtC foci were based on the conditions that the diameter of an enterococcal cell is ∼500 nm, that the bacteria are sectioned into 70-nm sections (see above), and that the microdomain where SrtC localizes is small relative to the section size and cell size. If the SrtC microdomain exists as only a single spot, it should be present in one-seventh (14%) of the sections analyzed and multiple spots should never be observed. If the SrtC microdomain exists as two spots, the number of foci predicted to be observed depends on how far apart the foci are. The maximum predicted number would be for diametrically opposed foci; the minimum predicted number would be observed for foci that are very close together, where the numbers will eventually converge to the case of just one spot. If there are two spots diametrically opposed, then the probability of seeing any spot in a given section is p(spot 1) + p(spot 2) − p(spot 1 and spot 2) ≤ 28.56%. p(spot 1 and spot 2) is at most 1/7 if they are very close together. p(spot 1 and spot 2) decreases to p(equatorial slice) × p(spots rotated into slice) = 1/7 × arcsin(70/250)/180° = 1.2%. As the two spots get closer together, the probability of seeing two spots increases to 14.28% and the probability of seeing a single spot decreases to 14.28%. It is likely that if a proportion of cells have two SrtC foci during replication (see Discussion), they will be somewhere between adjacent and diametrically opposed. If there is a mixture of cells with one spot and two spots, then these probabilities scale linearly with the proportion of cells with one spot and two spots.
Pilus localization and quantification.
For studies of pilus expression and localization, bacteria were grown in TSBG to enhance pilus production and immunoblotting was performed as described previously (18, 27, 39, 42), with the following modifications. Ten milliliters of bacteria (equivalent units of OD at 600 nm) were pelleted, and the supernatants were filtered through 0.2-μm filters and subsequently concentrated 100× by trichloroacetic acid precipitation. The pellet from 1 ml of each culture was resuspended in 1 ml of 50 mM Tris-HCl (pH 6.8) containing 125 units of mutanolysin (Sigma), incubated 2 h at 37°C with gentle rotation, and centrifuged for 15 min at 14,000 rpm. The supernatant containing cell wall constituents was collected.
Quantification of pilus expression on whole cells was carried on out negatively stained, immunolabeled bacteria. At least 100 bacteria per strain per experiment were scored for pilus expression, as determined by the presence of gold particles on the cell surface. Quantification of pilus subunit focal localization was assessed by electron microscopy on immunolabeled thin sections as described for SecA and sortases above.
Immunofluorescence microscopy was performed as described previously (14) with the following modifications: Bacteria were grown to stationary phase in TSBG, washed once in PBS, applied to glass slides, allowed to air dry, fixed in 3% paraformaldehyde for 10 minutes, washed again, and incubated for 1 hour with a 1:10,000 dilution of rabbit anti-EbpA or anti-EbpC in PBS-1% bovine serum albumin. Slides were washed, incubated with Cy3-labeled anti-rabbit secondary antibody, washed, and stained with DAPI (4′,6′-diamidino-2-phenylindole) (1 mg/ml) and wheat germ agglutinin for 10 min. All imaging was performed at the Karolinska Institutet Core Visualization Facility at Microbiology, Tumor and Cell Biology on a Leica (Wetzlar, Germany) fluorescence microscope equipped with Hamamatsu digital cameras operated by HiPic software (Hamamatsu). Images were prepared and processed in Adobe Photoshop. Pilus subunit localization was assessed for >200 bacteria per strain per experiment, where a single fluorescent spot on a cell was scored as a single focus and multiple spots or circumferential staining around the cell surface was scored as nonfocal.
RESULTS
Sortase A localizes in single foci in E. faecalis.
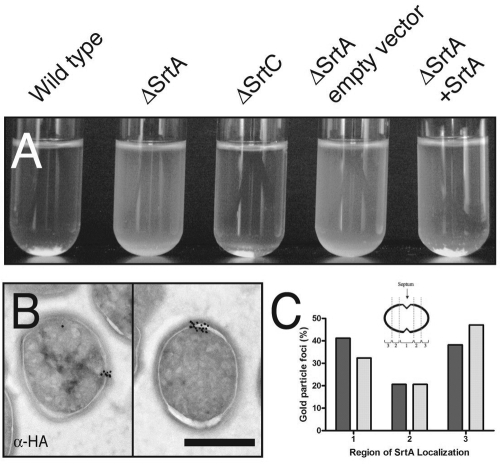
To investigate the overlap of secretion with cell wall assembly, we first constructed in-frame deletions of both sortases A and C (SrtA and SrtC), which are present in all examined strains of E. faecalis (27), creating strains ΔSrtA and ΔSrtC. To verify the loss of SrtA activity in E. faecalis, we monitored the phenotypic expression of the SrtA CWS-containing substrate, AS (15). AS expression leads to a marked, SrtA-dependent clumping of bacteria (11, 19). E. faecalis ΔSrtA, but not ΔSrtC, lost the ability to aggregate compared to the wild type (Fig. 1A). In complementation analyses, plasmids expressing either wild-type srtA (ΔSrtA + SrtA) or srtA constructed to express SrtA fused with a dual influenza virus HA epitope tag (SrtA-HA) restored AS-mediated aggregation in ΔSrtA to levels identical to wild type (Fig. 1A and data not shown, respectively). Introduction of the empty vector did not restore the aggregation phenotype to ΔSrtA. Immunoblot analysis of fractionated ΔSrtA cells verified that AS was no longer efficiently incorporated into the cell wall fraction (data not shown) (13).
FIG. 1.
Sortase A is focally localized in E. faecalis. (A) Broth clumping assay on OG1SS/pCF10-derived strains. Proper sorting of AS and subsequent clumping of E. faecalis is dependent on SrtA but not SrtC. In strains containing SrtA (first, third, and fifth tubes) the bacteria form large aggregates that settle to the bottom of the tube. Strains that lack SrtA (second and fourth tubes) remain turbid. (B) Anti-HA immunoelectron microscopy on OG1SS/pCF10 ΔSrtA complemented with an HA epitope-labeled SrtA expressed under its native promoter. SrtA-HA localizes to single domains on the surface of the bacterium. (C) Quantitative localization analysis of SrtA immunoelectron micrographs. Bacteria were equally divided into three regions from youngest visible septum to pole. Bacteria without a visible septum were excluded from this analysis. Dark gray bars represent log-phase bacteria, and light gray bars represent stationary-phase bacteria.
Immunogold electron microscopy of E. faecalis grown to early stationary phase and probed for SrtA-HA revealed that SrtA appeared clustered in foci in 71% (39/55) of labeled cells (defined as the presence of three or more gold particles on a single bacterium). SrtA foci in stationary-phase bacteria were always observed in a single domain on the surface of the bacteria (Fig. 1B). Sixty-four percent (37/58) of labeled bacteria grown to mid-log phase displayed focal SrtA localization. The majority (96%) of log-phase cells in which SrtA was observed to be localized displayed single SrtA foci (54/56), and 4% of the bacteria (2/56) had two SrtA foci each. Quantitative analysis of the location of SrtA foci on single bacteria was performed by dividing each bacterium into three equal regions spanning from the most recent visible septum to the pole, a method previously established for the localization of M protein and protein F on the surface of S. pyogenes (5). Region 1 corresponded to the equatorial region, which spanned the site of the current division plane. Region 2 was located where the next division plane would arise, which in enterococci is called the midcell, since this class of organisms divides in parallel chains. Region 3 included the polar region which corresponds to the septal area from the previous round of cell division (Fig. 1C). A significant majority, 79.4% (P < 0.0001), of SrtA foci observed in both log- and stationary-phase cultures were found either in region 1 (41.3% or 32.4%, respectively, for log- or stationary-phase cultures) or in region 3 (38.3% or 47.1%, respectively, for log- or stationary-phase cultures). Only 21% were localized to region 2. Together these data suggest that SrtA localizes to the active division plane but also remains associated with polar regions, the sites of previous cell division. This pattern of localization was not altered when actively dividing cells in mid-logarithmic growth were examined, indicating that our observations of SrtA foci were not growth stage dependent (Fig. 1C).
SecA localizes to a distinct membrane domain of Enterococcus faecalis.
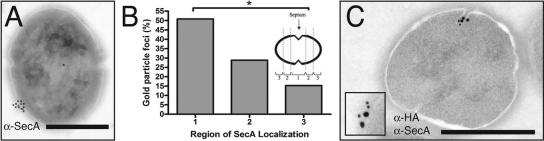
Frequent localization of SrtA to the equatorial region, the site of the nascent cell division septum, suggested a link between secretion, cell division, and cell wall synthesis. To address whether localized sites of Sec-mediated membrane translocation occur in E. faecalis, we first examined the distribution of SecA translocons in the membrane. The SecA antibody used in these studies was raised against Bacillus subtilis SecA and was shown to cross-react in immunoblots with a band of the size expected for SecA in both S. pyogenes (35) and E. faecalis (data not shown) whole-cell lysates. When examined by immunogold electron microscopy of thin-sectioned bacteria, SecA-specific staining of E. faecalis grown to early stationary phase was observed in single membrane domains in 3% of sections; the remaining 97% of cells were unlabeled. As thin sections reflect only a fraction of the cell, this frequency does not reflect the actual percentage of cells in which foci occur. To examine the site of SecA localization in each bacterium, we collected an additional 50 bacterial cells from the population labeled with three or more gold particles corresponding to SecA. Fifty-four percent (30/56) of SecA foci were located in the equatorial region 1 of the bacteria, compared to 30.4% (17/56) and 16.1% (9/56) in regions 2 and 3, respectively (Fig. 2B). Multiple foci were never observed (0/56) under the early-stationary-phase growth conditions examined here.
FIG. 2.
SecA localizes at single domains in E. faecalis. (A) Anti-SecA immune electron microscopy of OG1SS/pCF10. SecA localizes to a single domain on the surface of the bacterium near the equatorial region. (B) SecA focal localization, quantified as described for Fig. 1C. *, P < 0.005 by Fisher's exact test. (C) Double-label immunoelectron microscopy using anti-SecA (large particles) and anti-HA (small particles) antibodies. SecA and SrtA colocalize to the same region on the surface of E. faecalis. Scale bars, 0.5 μm. Inset, twofold magnification of a representative area of colocalization.
To address the possibility that SecA localization was dependent on growth phase, we next directly compared SecA localization in mid-logarithmic phase versus stationary phase. Among bacteria positively labeled with three or more gold particles, single SecA foci were observed at approximately the same frequency in a log-phase population (72.2%, 39/54) as in a stationary-phase population (68.1%, 32/47). In the remaining 15/54 and 15/47 SecA-labeled bacteria from each growth phase, respectively, the gold particles were not clustered. These observations indicate that SecA focus formation is not dependent on growth phase. Localization of SecA resembled the pattern observed for SrtA. Thus, we tested the hypothesis that both SrtA and SecA are localized at the same membrane microdomain in colabeling experiments. We observed consistent colocalization of both SrtA-HA and SecA to a single membrane site (Fig. 2C).
Disruption of sorting leads to focal substrate accumulation.
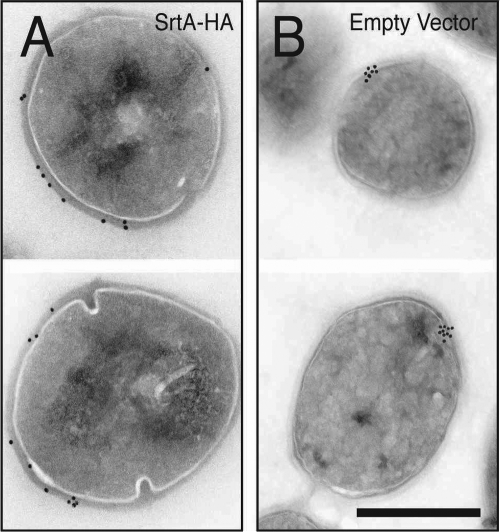
Following translation, sortase substrates such as AS are targeted to the membrane for Sec-mediated translocation across the membrane in a process facilitated by the Sec signal sequence. Current models predict that after translocation, sortase substrates are retained in the membrane by their transmembrane helix and flanking positively charged tail until sortase cleavage removes the helix and tail and covalently couples the substrate to the cell wall (41). Once secreted through the Sec pathway and processed by sortase, the distribution of sortase substrates around the cell is driven by incorporation into nascent cell wall components used in peptidoglycan biogenesis. Colocalization of SecA with SrtA at a single domain suggested that the secretion and processing machineries for CWS proteins are spatially coupled. This hypothesis was investigated by studying the localization pattern of AS using immunogold electron microscopy. In wild-type (30) or in ΔSrtA + SrtA-HA E. faecalis, AS was distributed around the bacterial surface (Fig. 3A). In contrast, in E. faecalis ΔSrtA, AS was no longer assembled around the periphery of the cell wall but instead localized to a single site (Fig. 3B), as was observed for SecA and SrtA (Fig. 2C). These results suggested that AS is retained at membrane microdomains when it is not properly incorporated into the cell wall by SrtA.
FIG. 3.
Sortase A substrate accumulates focally in the absence of sortase. Immunolocalization of AS in OG1SS/pCF10 ΔSrtA exposed to pheromone in the presence (A) or absence (B) of SrtA-HA is shown. E. faecalis was induced to express AS as described in Materials and Methods. Scale bar, 0.5 μm.
Pilus-associated sortase C and substrates localize to single sites.
While many SrtA enzymes are considered “housekeeping” sortases responsible for the surface attachment of most cell wall proteins, a subset of sortases function in pilus biogenesis (6, 9). E. faecalis SrtC is required for the polymerization of pili that consist of three proteins: EbpA, EbpB, and EbpC (27). We hypothesized that efficient pilus biogenesis is facilitated by focal localization of SrtC. To assess SrtC localization, we constructed a strain in which srtC is deleted and complemented with a plasmid expressing an HA-tagged SrtC (ΔSrtC + SrtC-HA). The HA epitope tag on SrtC did not alter its function in pilus production, since immunolabeling of negatively stained bacteria for the major pilus subunit EbpC showed that approximately 35% of the wild-type cells expressed pili, similar to levels for ΔSrtC + SrtC-HA (Fig. 4A) and similar to previously reported levels for the same growth conditions (27).
FIG. 4.
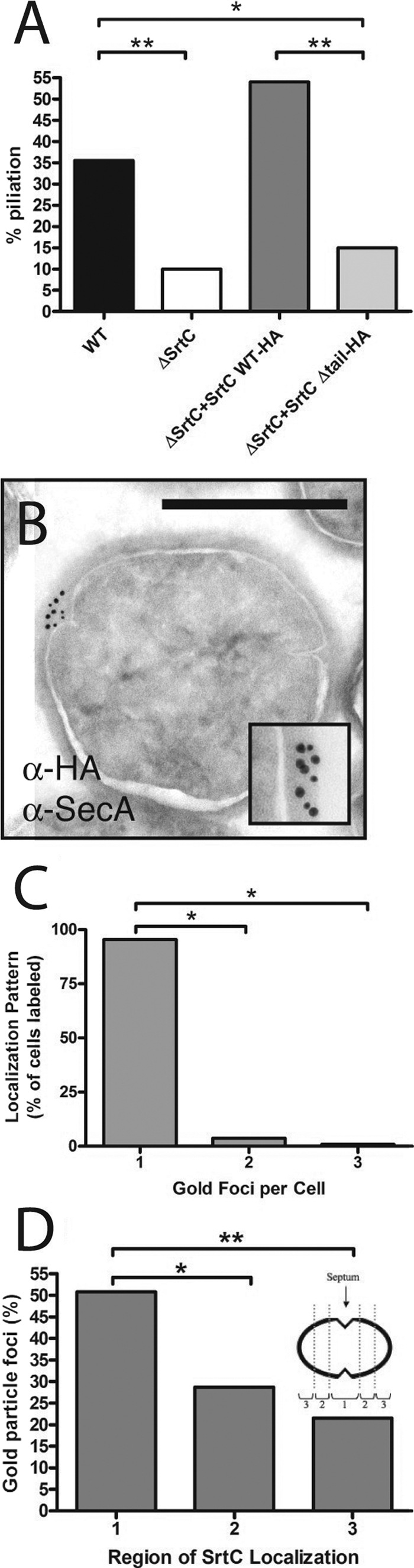
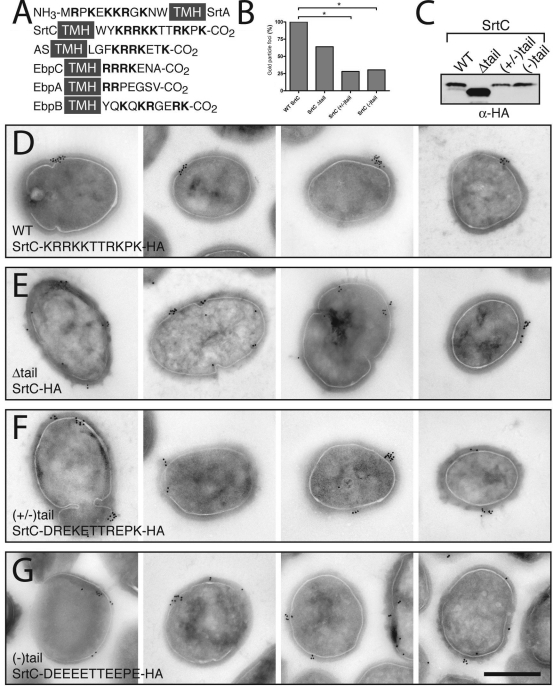
Sortase C localizes to single foci in E. faecalis. (A) Piliation levels of wild-type OG1X and sortase mutants. Results are from a representative experiment in which ≥100 cells/strain/experiment were counted. Statistical significance measured by chi-square test: *, P < 0.001. **, P < 0.0001. (B) Coimmunolocalization of SecA (large particles) and SrtC (small particles) found together in foci. Inset, close-up of colocalized SecA and SrtC. Scale bar, 0.5 μm. (C) Quantitative analysis of SrtC immunoelectron micrographs. *, P < 0.0001 by chi-square test. (D) Location of SrtC foci in bacteria equally divided into three regions from youngest visible septum to pole. Statistical significance measured by Fisher's exact test: *, P < 0.05; **, P < 0.001.
To determine if SrtC, like SrtA, is found at sites of secretion, we examined localization of SrtC-HA in early stationary phase. We found that SrtC-HA colocalized with SecA in discrete foci at the membrane (Fig. 4B). Examination of immunolabeled thin sections revealed focal SrtC localization on 18.4% (109/593) of these cells, while the rest were unstained. Of the 109 stained cells, 95.4% (104 bacteria) had a single focus, 3.7% (4 bacteria) had two foci, and 0.9% (1 bacterium) had three foci (Fig. 4C). This last class represented 0.2% (1/593) of all cells expressing SrtC-HA and was below the background frequency for single foci of 0.7% (4/577) observed in wild-type cells that do not express SrtC-HA (data not shown). Since a ring structure would be expected to produce a majority of cells with two foci and since the frequency of single foci is consistent with that predicted for 0.5-μm cells cut in 70-nm sections (17.5% observed versus 14.3% predicted; see Materials and Methods), these data support the conclusion that SrtC is localized to a single membrane domain rather than organized into a circumferential ring; however, we cannot exclude the possibility that more than one focus of localized protein exist on a cell. In addition, quantitative analysis of the locations of foci indicated a tendency toward localization in the vicinity of the septum versus the periphery or poles (Fig. 4D, region 1).
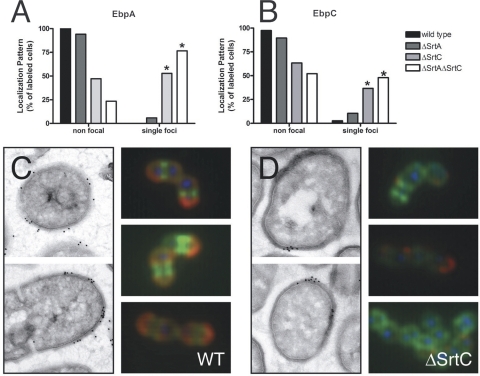
To examine whether other sortase substrates accumulate in foci in the absence of their cognate sortase, we analyzed the fate of the pilin subunit EbpA in the absence of SrtC. In a ΔSrtC strain, EbpA is not incorporated into pilus fibers in wild-type cells (27), and immunoblot analysis showed that EbpA monomers are instead both cell wall associated and also secreted into the culture supernatant in the absence of SrtC (see Fig. S1 in the supplemental material). Similar to the accumulation of AS in the absence of SrtA (Fig. 3B), immunoelectron microscopy showed that the cell wall-associated EbpA monomers formed foci in 53% of the EbpA-labeled cells in the absence of SrtC (Fig. 5A and C, left panel) while no foci were observed in wild-type cells (Fig. 5A and D, left panel), a statistically significantly enrichment (P < 0.00001). One explanation for the incomplete focal localization of EbpA in the ΔSrtC strain is that SrtA is able to attach a subset of EbpA subunits to the cell wall as has been described for Corynebacterium diphtheriae and Streptococcus pneumoniae (22, 28). Thus, to further validate focal pilin subunit accumulation in the absence of SrtC, immunofluorescence microscopy on whole bacterial cells was performed. EbpA and EbpC were shown to be distributed in a uniform pattern on the surface of wild-type cells (Fig. 5A, B, and C, right panel). Similarly, in ΔSrtA cells, pilin subunits exhibited uniformly distributed staining (Fig. 5A and B). In contrast, both the ΔSrtC and ΔSrtA ΔSrtC double mutant strains were significantly enriched for EbpA staining of singular foci (P < 0.0001) (Fig. 5A). Similar observations were made for EbpC (Fig. 5B). Thus, we conclude that the surface distribution of pilin subunits is altered in strains lacking the SrtC enzyme necessary for pilus polymerization.
FIG. 5.
Pilus subunits accumulate focally in the absence of SrtC. (A and B) Quantification of EbpA (A) or EbpC (B) immunofluorescent labeling of whole E. faecalis OG1X wild-type or sortase mutant cells grown to stationary phase. *, P < 0.0000001 by Fisher's exact test. (C and D) EbpA labeling of wild-type (WT) (C) or ΔSrtC bacteria (D) and localization by electron microscopy (left panels). Representative images of whole-cell immunofluorescence labeling of EbpA (red), DNA (blue), and cell wall (green) are also shown (right panels).
A positively charged sequence is important for focal sortase localization retention.
The clustering of SrtA and SrtC in singular foci, as well as sortase substrate clustering in the absence of the cognate sortase, suggested that sortases and CWS proteins may contain a signature domain that retains them within membrane microdomains prior to processing. Sequence comparisons revealed that each of these proteins has a single transmembrane domain that is flanked by a highly positively charged region. Positive charges flanking a transmembrane helix have been well characterized as a determinant of membrane topology (49). In SrtA, this positively charged domain is in the uncleaved secretion sequence of the protein (which also serves as the membrane-spanning domain), whereas in SrtC this domain is at the C terminus. Sequence comparisons of sortase substrates show that cell wall sorted proteins, including AS and the Ebps, contain a positively charged tail as part of their CWS (Fig. 6A and data not shown) (40, 41). Therefore, we hypothesized that in these proteins, a higher positive charge not only determines topology but also mediates localization to and retention within membrane microdomains.
FIG. 6.
Sortase C localization is dependent on a positively charged cytoplasmic tail. (A) Alignment of proteins observed in localized foci in E. faecalis. The cartoon depicts amino acids adjacent to the transmembrane helices (TMH) of E. faecalis SrtA, SrtC, AS, and Ebp pilus subunits. NH3 and CO2 indicate the N and C termini of the proteins, respectively. Boldface amino acids are positively charged. (B) Quantitative analysis of SrtC immunoelectron micrographs. Bacteria labeled with three or more gold particles were assessed for the presence or absence of focal localization. The percentage of cells displaying SrtC foci is expressed relative to wild-type (WT) value. Statistical significance measured by Fisher's exact test: *, P < 0.05; **, P < 0.001. (C) Anti-HA immunoblot of whole OG1RF, demonstrating stability of SrtC tail mutants. (D) Expression of SrtC-HA under control of the RofA promoter results in localization to single domains on the surface of the bacterium in wild-type E. faecalis strain OG1RF. (E to G) Immunolocalization of SrtC tail mutants (the amino acid sequence of the mutagenized tail is indicated). Scale bar, 0.5 μm.
Since the SrtC charged domain is at the C terminus and thus is not part of the signal peptide, we used it as a model protein to investigate whether the positively charged residues play a role in localizing membrane proteins to discrete foci, distinct from the signal peptide. This was examined by mutating the positively charged C-terminal domain in SrtC. Three different mutants were constructed: a mutant with a deletion of all positively charged residues from the C terminus, a mutant with every other positively charged amino acid replaced by a negatively charged residue, and a mutant with every positively charged residue replaced by a negative one. The effect of these mutations on SrtC localization was determined by immunogold electron microscopy. In contrast to the discrete localization observed for wild-type SrtC (Fig. 4B and 6D), all three mutants displayed an altered pattern of localization (Fig. 6E to G) corresponding to a decrease in the frequency of focal localization of gold particles and an increase in random gold labeling. Quantifying these results, we observed 36% fewer foci for SrtCΔtail, 72% fewer for SrtC(+/−)tail, and 70% fewer for SrtC(−)tail compared to wild-type SrtC (Fig. 6B). Mislocalized SrtC typically appeared as multiple smaller SrtC clusters along with several single gold particles. SrtC mislocalization is not due to instability of the mutated proteins, as verified by Western blot analysis (Fig. 6C). The clustering pattern of SrtC is unlikely to be mediated by the HA tag, since the clustering is dispersed in the SrtC mutants (which also have an HA tag). Thus, we conclude that the positively charged domain flanking the transmembrane helix is necessary for sortase localization.
Focal localization of SrtC is necessary for efficient pilus formation.
If SrtC localization to single foci is important for coupling secretion of pilus subunits and their subsequent processing into pilus fibers, then disruption of SrtC localization should also disrupt pilus biogenesis. We examined the location and extent of pilus expression on negatively stained bacteria with anti-EbpC antibodies. As reported for pili of other gram-positive organisms (14, 27), only a subset of the cells in any culture expressed pili, and for those cells that did express pili, no differences were observed in either the structure of the pili or their localization pattern on the cell surface for the wild-type, ΔSrtC + SrtCWT, and ΔSrtC + SrtCΔtail strains (see Fig. S2 in the supplemental material). In contrast, the overall proportion of piliated cells in each culture differed markedly. Complementation of ΔSrtC with a plasmid-encoded copy of srtC restored piliation to wild-type levels (Fig. 4A). However, mislocalization of SrtC through expression of SrtCΔtail, resulted in a significantly reduced level of piliation (15% piliation for ΔSrtC + pSrtCΔtail versus 55% piliation for ΔSrtC + pSrtCwt, P < 0.0001) (Fig. 4A), suggesting that focal subcellular localization of SrtC is required for efficient pilus biogenesis. Taken together, these data indicate that while SrtC mislocalization has no effect on the final destination of the sortase substrate after attachment to the cell wall, proper SrtC placement in the cell membrane facilitates its efficient function.
DISCUSSION
Bacterial cells display exquisite subcellular organization in the processing of virulence factors for display on the cell surface. Localization of presecretory proteins facilitates efficient secretion, processing, and assembly of macromolecular structures necessary for pathogenic interactions. This process has been enigmatic in gram-positive organisms due to incomplete understanding of basic molecular secretion mechanisms. Recently, two models have been proposed for how proteins are secreted in gram-positive cocci. In the first model, components of the general secretory pathway (Sec machinery) have been shown to localize to distinct domains in S. pyogenes and Streptococcus mutans (17, 35), leading to the proposal that protein secretion and processing may be spatially coupled. Supporting this model, two secreted proteins from S. pyogenes, SpeB and HtrA, colocalize with this secretion domain, termed the ExPortal. In the second model, domains within the secreted proteins themselves, and not the location of the Sec machinery, are proposed to direct the localization of secreted proteins in gram-positive cocci. A domain of the N-terminal secretion signal was shown to differentially influence the sites of protein F and M protein appearance on the cell surface of S. pyogenes, as well as that of a number of LPXTG-containing proteins in S. aureus (5, 7). Consistent with the model for localized secretion machinery, Sec components were found to localize in a helical pattern along the lengths of both gram-positive B. subtilis and gram-negative E. coli rods (3, 43), suggesting that secretion localization may be a conserved phenomenon in similarly shaped bacteria. Nevertheless, the molecular details governing the spatial subcellular distribution of the Sec apparatus in gram-positive cocci is not yet understood. Reconciliation of these models awaits careful examination of the entire secretion apparatus locale, including the SecYEG translocation channel, throughout the cell cycle.
Here we show that both secretion and sortase processing are spatially coupled in E. faecalis. The observation that SecA can localize to single domains in both log and stationary phases is consistent with the ExPortal model of localized protein secretion and indicates that sortase proteins are also found at this subcellular location. The significant enrichment of SecA and sortase enzyme domains in the vicinity of the cell division plane is consistent with cell wall synthesis in enterococci and streptococci occurring at the midcell (reviewed in reference 51). Localization of SrtC was facilitated by a positively charged sequence within the C terminus of the protein, distinct from the N-terminal secretion signal. Efficient assembly of sortase-dependent pili required proper subcellular localization of sortase. Our results suggest a model of coordinated localized secretion and sorting of cell wall proteins and are consistent with a model that in E. faecalis, proteins do not always traverse the cell membrane in a random manner but instead have at least one pathway that coordinates protein secretion and subsequent processing in localized regions across the cell membrane. It is likely that gram-positive cocci have evolved multiple mechanisms for subcellular localization. It will be interesting to determine whether signal sequence domains, analogous to YSIRK motifs identified in S. pyogenes and S. aureus (5, 7), also play a role in cell wall protein deposition in E. faecalis. Notably, none of the enterococcal proteins examined in this study and only one of 57 predicted CWS proteins in the sequenced E. faecalis V583 genome possesses a signal sequence bearing a canonical YSIRK motif (reference 32 and data not shown).
The cell wall of gram-positive bacteria is responsible for scaffolding its surface-exposed proteins but is also a significant barrier to secretion (44). This likely requires unique mechanisms for efficient transit and processing of virulence factors across the membrane and into the extracellular space. The assembly and attachment of gram-positive pili to the cell wall constitute an excellent model system for study of such complexity. The genes encoding pilus subunits are found genetically clustered with a sortase that is involved in the covalent polymerization of subunits during pilus biogenesis. After pilus polymerization, anchorage of the pilus to the growing cell wall is facilitated either by a “housekeeping” sortase encoded elsewhere on the chromosome (2, 29, 45) or by the pilus-associated sortase enzyme itself, as has been reported for S. pneumoniae (20). We show that the enterococcal pilus subunits EbpA and EbpC are significantly enriched in single foci in the absence of SrtC. These observations suggest a model in which a microdomain for secretion and sortase action may facilitate a high local concentration of subunits that are primed for pilus assembly. Interestingly, recent findings with S. pneumoniae show pilus localization at multiple, symmetric foci in wild-type cells (14). Pilus biogenesis in pneumococci appears to be a more complex process than in enterococci, with three pilin-associated sortase enzymes that not only display substrate specificity but also are required for focal presentation of pili (14). These findings are consistent with a model in which pilus formation in gram-positive bacteria involves the coordination of subunit secretion, processing by multiple sortase enzymes, and cell wall synthesis. For secreted proteins such as pilin subunits, the enrichment of SecA and SrtC foci at or near the division septum where peptidoglycan synthesis is occurring may also reflect the most energetically favorable site for secretion and processing, where the cell wall barrier is thinnest. Organization of multiple cellular processes at a single site not only facilitates spatial and temporal coordination of these processes but also promotes efficiency in their function.
An interesting prediction of any localized secretion model is that the membrane contains an asymmetric distribution of proteins, with one subset retained in foci while another becomes routed into the peripheral membrane. Thus, the membrane proteins themselves should contain specific motifs that are responsible for their trafficking to their appropriate destination following insertion into the membrane. Our data suggest that a high positive charge flanking a membrane-spanning region can function as a retention sequence for SrtC localization. Interestingly, the CWS sequences of sortase substrates in S. aureus and E. faecalis also possess a positively charged tail that is necessary for efficient cell wall sorting (19, 41). We hypothesize that this positively charged region may act to retain a sortase substrate within membrane microdomains following its translocation by the Sec pathway in order to promote interaction with the similarly localized sortase enzyme. Thus, in the absence of their cognate sortases, the sortase substrates AS and Ebp pilin subunits were retained in membrane microdomains.
Understanding how positive charge mediates localization of sortase C in E. faecalis will lead to important insights into the molecular underpinnings of gram-positive pathogen secretion and protein processing. One explanation is that cytoskeletal proteins or other subcellularly localized proteins form a scaffold for protein localization. Currently, the streptococci and enterococci lack any of the known bacterial cytoskeletal proteins described to date. Alternatively, a lipid-stabilized domain similar to lipid rafts in eukaryotic cells (34) may facilitate the retention of proteins by protein-lipid interactions. The latter possibility is consistent with findings that negatively charged lipids are necessary for SecA localization in B. subtilis and that anionic lipid domains occur at single sites that are consistent with the location of the ExPortal in S. pyogenes (3, 37).
In this study, we show that secretion and sortase processing occur together in E. faecalis. Sortase localization is facilitated by a positive charge that is necessary for efficient pilus biogenesis. These findings present a novel mechanism for coordinate secretion and processing of cell wall proteins in gram-positive cocci. Together, these data increase our understanding of basic molecular processes in this important category of pathogens and could lead to the identification of novel targets for therapeutic agents.
Supplementary Material
Acknowledgments
We thank members of the Hultgren, Caparon, and Normark labs for helpful discussion, critique, and critical reading of the manuscript; Darcy Gill for assistance with negatively stained immunoelectron microscopy; and G. Dunny and D. Manias for providing Asc10 antibody.
This work was supported by National Institutes of Health and Office of Research on Women's Health Specialized Center of Research grant P50DK6454002 with the Food and Drug Administration (to S.J.H.); NIH grants AI46433 (M.G.C.), AI47923 (B.E.M.), and AI068362 (S.L.C.); Medical Scientist Training Program grant T32 GM07200 (A.L.K.); and AHA postdoctoral fellowship 0625736Z (K.A.K.).
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article is available at http://jb.asm.org/.
REFERENCES
- 1.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 1842181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budzik, J. M., L. A. Marraffini, P. Souda, J. P. Whitelegge, K. F. Faull, and O. Schneewind. 2008. Amide bonds assemble pili on the surface of bacilli. Proc. Natl. Acad. Sci. USA 10510215-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campo, N., H. Tjalsma, G. Buist, D. Stepniak, M. Meijer, M. Veenhuis, M. Westermann, J. P. Muller, S. Bron, J. Kok, O. P. Kuipers, and J. D. Jongbloed. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 531583-1599. [DOI] [PubMed] [Google Scholar]
- 4.Caparon, M. G., B. Poolman, and A. Podbielski. 2007. Streptococcal peptide trans-membrane transport, p. 327-358. In R. Hakenbeck and S. Chhatwal (ed.), Molecular biology of the streptococci. Horizon Scientific Press, Norwich, United Kingdom.
- 5.Carlsson, F., M. Stalhammar-Carlemalm, K. Flardh, C. Sandin, E. Carlemalm, and G. Lindahl. 2006. Signal sequence directs localized secretion of bacterial surface proteins. Nature 442943-946. [DOI] [PubMed] [Google Scholar]
- 6.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 722710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeDent, A., T. Bae, D. M. Missiakas, and O. Schneewind. 2008. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 272656-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 393725-3733. [DOI] [PubMed] [Google Scholar]
- 9.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156289-297. [DOI] [PubMed] [Google Scholar]
- 10.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6270-278. [DOI] [PubMed] [Google Scholar]
- 11.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 753479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2454-465. [DOI] [PubMed] [Google Scholar]
- 13.Erlandsen, S. L., C. J. Kristich, G. M. Dunny, and C. L. Wells. 2004. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J. Histochem Cytochem. 521427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falker, S., A. L. Nelson, E. Morfeldt, K. Jonas, K. Hultenby, J. Ries, O. Melefors, S. Normark, and B. Henriques-Normark. 2008. Sortase mediated assembly and surface topology of adhesive pneumococcal pili. Mol. Microbiol. 70595-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galli, D., F. Lottspeich, and R. Wirth. 1990. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4895-904. [DOI] [PubMed] [Google Scholar]
- 16.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 1821529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, P., Z. Bian, M. Fan, M. Huang, and P. Zhang. 2008. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch. Oral Biol. 53150-154. [DOI] [PubMed] [Google Scholar]
- 18.Kemp, K. D., K. V. Singh, S. R. Nallapareddy, and B. E. Murray. 2007. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect. Immun. 755399-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristich, C. J., D. A. Manias, and G. M. Dunny. 2005. Development of a method for markerless genetic exchange in Enterococcus faecalis and its use in construction of a srtA mutant. Appl. Environ. Microbiol. 715837-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeMieux, J., S. Woody, and A. Camilli. 2008. Roles of the sortases of Streptococcus pneumoniae in assembly of the RlrA pilus. J. Bacteriol. 1906002-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 176263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matias, V. R., and T. J. Beveridge. 2005. Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol. Microbiol. 56240-251. [DOI] [PubMed] [Google Scholar]
- 24.Matias, V. R., and T. J. Beveridge. 2006. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 1881011-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 975510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285760-763. [DOI] [PubMed] [Google Scholar]
- 27.Nallapareddy, S. R., K. V. Singh, J. Sillanpaa, D. A. Garsin, M. Hook, S. L. Erlandsen, and B. E. Murray. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 1162799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson, A. L., J. Ries, F. Bagnoli, S. Dahlberg, S. Falker, S. Rounioja, J. Tschop, E. Morfeldt, I. Ferlenghi, M. Hilleringmann, D. W. Holden, R. Rappuoli, S. Normark, M. A. Barocchi, and B. Henriques-Normark. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nobbs, A. H., R. Rosini, C. D. Rinaudo, D. Maione, G. Grandi, and J. L. Telford. 2008. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect. Immun. 763550-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olmsted, S. B., S. L. Erlandsen, G. M. Dunny, and C. L. Wells. 1993. High-resolution visualization by field emission scanning electron microscopy of Enterococcus faecalis surface proteins encoded by the pheromone-inducible conjugative plasmid pCF10. J. Bacteriol. 1756229-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6519-527. [DOI] [PubMed] [Google Scholar]
- 32.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 2992071-2074. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8809-819. [DOI] [PubMed] [Google Scholar]
- 34.Rajendran, L., and K. Simons. 2005. Lipid rafts and membrane dynamics. J. Cell Sci. 1181099-1102. [DOI] [PubMed] [Google Scholar]
- 35.Rosch, J., and M. Caparon. 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 3041513-1515. [DOI] [PubMed] [Google Scholar]
- 36.Rosch, J. W., and M. G. Caparon. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol. Microbiol. 58959-968. [DOI] [PubMed] [Google Scholar]
- 37.Rosch, J. W., F. F. Hsu, and M. G. Caparon. 2007. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J. Bacteriol. 189801-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz, N., B. Wang, A. Pentland, and M. Caparon. 1998. Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A streptococci. Mol. Microbiol. 27337-346. [DOI] [PubMed] [Google Scholar]
- 39.Schlievert, P. M., P. J. Gahr, A. P. Assimacopoulos, M. M. Dinges, J. A. Stoehr, J. W. Harmala, H. Hirt, and G. M. Dunny. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneewind, O., K. F. Jones, and V. A. Fischetti. 1990. Sequence and structural characteristics of the trypsin-resistant T6 surface protein of group A streptococci. J. Bacteriol. 1723310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70267-281. [DOI] [PubMed] [Google Scholar]
- 42.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 694366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiomi, D., M. Yoshimoto, M. Homma, and I. Kawagishi. 2006. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol. Microbiol. 60894-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simonen, M., and I. Palva. 1993. Protein secretion in Bacillus species. Microbiol. Rev. 57109-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swaminathan, A., A. Mandlik, A. Swierczynski, A. Gaspar, A. Das, and H. Ton-That. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 66961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12420-430. [DOI] [PubMed] [Google Scholar]
- 47.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 9612424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ton-That, H., and O. Schneewind. 1999. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J. Biol. Chem. 27424316-24320. [DOI] [PubMed] [Google Scholar]
- 49.von Heijne, G. 1989. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341456-458. [DOI] [PubMed] [Google Scholar]
- 50.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49253-262. [DOI] [PubMed] [Google Scholar]
- 51.Zapun, A., T. Vernet, and M. G. Pinho. 2008. The different shapes of cocci. FEMS Microbiol. Rev. 32345-360. [DOI] [PubMed] [Google Scholar]
- 52.Zuber, B., M. Haenni, T. Ribeiro, K. Minnig, F. Lopes, P. Moreillon, and J. Dubochet. 2006. Granular layer in the periplasmic space of gram-positive bacteria and fine structures of Enterococcus gallinarum and Streptococcus gordonii septa revealed by cryo-electron microscopy of vitreous sections. J. Bacteriol. 1886652-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.