Abstract
The anaerobe Bacteroides fragilis is a gram-negative, opportunistic pathogen that is highly aerotolerant and can persist in aerobic environments for extended periods. In this study, the six B. fragilis thioredoxins (Trxs) were investigated to determine their role during oxidative stress. Phylogenetic analyses of Trx protein sequences indicated that four of the six Trxs (TrxA, TrxC, TrxD, and TrxF) belong to the M-type Trx class but were associated with two different M-type lineages. TrxE and TrxG were most closely associated to Y-type Trxs found primarily in cyanobacteria. Single and multiple trx gene deletions were generated to determine functional differences between the Trxs. The trxA gene was essential, but no anaerobic growth defects were observed for any other single trx deletion or for the ΔtrxC ΔtrxD::cfxA ΔtrxE ΔtrxF ΔtrxG quintuple mutant. Regulation of the trx genes was linked to the oxidative stress response, and all were induced by aerobic conditions. The ΔtrxC ΔtrxE ΔtrxF ΔtrxG and the ΔtrxC ΔtrxD::cfxA ΔtrxE ΔtrxF ΔtrxG multiple deletion strains were impaired during growth in oxidized media, but single trx gene mutants did not have a phenotype in this assay. TrxD was protective during exposure to the thiol oxidant diamide, and expression of trxD was induced by diamide. Diamide-induced expression of trxC, trxE, and trxF increased significantly in a trxD mutant strain, suggesting that there is some capacity for compensation in this complex Trx system. These data provide insight into the role of individual Trxs in the B. fragilis oxidative stress response.
Protective mechanisms for dealing with oxidative stress are an integral part of any organism that lives in, or is exposed to, an aerobic environment. While aerobic organisms have developed robust systems to contend with the constant threat of destructive oxygen radicals, anaerobic organisms introduced to an aerobic environment are at an elevated risk for damage. Oxygen toxicity in anaerobes is a complex phenomenon involving many aspects of cellular physiology that are impaired as oxidative damage occurs. For example, aerobic exposure of the aerotolerant Bacteroides thetaiotaomicron inhibits growth, in part due to the oxidation of iron-sulfur clusters located within metabolic enzymes (30). To combat this problem, some anaerobic bacteria have evolved multifaceted strategies to manage the production and effects of reactive oxygen species (38). Bacteroides fragilis is a commensal anaerobe found in the human intestine, but it also is the most frequently isolated anaerobe from human infections (10). B. fragilis is unable to multiply in the presence of air (21% O2); however, it is highly resistant to oxidative stress and can survive for extended periods in a fully aerobic environment. In this regard, B. fragilis is one of the most aerotolerant anaerobes known, and it is able to survive for at least 72 h in the presence of atmospheric oxygen. In contrast, intolerant anaerobes survive for less than 2 h in air (49). This remarkable resistance to oxidative stress is mediated by an oxidative stress response (OSR) which involves a wide array of genes activated during exposure to air or H2O2. An ever-growing set of genes and proteins that are induced in response to aerobic exposure have been discovered, and while the function of some have been deduced, many of their contributions to aerotolerance remain to be clarified (15, 36-38, 48). In this regard, a recent expression microarray showed that the B. fragilis thioredoxin (Trx) genes were induced by aerobic conditions, but their role in the OSR has not been adequately explored (48).
Trxs are small redox-active proteins (∼12 kDa) found in all phylogenetic branches. Trxs contain a highly conserved Cys-X-X-Cys motif at their active sites, allowing for catalysis of thiol-disulfide reactions (1, 40). The reduction of Trxs is mediated by flavin adenine dinucleotide-dependent Trx reductases (TrxB) which convert oxidized Trxs to their free thiol forms (1). Since the discovery of their role in DNA synthesis and in maintenance of the reduced state of intracellular protein disulfides, Trxs have been shown to be involved in defense against oxidative stress (17). Trxs regenerate oxidatively damaged proteins, modulate the activity of redox stressors, and act as hydrogen donors for detoxification enzymes important during the OSR (7, 9, 25, 27, 28).
Analysis of the B. fragilis genome revealed the presence of a single Trx reductase (TrxB) and six Trx homologs. This large repertoire of trx genes appears unusual compared to the typical smaller number of trx genes (two or three) found in other anaerobes (13, 19-21, 33, 42, 29). Previously, Rocha et al. (40) showed that the TrxB/Trx system is the primary thiol/disulfide redox system in B. fragilis; it has an important role in aerotolerance and is essential for survival in an in vivo mouse abscess model. These findings prompted us to propose that while TrxB is required for the function of the system overall, each Trx has important, specific roles in survival and defense against oxidative stress. In this study we present evidence that B. fragilis possesses a complex Trx system in which individual trx genes are differentially regulated but have some capacity to compensate for other trx genes under stress conditions. We also present evidence suggesting that TrxD has a major role in managing thiol oxidation and that trxA is an essential gene.
MATERIALS AND METHODS
Bacterial strains and growth.
B. fragilis strains used in this study are listed in Table 1. The trx gene homologs in strain 638R correspond to the following genes in the genome sequence (http://www.sanger.ac.uk/Projects/B_fragilis/): trxA, BF638R-0680, trxC, BF638R-2717, trxD, BF638R-2296; trxEF, BF638R-3044, 3045; and trxG, BF638R-1282. Strains were grown anaerobically in brain heart infusion broth supplemented with hemin and cysteine (BHIS) for routine cultures. Rifampin (20 μg/ml), gentamicin (100 μg/ml), tetracycline (5 μg/ml), erythromycin (10 μg/ml), and cefoxitin (25 μg/ml) were added to the medium when required. Disk diffusion assays to test for sensitivity to oxidative stress were performed by spreading overnight cultures on plates of either BHIS (without cysteine) or defined minimal medium (51), allowing the plates to dry, and then adding a sterile 6-mm filter disk to the center of the plate (47). Ten microliters of 2 M diamide was added to the disk, and then the plates were either placed in the anaerobic incubator or exposed to air (at 37°C) for 6 h prior to anaerobic incubation. Following overnight incubation, the diameters of the zones of growth inhibition were measured, and the results are the averages from three independent experiments done in triplicate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Phenotype and/or genotypea | Reference or source |
|---|---|---|
| B. fragilis strains | ||
| 638R | Clinical isolate, Rifr | 32 |
| ADB77 | 638R ΔthyA Rifr Tpr | 3 |
| IB298 | 638R ΔoxyR::tetQ Tetr Rifr | 37 |
| IB458 | ADB77 reverted to thyA+, ΔtrxC Rifr | This study |
| IB469 | ADB77 reverted to thyA+, ΔtrxD Rifr | This study |
| IB471 | ADB77 reverted to thyA+, ΔtrxG Rifr | This study |
| IB473 | ADB77 ΔthyA, ΔtrxA single crossover integrated into trxA gene, Rifr Tetr | This study |
| IB483 | ADB77 reverted to thyA+, ΔtrxC ΔtrxD::cfxA ΔtrxE ΔtrxF ΔtrxG Rifr Cfxr | This study |
| IB490 | ADB77 reverted to thyA+, ΔtrxE Rifr | This study |
| IB491 | ADB77 reverted to thyA+, ΔtrxF Rifr | This study |
| IB492 | ADB77 reverted to thyA+, ΔtrxE ΔtrxF Rifr | This study |
| IB498 | ADB77 reverted to thyA+, ΔtrxC ΔtrxE ΔtrxF ΔtrxG Rifr | This study |
| IB499 | IB498 ΔoxyR::tetQ Rifr Tetr | This study |
| IB500 | IB483 ΔoxyR::tetQ Rifr Cfxr Tetr | This study |
| E. coli strains | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG tonA | Invitrogen |
| HB101::RK231 | HB101 containing RK231, Kmr Tcr Str | 12 |
| Plasmids | ||
| pFD340 | Bacteroides-E. coli expression shuttle vector, (Ampr), Ermr | 44 |
| pFD516 | Bacteroides suicide vector, derived by deletion of Bacteroides replicon pBI143 from pFD288, (Spr), Ermr | 45 |
Ermr, erythromycin resistance; Cfxr, cefoxitin resistance; Rifr, rifampin resistance; Tetr, tetracycline resistance; Tpr, trimethoprim Spr, spectinomycin resistance; Ampr, ampicillin resistance. For Bacteroides-E. coli shuttle vectors, parentheses indicate antibiotic resistance expression in E. coli.
For the oxidized medium experiments, sterile BHIS broth with no cysteine was split and either maintained in the anaerobe chamber (anaerobic medium) or shaken in an aerobic incubator at 250 rpm for 24 h (oxidized medium). Overnight cultures grown in BHIS were diluted 1:20 in fresh anaerobic BHIS, grown to mid-log phase (A550 of 0.3 to 0.5), inoculated at 1:50 into 5 ml of “anaerobic” or “oxidized” medium, and placed in the anaerobic incubator at 37°C. The A550 of the cultures was followed, and the results represent two independent experiments performed in triplicate.
Construction of trx deletion mutants.
Briefly, chromosomal fragments containing an N-terminal portion of the trx gene, (upstream from the conserved Trx cysteine residues) was amplified by PCR with oligonucleotides containing nucleotide modifications to create sites for BamHI at the 5′ end and PstI at the 3′ end and then cloned into pUC19. The same approach was applied to create fragments for the C-terminal ends of the constructs, except PstI was at the 5′ end and HindIII at the 3′ end. The amplified fragments were then ligated together to create the mutated gene fragment, which was inserted into the Bacteroides suicide vector pYT102 (3). These plasmids were mobilized into B. fragilis ADB77, and exconjugates were selected on BHIS plates containing rifampin, gentamicin, and tetracycline (3). Sensitivity to tetracycline, resistance to trimethoprim, and PCR were used to confirm the double-crossover allelic exchange into the B. fragilis chromosome to create the in-frame, unmarked trx deletion mutants. Table S1 in the supplemental material shows the amino acids deleted for each mutant generated. Multiple trx mutations were constructed by subsequent rounds of mutagenesis, resulting in strain IB492 and strain IB498 (Table 1). All ΔthyA strains were reverted to thyA+ prior to phenotypic characterization as described previously (3).
Construction of a marked trxD deletion mutant.
Briefly, a 308-bp chromosome fragment containing the C-terminal portion of trxD was amplified by PCR using oligonucleotides containing restriction sites for SstI and EcoRI and cloned into the Bacteroides suicide vector pFD516 (45). Next, a 993-bp chromosome fragment containing the N-terminal portion of trxD was amplified by PCR using oligonucleotides containing restriction sites for BamHI and SstI and then cloned into the plasmid. The resulting plasmid contained a 215-bp ΔtrxD allele with a 139-bp deletion which encompassed the conserved cysteine residues. Next, a 1.1-kb SstI cefoxitin (cfxA) resistance gene cassette was cloned into the unique SstI site to create the plasmid pFDtrxDcfx. This plasmid was mobilized into B. fragilis strain IB498, and exconjugants were selected on BHIS containing rifampin, gentamicin, and cefoxitin. Sensitivity to erythromycin was determined, and PCR was performed to confirm the double-crossover allelic exchange of the trxD::cfxA mutation into strain IB498 to create the quintuple mutant designated strain IB483.
The ΔtrxC ΔtrxE ΔtrxF ΔtrxG ΔoxyR::tetQ and ΔtrxC ΔtrxD::cfxA ΔtrxE ΔtrxF ΔtrxG ΔoxyR::tetQ mutants were constructed by mobilizing suicide vector pFD754 containing the ΔoxyR::tetQ mutant allele (37) into B. fragilis as described above. Exconjugants were selected on BHIS containing rifampin, gentamicin, tetracycline, and cefoxitin (when necessary). Sensitivity to either tetracycline/cefoxitin (when necessary) or erythromycin was used to identify recombinants that were tetracycline and cefoxitin resistant and erythromycin sensitive. These two strains were designated strain IB499 and strain IB500, respectively.
Trx overexpression constructs.
Plasmids constitutively expressing specific trx genes were constructed by PCR amplification of promoterless trx genes. The promoterless trx gene fragments containing the ribosome binding site were cloned into the BamHI and SstI sites of the Bacteroides-Escherichia coli shuttle expression vector pFD340 (44) in the same orientation as the IS4351 constitutive promoter. The new constructs, ptrxA, ptrxC, ptrxD, ptrxE, ptrxF, ptrxG, and ptrxEF, were individually mobilized into B. fragilis strains as described above. Transconjugants were selected on BHIS containing rifampin, gentamicin, and erythromycin. The primers used for these plasmid overexpression constructs are listed in Table S2 in the supplemental material.
RNA isolation and cDNA synthesis.
RNA was isolated using the hot acid-phenol method (39). Fifty micrograms of total RNA was precipitated with ethanol, and contaminating DNA was removed by treatment with Turbo DNA-free DNase (Ambion). The RNA concentration was determined by measuring the A260/A280 ratio. Synthesis of cDNA was as follows: 0.75 μg of RNA was added to reaction mixtures containing 10 ng/μl random hexamers, 0.5 mM deoxynucleoside triphosphates, first-strand buffer (Invitrogen, Carlsbad, California), and 1 μl Superscript II RNase H reverse transcriptase I; reaction mixtures were incubated at 42°C for 50 min; and Superscript II was heat inactivated by incubating the reaction mixtures at 70°C for 15 min.
Quantitative PCR.
Quantitative real-time PCR was performed essentially as described previously using a Bio-Rad iCycler with the real-time PCR detection system (Bio-Rad, Hercules, California) (47). The primers used were designed to amplify products of 100 to 150 bp. All products were verified by agarose gel electrophoresis and by melting point analysis according to the Bio-Rad iCycler software. The reaction mixture contained 12.5 μl 2× iQ Sybr green Supermix, 1.5 μl of 5 μM of forward primer, 1.5 μl 5 μM of reverse primer, 8.5 μl H2O, and 1 μl of cDNA template (diluted 1/100) per well. All samples were run in triplicate, and RNA with no reverse transcriptase was run as a control to monitor for genomic DNA contamination. Relative expression values were calculated using the Pfaffl method (31). Fold induction relative to the wild type under anaerobic conditions was determined for each gene using the sigma-54 modulation protein gene as the reference gene, which does not vary significantly under conditions tested (48). All results were the averages from at least two independent experiments in triplicate with freshly isolated RNA.
Northern hybridization.
Cultures were grown in BHIS to early logarithmic phase and either maintained anaerobically for 15 min, treated with 50 μM H2O2 for 15 min, or shaken aerobically at 250 rpm for 1 hour as previously described (48). RNA was then isolated, and Northern blot analysis was carried out as previously described (39). The entire open reading frame of each trx gene was radiolabeled with [32P]dCTP and used as the hybridization probe. Densitometry analysis of the autoradiograph was normalized to the relative intensity of total 23S and 16S rRNAs detected on the ethidium bromide-stained agarose gel to correct for any loading differences.
RESULTS
Trx system in B. fragilis.
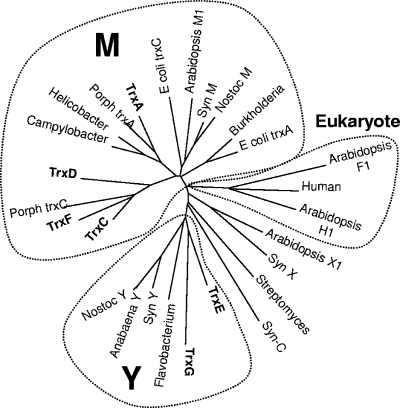
Previous studies demonstrated that B. fragilis possesses an extensive Trx system consisting of Trx reductase (TrxB) and six Trx orthologs, each of which has the classic CXXC redox-active center (40). The work showed that this is the primary mechanism for controlling the intracellular thiol/disulfide equilibrium and that in the absence of TrxB, cells required an exogenous reducing agent for growth (40). Since there is no glutathione system, it is likely that B. fragilis Trx proteins play diverse roles in cellular metabolism, some of which may overlap with classic functions associated with glutaredoxins or glutathione in other organisms. The presence of six distinct Trx proteins in a heterotrophic prokaryote is quite unusual. Therefore, we used phylogenetic comparisons to gain insight into the evolution and potential roles of these Trxs. The results in Fig. 1 suggest that the B. fragilis Trxs are comprised of several discrete types, analogous to the case for cyanobacteria, where six or more Trxs are common and at least four types, M, X, Y, and C have been described (11, 16). Four of the B. fragilis Trxs grouped with the archetypical prokaryotic/mitochondrial Trxs referred to as the M type, but within this group they formed distinct subgroups. TrxA was associated with Campylobacter, Helicobacter, and Porphyromonas gingivalis in one divergent group, whereas TrxC, TrxD, and TrxF, which appear to share a common ancestor, formed a second M subgroup that included a second P. gingivalis Trx. The greatest divergence was observed for TrxE and TrxG, which usually clustered with the Y type of prokaryotic Trx isoforms. The Y isoforms have been found in cyanobacteria, single-cell algae, and plants and are known to have unique in vitro target specificity (22).
FIG. 1.
Phylogenetic comparison of 27 Trx proteins from diverse sources. ClustalW was used to align protein sequences for 27 Trx proteins. The unrooted bootstrap consensus tree was inferred using the minimum-evolution method with 500 bootstrap replicates (41). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Phylogenetic analyses were conducted in MEGA4 (50). The following sequences with accession numbers were used: Burkholderia, YP_333769; human, NP_003320; E. coli TrxA, AAA67582; Porph TrxC, P. gingivalis AAQ65495; Syn M, Synechococcus ZP_01124485; Anabaena Y, ABA23368; Flavobacterium, CAL43878; Nostoc Y, NP_485933; Syn Y, Synechocystis NP_442168; Campylobacter, YP_178167; Helicobacter, NP_223481; Porph TrxA, P. gingivalis NP_904389; Streptomyces, CAB72414; TrxA, YP_210347; TrxC, YP_212311; TrxD, YP_211860; TrxE, YP_212629; TrxF, YP_212630; TrxG, YP_210941; Arabidopsis H1, CAA78462; Arabidopsis F1, AAD35003; Arabidopsis X1, NP_564566; Syn X, Synechocystis NP_440611; Syn C, Synechocystis NP_439965; E. coli TrxC, NP_417077; Arabidopsis M1, AAF15948; Nostoc M, NP_485906.
Several of the branches in the tree shown in Fig. 1 had low bootstrap values (see Fig. S1 in the supplemental material), presumably because of the small size of the Trx proteins. However, we have a high degree of confidence in the tree topology for the following reasons: (i) consensus trees constructed by four different methods yielded very similar results (the methods used were neighbor joining, minimum evolution, unweighted-pair group method using average linkages, and maximum parsimony); (ii) consensus trees constructed using 81 Trxs and protein disulfide isomerases yielded similar results; and (iii) results from interior branch tests were 98% confidence probability for the TrxA group, 99% for the TrxC,D,F group, and 84% for the TrxE,G group (data not shown). In summary, the B. fragilis Trxs fall into three divergent groups resulting from at least two independent lines of descent. The phylogenetic relationships and sequence divergence of the Trxs are consistent with evolution toward specialized functions which will need to be determined for a better understanding of their roles in the physiology of anaerobic bacteria.
Generation of trx mutants of B. fragilis.
The first step for analysis of the Trxs was construction of unmarked deletion mutants using a two-step positive-selection vector, pYT102 (Fig. 2). Deletions were successfully obtained for all genes except trxA, in which case it was possible to obtain the initial single-crossover event but selection for the double-crossover event always resulted in the isolation of colonies with the wild-type locus (Table 2). By comparison, between 20 and 40% of trxC, trxD, trxE, trxF, and trxG double crossovers were deletion mutants. This result suggested that trxA might be essential, so we set out to determine if trxA cloned into a constitutive expression vector could rescue deletion mutants. As shown in Table 2, in the presence of ptrxA, deletion mutants were recovered at a frequency similar to that observed for the other trx genes. In contrast, none of the other cloned trx genes were able to rescue trxA deletion formation. This was the first indication of Trx target specificity in B. fragilis.
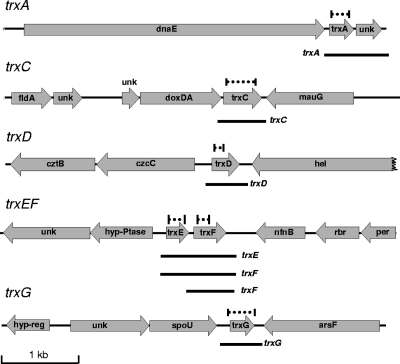
FIG. 2.
Genetic loci of the six B. fragilis trx genes. The maps are drawn to scale; the dashed lines above the trx genes show the regions deleted in each trx mutant, and the black lines under the trx genes represent the approximate sizes of the mRNAs observed in Fig. 3. Genes: unk, unknown with no matches in database; dnaE, DNA polymerase III; fldA, flavodoxin; doxDA, thiosulfate quinone oxidoreductase; mauG, tryptophan tryptophylquinone synthesis; cztBC, heavy metal efflux pump; hel, DNA helicase; hyp-Ptase, hypothetical phosphatase; nfnB, oxygen-insensitive nitroreductase; rbr, rubrerythrin-like; per, peroxide response regulator homolog; hyp-reg, hypothetical DNA binding protein; spoU, SpoU-like RNA methylase; arsF, sulfatase.
TABLE 2.
Isolation of trxA deletion mutations in the presence of complementing plasmidsa
| Plasmid | No. (%) of trxA deletions observed/total no. tested |
|---|---|
| None | 0/468 |
| ptrxA | 27/84 (32) |
| ptrxC | 0/23 |
| ptrxD | 0/20 |
| ptrxE | 0/51 |
| ptrxF | 0/47 |
| ptrxG | 0/72 |
Strain IB473 containing the trxA deletion construct single crossover integrated into the trxA gene was the recipient for all trx-containing plasmids. Deletions were identified by PCR analysis of the genomic DNA isolated from individual colonies that had resolved the trxA deletion construct (see Materials and Methods for more details).
Multiple trx gene knockouts were constructed from single deletion mutants by applying multiple rounds of the pYT102/ABD77 strategy. In this way an unmarked ΔtrxC ΔtrxE ΔtrxF ΔtrxG quadruple mutant (strain IB498) was constructed, but multiple attempts to construct the quintuple mutant using the unmarked ΔtrxD were not successful with IB498. Finally, the quintuple mutant was constructed by double-crossover insertion of a ΔtrxD::cfxA construct containing a cefoxitin resistance cassette. This mutant, strain IB483, had only an intact trxA gene, but it did not display any anaerobic growth defects in either complex or defined media (data not shown).
Induction of trx genes by oxidative stress.
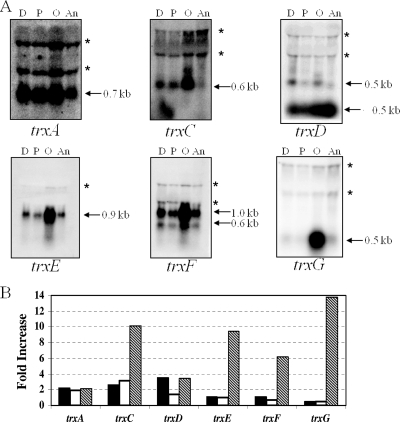
Trxs have been shown to be important during oxidative stress as a source of reducing power for detoxification reactions and the regeneration of inactivated proteins (7, 9, 25, 27, 28). Consistent with this, a previous study using expression microarray data showed trx gene induction in B. fragilis exposed to aerobic conditions (48). To verify this induction and determine trx gene organization, we performed Northern blot hybridizations using RNA isolated from cultures exposed to atmospheric oxygen, hydrogen peroxide, and the thiol-specific oxidant diamide. The analysis revealed differential expression of the trx genes under each of the conditions tested (Fig. 3). Aerobic conditions induced the expression of all trx genes. The trxC, trxD, trxF, and trxG transcripts were monocistronic, whereas the trxA transcript was part of an operon with a hypothetical gene and trxEF was a bicistronic mRNA. The trxG transcript showed the highest fold induction at nearly 14-fold over the anaerobic control. The trxC and trxD genes were induced during diamide exposure, and trxC showed substantial induction (threefold) during hydrogen peroxide exposure. Interestingly, a second RNA species, which was less than 200 bp, was observed to hybridize strongly to the trxD probe. This RNA was in greatest abundance during anaerobic growth and may be a small RNA species. Alternatively, this fast-migrating RNA band may be the product of premature trxD termination or posttranscriptional regulation. The trxA transcript was constitutively expressed under anaerobic conditions and increased only about twofold under the stress conditions tested. Finally, the Northern blot hybridizations confirmed previous in silico analysis that trxE and trxF are in a two-gene operon and are expressed primarily in a polycistronic message (Fig. 3).
FIG. 3.
Northern hybridization analysis of total RNA of B. fragilis strain 638R (wild type). RNA was isolated from cells grown to mid-logarithmic phase in BHIS and then treated as described in the text: 500 μM diamide (D), 50 μM hydrogen peroxide (P), exposed to air (O), or untreated (An). (A) Autoradiographs of blots hybridized to radiolabeled probes containing the entire open reading frame of each trx gene as indicated. The approximate sizes of the transcripts are shown. The apparent bands (*) at about 1.5 and 2.5 kb are a commonly observed compression artifact caused by the 16S and 23S rRNAs. (B) Fold increase of transcript levels under each condition compared to the anaerobic control based on densitometric values. Black bars, 500 μM diamide; white bars, 50 μM H2O2; hatched bars, aerobic exposure.
Growth in oxidized medium.
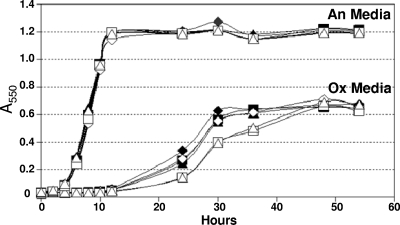
In order to determine if the Trxs were important for growth during oxidative stress, we examined the ability of trx mutants to initiate growth in oxidized medium. This assay is used to determine if there is a defect in the ability to rapidly reduce the medium and initiate growth in the presence of low levels of oxygen. Typically, the expected phenotype is an extended lag period prior to the start of growth. Compared to the wild-type strain, the single trx mutant strains had no significant defect in either anaerobic growth or the ability to initiate growth in the oxidized medium (data not shown). However, as in seen Fig. 4, the ΔtrxC ΔtrxE ΔtrxF ΔtrxG (strain IB498) and ΔtrxC ΔtrxD::cfxA ΔtrxE ΔtrxF ΔtrxG (strain IB483) mutants were somewhat impaired in the ability to grow in the oxidized medium. We hypothesized that the OxyR regulon might have masked a greater growth defect of the trx mutants due to its role in the rapid removal of oxygen radicals (48). Thus, oxyR deletion derivatives of both strain IB498 and strain IB493 were constructed by allelic exchange and tested in the oxidized medium. These mutants had a reproducibly longer lag period in oxidized medium than the wild type, multiple trx mutant strains, or the oxyR single mutant, but there were no anaerobic growth defects (Fig. 4). When in combination with oxyR, the multiple trx mutants also grew at a lower rate, taking longer to reach maximum growth, suggesting a cumulative decrease in the ability to combat oxidative stress when both the Trx and OxyR systems are impaired.
FIG. 4.
Growth analysis of B. fragilis trx and oxyR mutant strains in anaerobic and oxidized media. Strains were grown overnight in BHIS and then inoculated into either fully oxidized medium (Ox) or anaerobic medium (An). Growth was measured on a spectrophotometer at 550 nm. The results shown are the averages from triplicate observations in two growth experiments. Strains IB101 (wild type, ⧫), IB298 (ΔoxyR, ⋄), IB498 (ΔtrxC ΔtrxE ΔtrxF ΔtrxG ▪), IB499 (ΔtrxC ΔtrxE ΔtrxF ΔtrxG ΔoxyR::tetQ □), IB483 (ΔtrxC ΔtrxD::cfxA ΔtrxE ΔtrxF ΔtrxG ▴), and IB500 (ΔtrxC ΔtrxD::cfxA ΔtrxE ΔtrxF ΔtrxG ΔoxyR::tetQ ▵) were used.
Sensitivity to diamide.
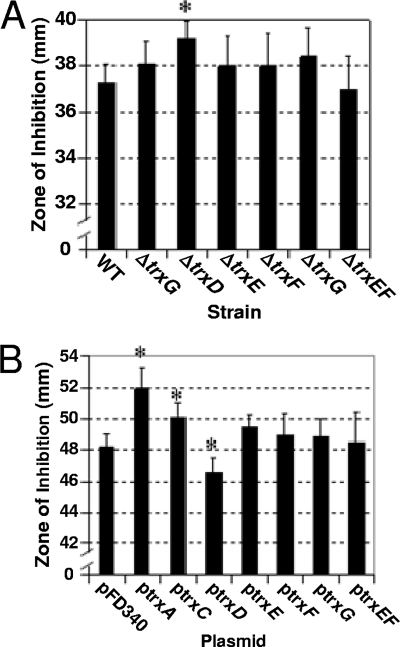
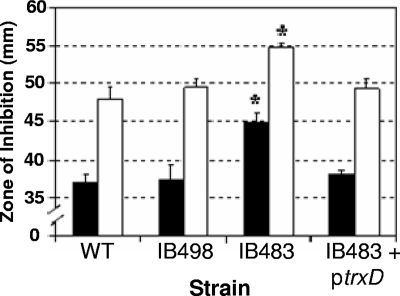
Diamide is a thiol-oxidizing agent that mimics damage due to oxygen exposure (26). Therefore, sensitivity to diamide was used to establish if any of the Trxs were important for thiol/disulfide homeostasis. As shown in Fig. 5A, the ΔtrxD mutant was more sensitive to diamide in disk diffusion assays than the parent strain, other single trx mutants, and the ΔtrxE ΔtrxF double mutant. Furthermore, the parent strain harboring the multicopy plasmid with trxD expressed from the constitutive IS4351 promoter (ptrxD) was less sensitive to diamide than the parent strain or strains with any of the other trx gene-containing expression plasmids (Fig. 5B). Although the effect of the single ΔtrxD mutation alone was small, but statistically significant, a more dramatic difference was observed with strain IB483, which lacked all functional trx genes except for trxA. In this mutant background, the addition of ptrxD also restored diamide sensitivity back to a wild-type level. This complementation with trxD is consistent with the observation that the quadruple mutant, IB498, which lacked all functional trx genes except for trxA and trxD, was not significantly more sensitive to diamide than the wild type (Fig. 6).
FIG. 5.
Effect of each Trx on survival during oxidative stress. (A) Wild-type strain 638R was compared to trx mutant strains in diamide disk diffusion assays on BHIS plates with no added cysteine. (B) Strain 638R harboring the empty expression vector pFD340 was compared to 638R strains harboring pFD340 containing B. fragilis trx genes in diamide disk diffusion assays on defined minimal medium. The values are mean diameters of growth inhibition zones measured in three independent experiments performed in triplicate and are given in millimeters. The error bars indicate standard deviations. *, P < 0.01 compared to wild-type strain. Strains in panel A: wild type (WT), 638R; ΔtrxC, IB458; ΔtrxD, IB469; ΔtrxE, IB490; ΔtrxF, IB491; ΔtrxG, IB471; and ΔtrxEF, IB492.
FIG. 6.
Rescue of strain IB483 diamide sensitivity phenotype by plasmid ptrxD. Diamide disk diffusion assays were used to compare sensitivities of the wild-type strain 638R, the quadruple trx mutant strain IB498, the quintuple trx mutant strain IB483, and IB483 expressing trxD on plasmid ptrxD. Black bars represent plates placed directly into an anaerobic incubator after plating, and open bars represent plates placed in an aerobic incubator for 6 h prior to being placed into the anaerobic incubator. The values are mean diameters of growth inhibition from three independent experiments performed in triplicate, and are given in millimeters. The error bars indicate standard deviations. *, P < 0.001 compared to wild-type strain.
Real-time RT-PCR analysis of trx gene expression during diamide exposure.
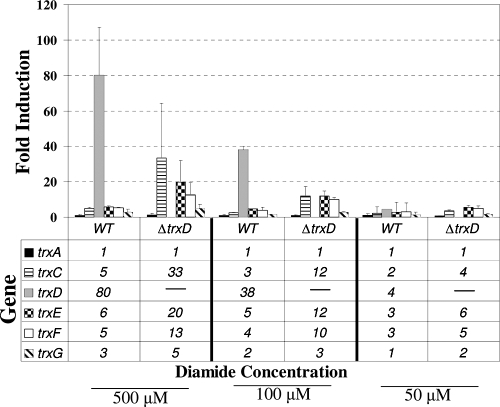
The presence of multiple trx genes in the genome suggested the possibility that there may be some overlap in their roles in managing thiol stresses. Consistent with this is the observation that the single trx mutations generally did not result in strong oxidative stress phenotypes (Fig. 5). To investigate the potential for compensatory regulation of trx genes, real-time reverse transcription-PCR (RT-PCR) analysis was performed to determine specific differences in activation of these genes after exposure to diamide oxidative stress. As shown in Fig. 7, the wild-type strain 638R demonstrates a pattern of trx gene expression after 5 min of exposure to diamide that is similar to what was observed in the Northern blot hybridization analysis (Fig. 3), with trxD having the highest induction of any trx gene under this stress. The fold induction of trxD was somewhat greater than that seen in the Northern blot experiments, but this is likely due to the greater sensitivity and dynamic range of real-time RT-PCR. Interestingly, in the ΔtrxD mutant, there was a dramatic increase in the induction of trxC, trxE, and trxF after exposure to both 500 μM and 100 μM diamide, suggesting that these genes were upregulated in response to the cell now lacking a functional TrxD.
FIG. 7.
Transcriptional analysis of trx genes. The parental strain (wild type [WT], B. fragilis strain 638R) and the isogenic trxD mutant (ΔtrxD, strain IB469) were exposed for 5 min to 500 μM, 100 μM, and 50 μM diamide or maintained under standard anaerobic conditions (0 μM control). For each condition, RNA was isolated and real-time RT-PCR was performed in triplicate. The sigma-54 modulation protein gene was used as a standard, and the results are expressed as fold induction relative to levels under the control condition. The values are means of fold induction, compared to the 0 μM control, from two independent experiments. The error bars indicate standard deviations.
DISCUSSION
It has been known for some time that Trxs play important roles in the virulence and survival of a wide array of pathogenic bacteria, yeast, and protozoa, but there has been very little information available on Trxs in anaerobes (5, 24, 52). In this regard, the genome sequence of B. fragilis revealed that the Trx system was unexpectedly complex, with six trx genes and at least 11 additional genes for proteins in the Trx superfamily of thiol-disulfide interchange proteins. Our phylogenetic analysis showed that the six bona fide B. fragilis Trxs did not arise from recent gene duplication events but had divergent lineages and fell into three distinct classes (Fig. 1). The diversity among the B. fragilis Trx protein sequences suggests some level of functional specialization, and this was borne out by their differential regulation and phenotypic analyses. In one clear example, TrxA appeared to be essential (Table 2), and it was highly expressed under all stress conditions tested (Fig. 3), suggesting that it plays a specific and crucial role in growth and during stress. Evidence for this is that the trxA gene constitutively expressed on a plasmid was able to rescue a trxA chromosomal deletion mutant, but neither trxC, trxD, trxE, trxF, nor trxG could substitute (Table 2). The role of TrxA is not known, but we cannot rule out that it may function in DNA replication. One scenario is based on the chromosomal location of trxA, which is just 62 bp downstream of dnaE, encoding the alpha subunit of DNA polymerase III (Fig. 2), although it is transcribed independently of dnaE. There is some precedent for this type of a role for Trx, which is required for the processivity of T7 DNA polymerase, but the active-site cysteines are not required for this activity (18). Alternatively, TrxA could function in its traditional role to reduce ribonucleotide reductase (RNR), but this is generally not required for anaerobic RNRs.
In a previous study we showed that the B. fragilis Trx system is dependent on a single Trx reductase, TrxB, and since there is no glutathione system, the deletion of trxB completely disrupts cellular redox homeostasis, resulting in sensitivity to oxidative stress (40). This study did not provide any insight into the roles of individual Trxs, but it did suggest that some Trxs should have specific roles in the OSR. In order to determine these roles, mutant strains harboring single and multiple trx deletions were compared with complemented strains in several oxidative stress assays. When comparing the phenotypes observed after the deletion of trxD in both the wild-type and ΔtrxC ΔtrxE ΔtrxF ΔtrxG backgrounds (Fig. 5A and 6) and the subsequent complementation of trxD on a plasmid (Fig. 5B and 6), we were able to demonstrate the importance of TrxD in protection against diamide-induced disulfide stress, a specific subset of oxidative stress. It is possible that TrxD is the preferred electron donor for the repair of inadvertent disulfides due to its transcriptional regulation, or it may be a partner in a specific disulfide repair pathway. Future studies to identify TrxD protein partners will be able to provide insight into this.
Results from the current study suggest that all of the Trxs have some role in the OSR, as they are induced by aerobic conditions. Further, there may be significant overlap in the Trx stress activities, since the cell needed to be depleted of at least four of the six Trxs before there was any effect on growth in the oxidized medium (Fig. 4). This suggests that these proteins can compensate for one another, and this was supported by observations on the regulation of their expression. For example, we observed substantial increases in diamide-induced expression of four trx genes in the ΔtrxD mutant compared to the wild type. However, we should point out that these studies did not directly address the actual levels of Trx proteins produced, and there could be forms of posttranscriptional or posttranslational regulation that contribute to the overall control of redox homeostasis. In this regard, there was the observation of a putative small RNA species associated with trxD transcription, and this could potentially be involved in some posttranscriptional regulation.
Overall, there are many roles that Trx proteins may play during oxidative stress, such as providing reducing power for methionine sulfoxide reductase and peroxidases. B. fragilis induces five peroxidases in the presence of oxygen, but only one of these, AhpC, has a known specific reductant (48). Another possible role for the Trxs may be during emergence from oxidative stress. Although B. fragilis is an obligate anaerobe, previous studies have shown that expression of an aerobic class Ia RNR is induced in response to aerobic exposure, and mutants lacking this RNR have an impaired recovery response following exposure to air (43). There also is the potential need for a class Ia aerobic RNR during growth of B. fragilis in the presence of low (nanomolar) concentrations of oxygen (4). Thus, there may be several opportunities for some of the Trx proteins to act as reductants for the aerobic RNR.
In other organisms, such as E. coli, different components of the cellular redox systems show some specificity yet there is significant redundancy as well (6, 29, 34, 35, 46, 53). The glutathione/glutaredoxin and TrxB/Trx systems share the ability to reduce many overlapping cytoplasmic substrates, including RNR (2, 23). However, E. coli also demonstrates significant specificity with some substrates such as the membrane-associated reducing protein DsbD, which requires Trx1, and methionine sulfoxide reductase is optimally reduced by Trx1 (46). Interestingly, the roles of the glutathione system may be tasked by the expanded Trx system in B. fragilis, since it lacks an alternative (8, 40).
It should be noted that the OSR in B. fragilis is not limited to trx genes alone. The OxyR regulon has been shown to be vital for dealing with oxidative stress in B. fragilis, and our data in Fig. 5 indicate that the Trx and OxyR systems have an additive effect on resistance to oxidative stress (37, 48). Previous work suggests that the Trx system acts independently on different oxidative stresses than the OxyR regulon (40, 48). Consistent with this, OxyR does not appear to control any of the Trx genes, including trxB, indicating that there is separation of the control of thiol metabolism from the peroxide response, which is similar to what occurs in Bacillus subtilis (14). However, there likely is an important link between Trx and OxyR in B. fragilis. This is suggested by previous studies with E. coli that have shown that a deficit in the reducing power of the cytoplasm can delay the deactivation of OxyR, enhancing the stress response (53). Therefore, if we are able to show in future experiments that a depletion of Trx in B. fragilis can likewise delay the modulation of OxyR activity, such data would help to provide evidence of a Trx-controlled OxyR pathway.
The continued examination of trx genes in B. fragilis illustrates the complexity of this system in this species compared to other organisms. The evolutionary benefits of acquiring and maintaining the wide array of trx genes may offer a partial explanation as to why this obligate anaerobe is able to endure temporary environmental exposures of atmospheric oxygen. Understanding the coordinate regulation of this system and other aspects of the OSR will be necessary to determine how B. fragilis is able to adapt to niches outside its normal intestinal environment.
Supplementary Material
Acknowledgments
We thank J. M. Gee for helpful discussions and suggestions.
This work was supported in part by Public Health Service grant AI40588 to C.J.S.
Footnotes
Published ahead of print on 13 March 2009.
Supplemental material for this article is available at http://jb.asm.org/.
REFERENCES
- 1.Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2676102-6109. [DOI] [PubMed] [Google Scholar]
- 2.Aslund, F., B. Ehn, A. Miranda-Vizuete, C. Pueyo, and A. Holmgren. 1994. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc. Natl. Acad. Sci. USA 919813-9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baughn, A. D., and M. H. Malamy. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 994662-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427441-444. [DOI] [PubMed] [Google Scholar]
- 5.Bjur, E., S. Eriksson-Ygberg, F. Aslund, and M. Rhen. 2006. Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 745140-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54439-461. [DOI] [PubMed] [Google Scholar]
- 7.Chae, H. Z., S. J. Chung, and S. G. Rhee. 1994. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 26927670-27678. [PubMed] [Google Scholar]
- 8.Fahey, R. C., and G. L. Newtwon. 1983. Occurrence of low molecular weight thiols in biological systems, p. 251-260. In A. Larsson, S. Orrenius, A. Holmgren, and B. Mannervik (ed.), Functions of glutathione: bio-chemical, physiological, toxicological, and clinical aspects. Raven Press, New York, NY.
- 9.Fernando, M. R., H. Nanri, S. Yoshitake, K. Nagata-Kuno, and S. Minakami. 1992. Thioredoxin regenerates proteins inactivated by oxidative stress in endothelial cells. Eur. J. Biochem. 209917-922. [DOI] [PubMed] [Google Scholar]
- 10.Finegold, S. M., and W. L. George. 1989. Anaerobic infections in humans. Academic Press, New York, NY.
- 11.Florencio, F. J., M. E. Perez-Perez, L. Lopez-Maury, A. Mata-Cabana, and M. Lindahl. 2006. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth. Res. 89157-171. [DOI] [PubMed] [Google Scholar]
- 12.Guiney, D. G., P. Hasegawa, and C. E. Davis. 1984. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc. Natl. Acad. Sci. USA 817203-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms, C., M. A. Meyer, and J. R. Andreesen. 1998. Fast purification of thioredoxin reductases and of thioredoxins with an unusual redox-active centre from anaerobic, amino-acid-utilizing bacteria. Microbiology 144793-800. [DOI] [PubMed] [Google Scholar]
- 14.Helmann, J. D., M. F. Wu, A. Gaballa, P. A. Kobel, M. M. Morshedi, P. Fawcett, and C. Paddon. 2003. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herren, C. D., E. R. Rocha, and C. J. Smith. 2003. Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene 316167-175. [DOI] [PubMed] [Google Scholar]
- 16.Hisabori, T., K. Motohashi, N. Hosoya-Matsuda, H. Ueoka-Nakanishi, and P. G. Romano. 2007. Towards a functional dissection of thioredoxin networks in plant cells. Photochem. Photobiol. 83145-151. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren, A. 1984. Enzymatic reduction-oxidation of protein disulfides by thioredoxin. Methods Enzymol. 107295-300. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, D. E., and C. C. Richardson. 2003. A covalent linkage between the gene 5 DNA polymerase of bacteriophage T7 and Escherichia coli thioredoxin, the processivity factor: fate of thioredoxin during DNA synthesis. J. Biol. Chem. 27823762-23772. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, T. C., N. A. Crawford, and B. B. Buchanan. 1984. Thioredoxin system of the photosynthetic anaerobe Chromatium vinosum. J. Bacteriol. 1581061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasaki, S., Y. Watamura, M. Ono, T. Watanabe, K. Takeda, and Y. Niimura. 2005. Adaptive responses to oxygen stress in obligatory anaerobes Clostridium acetobutylicum and Clostridium aminovalericum. Appl. Environ. Microbiol. 718442-8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuchi, Y., N. Ohara, K. Sato, M. Yoshimura, H. Yukitake, E. Sakai, M. Shoji, M. Naito, and K. Nakayama. 2005. Novel stationary-phase-upregulated protein of Porphyromonas gingivalis influences production of superoxide dismutase, thiol peroxidase and thioredoxin. Microbiology 151841-853. [DOI] [PubMed] [Google Scholar]
- 22.Lemaire, S. D., V. Collin, E. Keryer, A. Quesada, and M. Miginiac-Maslow. 2003. Characterization of thioredoxin y, a new type of thioredoxin identified in the genome of Chlamydomonas reinhardtii. FEBS Lett. 54387-92. [DOI] [PubMed] [Google Scholar]
- 23.Miranda-Vizuete, A., A. Rodriguez-Ariza, F. Toribio, A. Holmgren, J. Lopez-Barea, and C. Pueyo. 1996. The levels of ribonucleotide reductase, thioredoxin, glutaredoxin 1, and GSH are balanced in Escherichia coli K12. J. Biol. Chem. 27119099-19103. [DOI] [PubMed] [Google Scholar]
- 24.Missall, T. A., and J. K. Lodge. 2005. Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol. Microbiol. 57847-858. [DOI] [PubMed] [Google Scholar]
- 25.Mitsui, A., T. Hirakawa, and J. Yodoi. 1992. Reactive oxygen-reducing and protein-refolding activities of adult T cell leukemia-derived factor/human thioredoxin. Biochem. Biophys. Res. Commun. 1861220-1226. [DOI] [PubMed] [Google Scholar]
- 26.Morris, J. G. 1975. The physiology of obligate anaerobiosis. Adv. Microbiol. Physiol. 12169-246. [Google Scholar]
- 27.Nakamura, H., M. Matsuda, K. Furuke, Y. Kitaoka, S. Iwata, K. Toda, T. Inamoto, Y. Yamaoka, K. Ozawa, and J. Yodoi. 1994. Adult T cell leukemia-derived factor/human thioredoxin protects endothelial F-2 cell injury caused by activated neutrophils or hydrogen peroxide. Immunol. Lett. 4275-80. [DOI] [PubMed] [Google Scholar]
- 28.Natsuyama, S., Y. Noda, K. Narimoto, Y. Umaoka, and T. Mori. 1992. Release of two-cell block by reduction of protein disulfide with thioredoxin from Escherichia coli in mice. J. Reprod. Fertil. 95649-656. [DOI] [PubMed] [Google Scholar]
- 29.Ortenberg, R., S. Gon, A. Porat, and J. Beckwith. 2004. Interactions of glutaredoxins, ribonucleotide reductase, and components of the DNA replication system of Escherichia coli. Proc. Natl. Acad. Sci. USA 1017439-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan, N., and J. A. Imlay. 2001. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol. Microbiol. 391562-1571. [DOI] [PubMed] [Google Scholar]
- 31.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Privitera, G., A. Dublanchet, and M. Sebald. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 13997-101. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds, C. M., J. Meyer, and L. B. Poole. 2002. An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry 411990-2001. [DOI] [PubMed] [Google Scholar]
- 34.Rietsch, A., D. Belin, N. Martin, and J. Beckwith. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. USA 9313048-13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rietsch, A., P. Bessette, G. Georgiou, and J. Beckwith. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 1796602-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocha, E. R., C. D. Herren, D. J. Smalley, and C. J. Smith. 2003. The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe 9165-173. [DOI] [PubMed] [Google Scholar]
- 37.Rocha, E. R., G. Owens, Jr., and C. J. Smith. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 1825059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocha, E. R., T. Selby, J. P. Coleman, and C. J. Smith. 1996. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J. Bacteriol. 1786895-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha, E. R., and C. J. Smith. 1997. Regulation of Bacteriodes fragilis katB mRNA by oxidative stress and carbon limitation. J. Bacteriol. 1797033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rocha, E. R., A. O. Tzianabos, and C. J. Smith. 2007. Thioredoxin reductase is essential for thiol/disulfide redox control and oxidative stress survival of the anaerobe Bacteroides fragilis. J. Bacteriol. 1898015-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rzhetsky, A., and M. Nei. 1992. A simple method for estimating and testing minimum evolution tress. Mol. Biol. Evol. 9945-967. [Google Scholar]
- 42.Sarin, R., and Y. D. Sharma. 2006. Thioredoxin system in obligate anaerobe Desulfovibrio desulfuricans: identification and characterization of a novel thioredoxin 2. Gene 376107-115. [DOI] [PubMed] [Google Scholar]
- 43.Smalley, D., E. R. Rocha, and C. J. Smith. 2002. Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J. Bacteriol. 184895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, C. J., M. B. Rogers, and M. L. McKee. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27141-154. [DOI] [PubMed] [Google Scholar]
- 45.Smith, C. J., L. A. Rollins, and A. C. Parker. 1995. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid 34211-222. [DOI] [PubMed] [Google Scholar]
- 46.Stewart, E. J., F. Aslund, and J. Beckwith. 1998. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 175543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sund, C. J., W. G. Wells, and C. J. Smith. 2006. The Bacteroides fragilis P20 scavengase homolog is important in the oxidative stress response but is not controlled by OxyR. FEMS Microbiol. Lett. 261211-217. [DOI] [PubMed] [Google Scholar]
- 48.Sund, C. J., E. R. Rocha, A. O. Tzinabos, W. G. Wells, J. M. Gee, M. A. Reott, D. P. O'Rourke, and C. J. Smith. 2008. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol. Microbiol. 67129-142. [DOI] [PubMed] [Google Scholar]
- 49.Tally, F. P., P. R. Stewart, V. L. Sutter, and J. E. Rosenblatt. 1975. Oxygen tolerance of fresh clinical anaerobic bacteria. J. Clin. Microbiol. 1161-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 51.Varel, V. H., and M. P. Bryant. 1974. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl. Microbiol. 28251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, G., L. Nie, and W. Zhang. 2006. Predicted highly expressed genes in Nocardia farcinica and the implication for its primary metabolism and nocardial virulence. Antonie van Leeuwenhoek 89135-146. [DOI] [PubMed] [Google Scholar]
- 53.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 2791718-1721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.