Abstract
In addition to the ATP-binding cassette (ABC)-type nitrate/nitrite-bispecific transporter, which has a high affinity for both substrates (Km, ∼1 μM), Synechococcus elongatus has an active nitrite transport system with an apparent Km (NO2−) value of 20 μM. We found that this activity depends on the cynABD genes, which encode a putative cyanate (NCO−) ABC-type transporter. Accordingly, nitrite transport by CynABD was competitively inhibited by NCO− with a Ki value of 0.025 μM. The transporter was induced under conditions of nitrogen deficiency, and the induced cells showed a Vmax value of 11 to 13 μmol/mg of chlorophyll per h for cyanate or nitrite, which could supply ∼30% of the amount of nitrogen required for optimum growth. Its relative specificity for the substrates and regulation at transcriptional and posttranslational levels suggested that the physiological role of the bispecific cyanate/nitrite transporter in S. elongatus is to allow nitrogen-deficient cells to assimilate low concentrations of cyanate in the medium. Its contribution to nitrite assimilation was significant in a mutant lacking the ABC-type nitrate/nitrite transporter, suggesting a possible role for CynABD in nitrite assimilation by cyanobacterial species that lack another high-affinity mechanism(s) for nitrite transport.
Cyanobacteria are capable of nitrate assimilation, except for the marine strains of the Prochlorococcus group (9, 14, 33, 39, 41) and a few strains isolated from ammonium-rich hot springs (32). Nitrate is transported into cyanobacterial cells by an active transport system and reduced to nitrite by nitrate reductase (NR). Nitrite is further reduced to ammonium by nitrite reductase (NiR), and the resulting ammonium is assimilated as the amide group of Gln by glutamine synthetase. Nitrite thus arises in cyanobacterial cells as an intermediate of the assimilatory reduction of nitrate, but it can also serve as a good nitrogen source for cyanobacteria when added to the growth medium (13). In the ocean, nitrite seems to serve as a nitrogen source for those strains of Prochlorococcus marinus that have NiR but not NR. Similar to nitrate, nitrite needs to be transferred into the cell before its assimilation. Because nitrous acid (HNO2; the protonated form of nitrite) is a weak acid (pKa = 3.15), its transfer across the plasma membrane involves two distinct mechanisms, (i) active transport of nitrite and (ii) passive diffusion of HNO2. The contribution of passive HNO2 diffusion to the net uptake of nitrite decreases as the pH of the medium is raised and the nitrite concentration in the medium is lowered (12, 27).
To date, two distinct transporters involved in the active uptake of nitrite have been identified and characterized in cyanobacteria, (i) the ATP-binding cassette (ABC)-type bispecific nitrate/nitrite transporter (NRT) encoded by the four genes nrtA, -B, -C, and -D (35, 36, 38) and (ii) the major facilitator superfamily (MFS)-type NRT encoded by a single gene, nrtP (also designated napA) (42, 48). The ABC-type NRT is found in freshwater strains of cyanobacteria (1, 7, 30, 36) and transports both nitrate and nitrite with high affinity (28, 34). The MFS-type NRT has been found in marine strains other than those belonging to the Prochlorococcus group (6, 42, 48) and also in one of the freshwater strains, i.e., Nostoc punctiforme (2). Unlike the ABC-type NRT, the MFS-type NRT has much lower affinity for nitrite than for nitrate (2). Cyanobacterial strains capable of nitrate (and hence nitrite) assimilation have either of the two types of bispecific transporters. In addition to these, a putative nitrite transporter gene has been recognized in cyanobacteria: Some NrtP-containing Synechococcus strains and the NiR-containing Prochlorococcus strains have a focA-like gene that presumably encodes a nitrite transporter. Among cyanobacteria, this gene was first recognized in the Prochlorococcus strains that have NiR but lack NR (41). The gene forms a putative operon with the NiR structural gene nirA, except in Synechococcus sp. strain PCC7002, and the deduced protein is similar to the nitrite transporters from the green alga Chlamydomonas reinhardtii (40), the fungus Aspergillus nidulans (49), and Escherichia coli (8), strongly suggesting that it has a role in nitrite transport (41), although a functional characterization of the gene has yet to be performed.
The unicellular cyanobacterium Synechococcus elongatus has the ABC-type NRT. The four genes that encode the components of this transporter form an operon, nirA-nrtABCD-narB (designated the nirA operon), in which nirA and narB are the structural genes for NiR and NR, respectively (5, 23, 45). The NA3 mutant of S. elongatus strain PCC7942, which was constructed by deleting the nrtABCD genes from the nirA operon, was defective in the active transport of nitrate. However, it retained significant nitrite uptake activity (28). Detailed analysis of nitrite uptake by NA3 revealed that the cyanobacterium has an active transport system for nitrite (NIT), which has an apparent Km (NO2−) of 20 μM (27). Since S. elongatus does not have the focA-like gene (GenBank accession no. NC007604), its NIT activity has been ascribed to a novel transporter that has yet to be identified. In this work, we used a nitrogen-responsive promoter-reporter fusion to isolate a mutant defective in NIT activity. It is shown that the three genes cynABD, which encode an ABC-type transporter previously identified as a cyanate (NCO−) transporter (11), is responsible for active nitrite transport by NA3.
MATERIALS AND METHODS
Strains and growth conditions.
A derivative of S. elongatus strain PCC7942 that is cured of the resident small plasmid pUH24 (R2-SPc; hereafter referred to as the wild-type strain) (21) was the parental strain of all of the mutant strains used in this study. The wild-type and mutant strains were grown photoautotrophically as described previously (46). Ammonium-containing medium, nitrite-containing medium, and nitrate-containing medium were prepared by the addition of 3.75 mM (NH4)2SO4, 5 mM NaNO2, and 60 mM KNO3, respectively, to a nitrogen-free medium obtained by the modification of BG11 medium (44) as described previously (46). Solid media were prepared by adding 1.5% Bacto agar (Difco) to the liquid media. Media were buffered with 20 mM HEPES-KOH (pH 7.2 or 8.2) or 20 mM 2-(N-cyclohexylamino)ethanesulfonic acid (CHES)-KOH (pH 9.6). When appropriate, kanamycin and chloramphenicol were added to the media at 25 and 6 μg ml−1, respectively.
Random insertional mutagenesis and isolation of mutants.
A genomic library of S. elongatus was prepared with a modified pTrc99a vector, pBam99a, that was constructed by deleting the trc promoter, the lacIq gene, and all of the multiple cloning sites except the BamHI and XbaI sites as described previously (25). Genomic DNA was partially digested with Sau3AI, and fragments of 2 to 6 kb were inserted into the BamHI site of pBam99a. Aliquots of the genomic library were partially digested with PvuI, and fragments of 4 to 10 kbp were collected. A chloramphenicol resistance (Cmr)-encoding gene cassette (10) was amplified by PCR with primers carrying PvuI recognition sequences (GGGGGGCGATCGCTCGACTAGAGTCGATCTC and GGGGGGCGATCGCGTTTAAGGGCACCAA). After digestion with PvuI, the PCR product was ligated with the PvuI-digested genomic library and the resulting plasmids were used for transformation of E. coli JM109 to prepare a Cmr gene cassette-tagged library of S. elongatus.
Cells of the YKA1 mutant of S. elongatus, carrying a transcriptional fusion of the nitrogen-responsive promoter of nirA and the coding sequences of luxA and luxB (PnirA::luxAB) and lacking the ABC-type NRT genes nrtABCD (26), were transformed with the Cmr gene cassette-tagged library through double-sided homologous recombination, and the transformants were selected on plates containing 5 mM nitrite and chloramphenicol at pH 9.6. After cultivation for 10 days on agar plates, colonies emitting strong bioluminescence were selected as described previously and shown to be defective in growth on 0.5 mM nitrite at pH 9.6.
Retrieval and analysis of tagged genomic DNA fragments.
Genomic DNA isolated from the selected mutants was digested with BglII and fractionated by electrophoresis on a 0.7% agarose gel. DNA fragments of 2 to 10 kbp were eluted from the gel and ligated into the BamHI site of pUC19. The resulting plasmids were used for the transformation of E. coli JM109. Cmr E. coli transformants were isolated and shown to contain a plasmid carrying an S. elongatus genomic DNA fragment tagged with the Cmr gene cassette. Nucleotide sequences of the DNA regions flanking the Cmr gene cassette were determined to identify the gene(s) interrupted by, or replaced with, the Cmr gene cassette.
Construction of insertion and deletion mutants.
For the construction of cynA and cynB insertional mutants, a 3.3-kbp BglII-BglII DNA fragment carrying the cynABD region (from nucleotide position +128 of the cynA coding sequence to position +179 of the cynS coding sequence) was amplified by PCR and cloned into the BamHI site of pUC19. The Cmr gene cassette was inserted into the EcoRV and NheI sites located in cynA and cynB, respectively, in the cloned fragment. For the construction of cynD and cynS insertional mutants, a 2.6-kbp DNA fragment carrying the cynBDS coding regions (from nucleotide position +1542 of the cynA coding sequence to position +422 with respect to the cynS translation termination site) was amplified by PCR with primers carrying BamHI recognition sequences and cloned into the BamHI site of pUC19. The Cmr gene cassette was inserted into the NruI and BglII sites located in cynD and cynS, respectively, in the cloned fragment. The resulting plasmids were used to transform the NA3 mutant of S. elongatus to Cmr through homologous recombination. To obtain an NA3 derivative (NA4) carrying the same gene replacement as that found in the NIT1 mutants (Fig. 1), the plasmid carrying the Cmr gene cassette-tagged DNA fragment retrieved from NIT1-1 was used to transform NA3 to Cmr. The transformants were allowed to grow on solid medium containing ammonium and chloramphenicol. After serial streak purifications to segregate homozygous mutants, the genomic DNA from the selected clones was analyzed by PCR with whole cells as templates to confirm the presence and position of the Cmr gene cassette.
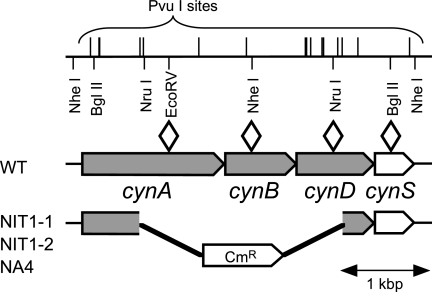
FIG. 1.
Physical map of the cynABDS genomic region of the S. elongatus wild-type (WT) strain and mutants deficient in NIT activity. Pentagons show the locations and directions of genes. NIT1-1 and NIT1-2 were the mutants obtained from luxAB reporter strain YKA1 by random gene tagging with a Cmr-encoding gene cassette. The same gene replacement found in these mutants was introduced into NA3 to obtain the NA4 strain. The open diamonds show the locations of Cmr gene cassette insertions at the indicated restriction endonuclease sites for the construction of the NA3A, NA3B, NA3D, and NA3S strains from NA3.
Expression of plasmid-encoded proteins in S. elongatus.
A shuttle expression vector (pSE1) (28) was used for the expression of cloned genes in S. elongatus. A 3.3-kbp DNA fragment carrying the coding regions of cynABD and a 1.7-kbp fragment carrying those of cynBD were amplified from S. elongatus chromosomal DNA by PCR. The reverse primer carried a BamHI recognition sequence immediately downstream of the termination codon of cynD. The forward primer used for the amplification of cynABD carried mismatches with the genomic sequence that created a BspHI recognition site at the translation start site of cynA. Due to the G4A replacement in the nucleotide sequence, the CynA protein encoded by the resulting plasmid (pcynABD) carried a V2I amino acid substitution. The forward primer used for the amplification of cynBD carried mismatches with the genomic sequence that created an NcoI recognition site at the translation start site without changing the encoded amino acid sequence. The resulting plasmids were introduced into NA4 cells after verification of the nucleotide sequences.
Measurements of nitrite uptake.
Cells grown in nitrate (60 mM)-containing medium (pH 8.2) were washed with the basal medium supplemented with 10 mM KHCO3, 5 mM K2CO3, and 20 mM CHES-KOH (pH 9.6) and suspended in the same medium at a chlorophyll (Chl) concentration of 10 μg ml−1. When the effects of ammonium on nitrite uptake were examined, 20 mM HEPES-KOH (pH 9.6) was used as the buffer because CHES interferes with the determination of ammonium. The reaction was started by addition of 30, 60, 100, or 200 μM NaNO2 to the cell suspensions, which were kept at 30°C in the light (100 μE m−2 s−1). Aliquots were withdrawn from the cell suspensions at 10- or 15-min intervals, and after immediate centrifugation for 60 s at 15,000 × g to sediment the cells, the nitrite concentration in the supernatant was determined.
Other methods.
NR and NiR activities were determined at 30°C by using toluene-permeabilized cells with dithionite-reduced methylviologen as the electron donor (16, 17). Chromosomal DNA was extracted from S. elongatus cells and purified as described by Williams (50). The in vivo bioluminescence from S. elongatus transformants carrying a PnirA::luxAB transcriptional fusion was measured with a luminometer (ARGUS-50; Hamamatsu Photonics) as described previously (26). Nitrite and ammonium were determined as described by Snell and Snell (43) and Anderson and Little (4), respectively. Chl was determined according to Mackinney (24). Manipulations and analyses of DNA were performed according to standard protocols.
RESULTS
Genes required for the nitrite transport activity of NA3.
To genetically identify the genes that encode the active NIT found in the NA3 strain, we used an NA3 derivative (YKA1) carrying a PnirA::luxAB transcriptional fusion (26) as the parental strain for random mutagenesis via gene tagging. The PnirA promoter is active under nitrogen-limited conditions. Although YKA1 could grow on 0.5 mM nitrite at pH 9.6, the cells were nitrogen limited due to the low NIT capacity (see below) and the luciferase expression level was high. When grown on 5 mM nitrite (pH 9.6), by contrast, the cells were nitrogen replete and the luciferase expression level was less than 1% of that observed on the medium containing 0.5 mM nitrite, indicating that the high nitrite concentration in the medium allowed passive HNO2 entrance to compensate for the low NIT capacity. Colonies emitting strong luminescence were selected under high-nitrite conditions (5 mM nitrite and pH 9.6) after random tagging mutagenesis of YKA1 with a Cmr gene cassette. Of the six mutants thus isolated, four that showed very strong luminescence grew very slowly with significant loss of pigmentation even under high-nitrite conditions but grew normally on ammonium-containing media. These mutants carried the Cmr gene cassette inserted in the NiR structural gene nirA. The other two mutants, termed NIT1-1 and NIT1-2, showed no obvious growth phenotype on the medium containing 5 mM nitrite (pH 9.6) but grew only poorly on medium containing 0.5 mM nitrite (pH 9.6), suggesting that they were defective in the utilization of low concentrations of nitrite. In the two mutants, the Cmr gene cassette was found to replace the same 2.3-kbp genomic region carrying the cynA, cynB, and cynD genes (Fig. 1). The cynABD genes encode components of an ABC-type transporter. cynB encodes a hydrophobic protein with structural similarities to the integral membrane components of ABC-type transporters, and cynD encodes a protein that resembles the ATP-binding protein of ABC-type transporters (15). The deduced CynA protein (GenBank AF001333) is 28 and 26% identical to the bicarbonate-binding protein CmpA (19, 29, 37) and the nitrate/nitrite-binding protein NrtA (20, 28, 35), respectively, and is hence supposed to be the substrate-binding protein of the ABC-type transporter. The cynABD genes are located upstream of the cynS gene for cyanase, forming an operon, cynABDS (15). Cyanase catalyzes the bicarbonate-dependent decomposition of cyanate (4), and the resulting ammonium and CO2 can be utilized as nitrogen and carbon sources, respectively, by cyanobacterial cells (31). Because a cynA insertional mutant was as defective in cyanate decomposition as a cynS insertional mutant was, the cynABD genes were identified as the genes for an cyanate transporter (11).
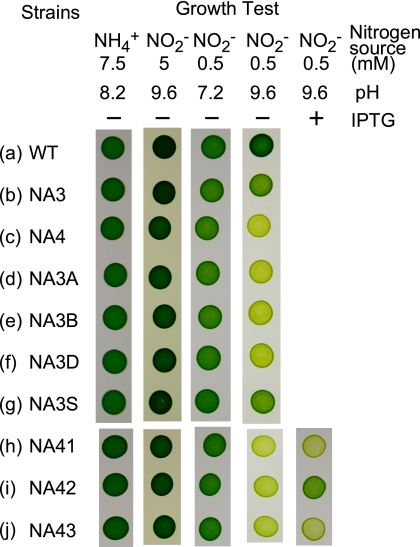
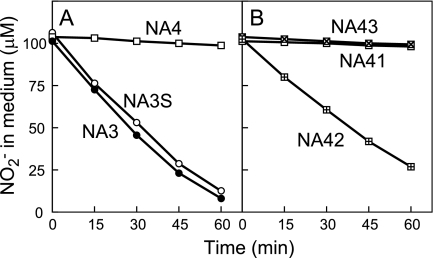
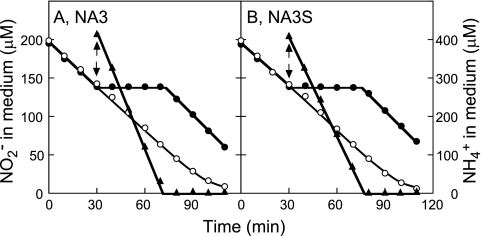
To examine the roles of the cynA, cynB, cynD, and cynS genes in nitrite uptake by the NA3 strain, an insertional mutant of NA3 was constructed for each of the four genes (Table 1). A cynABD deletion-insertion mutant, NA4, carrying the same gene replacement as the NIT1-1 and NIT1-2 mutants (Fig. 1), was also constructed from NA3. Figure 2 compares the growth characteristics of these mutants with those of parental strain NA3. All of the strains grew well on 5 mM nitrite at pH 9.6 or on 0.5 mM nitrite at pH 7.2, under which conditions nitrite enters cells mainly via passive diffusion of HNO2 (27). On the medium containing 0.5 mM nitrite and buffered at pH 9.6, under which conditions the contribution of passive HNO2 entrance is small, the cynA, cynB, and cynD insertion mutants and the cynABD deletion mutant NA4 failed to grow (Fig. 2, rows c to f) while NA3 grew at an appreciable rate (row b). The growth of the cynS insertion mutant was comparable to that of NA3 under these conditions (row g). The cynABD deletion mutant failed to take up low concentrations of nitrite from a liquid medium buffered at pH 9.6, but the cynS mutant NA3S was as active as the NA3 strain in nitrite uptake (Fig. 3A). With an initial nitrite concentration of 100 μM, NA3 and NA3S consumed nitrite at the same initial rate (11 μmol mg Chl−1 h−1) and with essentially the same time course thereafter (Fig. 3A). These results suggested that the cynABD genes are required for NIT activity but cynS is not.
TABLE 1.
Cyanobacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference |
|---|---|---|
| Strains | ||
| SPc | S. elongatus strain PCC7942 cured of plasmid pUH24, wild type | 21 |
| NA3 | SPc ΔnrtABCD lacking genes encoding ABC-type nitrate/nitrite transporter | 28 |
| YKA1 | NA3 PnirA::luxAB | 26 |
| NA3A | NA3 cynA::Cmr | This study |
| NA3B | NA3 cynB::Cmr | This study |
| NA3D | NA3 cynD::Cmr | This study |
| NA3S | NA3 cynS::Cmr | This study |
| NA4 | NA3 ΔcynABD::Cmr | This study |
| NA41 | NA4 harboring pSE1 | This study |
| NA42 | NA4 harboring pcynABD1 | This study |
| NA43 | NA4 harboring pcynBD1 | This study |
| Plasmids | ||
| pSE1 | Kmr, Synechococcus shuttle expression vector | 28 |
| pcynABD1 | pSE1 derivative encoding CynA, CynB, and CynD | This study |
| pcynBD1 | pSE1 derivative encoding CynB and CynD | This study |
FIG. 2.
Growth test on nitrite-containing media showing the effects of interruption and deletion of the cyn genes in the NA3 strain and expression of the cyn genes in the NA3 derivative defective in the cynABD genes (NA4). Synechococcus cells (n = 106) were spotted onto solid medium containing 7.5 mM ammonium, 5 mM nitrite, or 0.5 mM nitrite and buffered at the indicated pH value and incubated under illumination for 4 days. Where indicated, isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1 mM) was added to induce the expression of the plasmid-borne genes. WT, wild type.
FIG. 3.
Uptake of nitrite from medium by cells of the NA3 mutant and its derivatives. Changes in the concentration of nitrite in the medium after the addition of nitrite to cell suspensions containing 10 μg of Chl ml−1 are shown. NA3, NA4, and NA3S cells grown with 60 mM nitrate (A) and NA41, NA42, and NA43 cells grown with 60 mM nitrate plus 0.1 mM isopropyl-β-d-thiogalactopyranoside (B) were used for the measurements.
To further analyze the role of the cynABD genes in nitrite transport by NIT, a transcriptional fusion of the trc promoter and the cynABD open reading frames was introduced into cynABD deletion mutant NA4 to construct the NA42 strain by using the pSE1 shuttle expression vector. The NA43 strain, carrying a transcriptional fusion of Ptrc and two of the three cyn open reading frames (cynB and cynD), was constructed in a similar manner. The pSE1 vector was also introduced into NA4 to construct reference strain NA41. The induced NA42 cells grew on medium containing 0.5 mM nitrite at pH 9.6 (Fig. 2, row i) and took up low concentrations of nitrite from the medium at pH 9.6 (Fig. 3B). The NA41 and NA43 strains, by contrast, failed to grow on medium containing 0.5 mM nitrite (Fig. 2, rows h and j) and to utilize low concentrations of nitrite (Fig. 3B). These results confirmed that the CynA protein is essential for NIT activity.
Competitive inhibition of nitrite transport by cyanate.
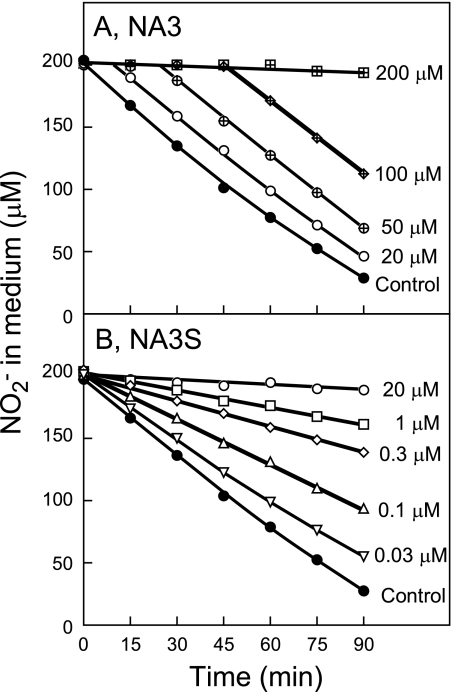
Given that the CynA, CynB, and CynD proteins had been identified as the components of an ABC-type cyanate transporter, the effects of cyanate on the uptake of nitrite by NA3 cells were examined (Fig. 4A). From a medium containing 200 μM nitrite and no cyanate, NA3 cells took up nitrite at an initial rate of 12 μmol mg Chl−1 h−1. When 200 μM cyanate was added simultaneously with nitrite, by contrast, no uptake of nitrite was observed within 90 min after the nitrite/cyanate addition. When 20, 50, and 100 μM cyanate was added to the medium simultaneously with 200 μM nitrite, the nitrite concentration in the medium began to decrease after 9, 23, and 45 min, respectively, with the initial rate being the same as that observed in the absence of cyanate (12 μmol mg Chl−1 h−1). These results indicate that the NA3 cells took up cyanate preferentially over nitrite and that nitrite uptake was effectively inhibited by cyanate. From the linear relationship between the amounts of cyanate added to the medium and the length of the time lag between the addition of nitrite/cyanate and the onset of nitrite uptake (see above), the rate of cyanate uptake was calculated as 13 μmol mg Chl−1 h−1.
FIG. 4.
Inhibition by cyanate of the nitrite uptake activity of NA3 and NA3S cells. Strain NA3 (A) and NA3S (B) cells grown with 60 mM nitrate were suspended in nitrogen-free medium to a Chl concentration of 10 μg ml−1. Nitrite (200 μM) and cyanate at the concentrations indicated were added to the cell suspensions at time zero. Changes in the nitrite concentration in the medium are shown.
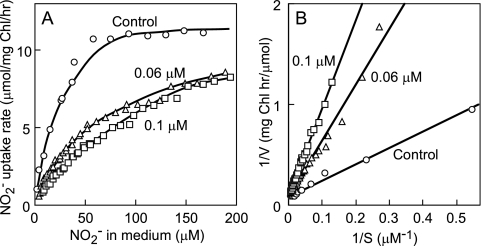
Unlike in NA3, 20 μM cyanate was sufficient to permanently inhibit nitrite uptake by NA3S cells lacking cyanase (Fig. 4B). This confirmed that cyanate per se inhibits the uptake of nitrite. Even when the cyanate concentration was as low as 0.03 μM, the inhibitory effect of cyanate was evident and permanent. The extent of inhibition of nitrite uptake by 0.03, 0.1, 0.3, and 1 μM cyanate was 12, 39, 62, and 76%, respectively. Figure 5 shows the effects of low concentrations of cyanate on the kinetics of nitrite uptake by NA3S cells. In the absence of cyanate, the nitrite transport activity of NA3S cells showed saturation kinetics with a Vmax value of 12.1 μmol mg Chl−1 h−1 and an apparent Km (NO2−) value of 19.4 μM, which were close to those previously reported for the NA3 strain (27). Nitrite uptake showed saturation kinetics in the presence of 0.06 and 0.1 μM cyanate as well, with Vmax values of 11.1 and 11.6 μmol mg Chl−1 h−1, respectively. These values were essentially the same as that observed in the absence of cyanate. The apparent Km values for nitrite transport activity in the presence of 0.06 and 0.1 μM cyanate were calculated as 64 and 96 μM, respectively, and the Ki value of cyanate for nitrite transport was calculated as 0.026 and 0.025 μM, indicating that cyanate competitively inhibited nitrite transport activity. These results indicated that the CynABD transporter has much higher affinity for cyanate than for nitrite, with the Km for cyanate being nearly 3 orders of magnitude smaller than that for nitrite.
FIG. 5.
Effects of cyanate on the nitrite uptake activity kinetics of NA3S cells. Cells of the NA3S mutant were grown with 60 mM nitrate and suspended to a Chl concentration of 10 μg ml−1 in nitrogen-free medium. The cell suspension was separated into three portions, two of which were supplemented with cyanate (0.06 and 0.1 μM, respectively). Each cell suspension was further separated into four portions, and nitrite uptake was measured at initial nitrite concentrations of 30, 60, 100, and 200 μM. The nitrite concentration in the medium was measured at 10-min intervals, and the rate of nitrite uptake after every other sampling time was calculated and plotted against the mean of the nitrite concentrations at the two relevant time points. (A) Plots of the nitrite uptake rate versus the nitrite concentration in the medium. (B) Double-reciprocal plots of the data shown in panel A.
Posttranslational regulation of NIT activity.
Similar to the activity of ABC-type NRT, NIT activity is reversibly inhibited by the addition of ammonium to the medium (27). Since the inhibitory effect of ammonium is abolished by treating cells with methionine sulfoximine, an inhibitor of glutamine synthetase, it was assumed that a metabolic change arising from the assimilation of ammonium leads to posttranslational regulation of the nitrite transporter. In the light of the involvement of the ABC-type cyanate transporter in NIT activity, however, the mechanism of ammonium inhibition of NIT had to be reinvestigated. Cyanate is supposed to arise in cyanobacterial cells from the spontaneous dissociation of carbamoylphosphate (22), a metabolite essential for both the arginine and pyrimidine biosynthetic pathways. Since carbamoylphosphate is synthesized from Gln, CO2, and ATP (31), its accumulation in the cell and hence the production of cyanate would therefore be stimulated by the addition of ammonium to nitrogen-limited cells, and the resulting cyanate may leak out of the cell and competitively inhibit NIT. To determine whether or not the inhibition of NIT by ammonium involves cyanate, the effects of ammonium on NIT activity were examined in the cyanase-deficient mutant NA3S. Like the NIT activity of NA3, that of NA3S was reversibly inhibited by ammonium (Fig. 6). If this inhibition by ammonium had been due to the accumulation of cyanate, it would have been permanent in NA3S, as shown in Fig. 5B. The reversibility of the ammonium inhibition of NIT indicated that cyanate was not involved.
FIG. 6.
Reversible inhibition by ammonium of nitrite uptake by NA3 and NA3S cells. NA3 cells grown with 60 mM nitrate were suspended in nitrogen-free medium to a Chl concentration of 10 μg ml−1. Nitrite (200 μM) was added to the cell suspensions at time zero, and ammonium (400 μM) was added at the time indicated by the arrows. Changes in the nitrite (circles) and ammonium (triangles) concentrations in the medium are shown. Open circles, control; closed symbols, plus ammonium.
NIT activity in wild-type cells.
Similar to the nirA operon that encodes the proteins involved in nitrate assimilation, the cynABDS operon is under the control of NtcA, a Crp-type transcriptional regulator (47), and is induced under the conditions of nitrogen deficiency (15). In this study, we used NA3 cells grown in nitrate (60 mM)-containing medium (pH 8.2) for the characterization of nitrite uptake by NIT (Fig. 3 to 6). The growth of the cells was slower under these conditions than in ammonium-containing medium, indicating that the cells were nitrogen limited. In nitrite (5 mM)-containing medium (pH 8.2), on the other hand, NA3 grew as rapidly as in ammonium-containing medium, showing that the cells were nitrogen replete. In accordance with the dependence of the cynABDS operon and the nirA operon on NtcA, the NIT, NR, and NiR activities in nitrate-grown NA3 cells were four, two, and three times higher than those in nitrite-grown cells, respectively (Table 2). Given that the wild-type cells are nitrogen replete in nitrate-containing medium, as well as in nitrite-containing medium, we inferred that the NIT, NR, and NiR activities of nitrate- or nitrite-grown wild-type cells would be as low as those of nitrite-grown NA3 cells. In wild-type cells, however, NIT activity could not be determined by following nitrite consumption by the cells due to the presence of the ABC-type NRT. We hence made use of the fact that nitrite uptake by the ABC-type NRT is competitively inhibited by nitrate (27). The NIT activity of wild-type cells was estimated, by measuring the rate of nitrite uptake at a 100 μM external nitrite concentration in the presence of 10 mM nitrate (Table 2), to be ∼3 μmol mg Chl−1 h−1 in both nitrate-grown and nitrite-grown cells, which is essentially the same as that of nitrite-grown NA3 cells, as expected. The NR and NiR activities of nitrate-grown and nitrite-grown wild-type cells were also as low as the corresponding activities of nitrite-grown NA3 cells.
TABLE 2.
NIT, NR, and NiR activities of nitrite-grown and nitrate-grown cells of the wild-type strain and the NA3 mutanta
| Strain and growth condition | NIT | NR | NiR |
|---|---|---|---|
| Wild type | |||
| NO2− | 2.9 ± 0.3 | 186 ± 22 | 82 ± 21 |
| NO3− | 2.7 ± 0.3 | 196 ± 27 | 80 ± 15 |
| NA3 | |||
| NO2− | 2.8 ± 0.4 | 192 ± 39 | 72 ± 25 |
| NO3− | 11.4 ± 0.9 | 361 ± 63 | 243 ± 36 |
Data are presented as the mean ± standard deviation of three experiments. Ammonium-grown cells of wild-type S. elongatus and the NA3 mutant were transferred to NO2− (5 mM)-containing and nitrate (60 mM)-containing media at pH 8.2, and the activities of NIT, NR, and NiR were assayed after 16 h of growth. Activities are shown in micromoles per milligram of Chl per hour.
DISCUSSION
In the present study, genetic characterization of the previously identified “nitrite-specific” transporter (27) revealed that the transport activity is ascribed to the ABC-type cyanate transporter encoded by the cynABD genes (11). In the previous study on the NIT (27), competition experiments performed with nitrate, sulfate, sulfite, chlorate, and chlorite led to the notion that the NIT is “nitrite specific,” but the present results demonstrate the competitive inhibition of NIT activity by cyanate (Fig. 4 and 5). The Ki (NCO−) value for the nitrite transport activity, and hence the Km (NCO−) value of the CynABD transporter, was estimated to be about 0.025 μM. This is nearly 800 times smaller than the Km (NO2−) of the same transporter, 20 μM (27). Thus, the CynABD transporter is a bispecific transporter with far greater affinity for cyanate than for nitrite. In the presence of millimolar concentrations of cyanate in the medium, passive permeation of cyanic acid (HNCO, the protonated form of cyanate) across the plasma membrane and equilibration between HNCO and cyanate account for the uptake of cyanate (see below). At submicromolar concentrations of cyanate, passive HNCO entrance into the cell would be negligible and active cyanate transport would play an essential role in the uptake and utilization of cyanate.
Similar to the nitrate assimilation operon nirA-nrtABCD-narB, the cynABDS operon is under the control of the global nitrogen regulator protein NtcA, being repressed in the presence of ammonium in the medium and activated under conditions of nitrogen deficiency (15). Because of the lack of an ABC-type NRT, the nitrate-grown NA3 mutant of S. elongatus is under the constant stress of nitrogen limitation and expresses higher activities of NR and NiR than do nitrate- or nitrite-grown wild-type cells (Table 2). Even in such “induced” cells, the CynABD activity was 11 to 12 μmol mg Chl−1 h−1 (Table 2), which corresponded to only ∼30% of the nitrogen flux required for nitrogen-replete growth of the cells in dilute liquid cultures (40 μmol mg Chl−1 h−1) (38). The high-affinity/low-flux nature of the cyanate transport activity seems to conform to the limited availability of cyanate in the natural environment (18). We suggest that the primary physiological role of CynABD is to take up low concentrations of cyanate from the medium to provide the cells with small but significant amounts of nitrogen under conditions of nitrogen limitation. In nitrogen-replete cells grown with nitrate or nitrite as the nitrogen source (i.e., nitrate- or nitrite-grown wild-type cells and nitrite-grown NA3 cells), on the other hand, the level of the CynABD activity is low, being ∼3 μmol mg Chl−1 h−1 (Table 2), indicating that the contribution of active cyanate uptake to the total nitrogen uptake would be less than 8% in nitrate- or nitrite-containing medium, even if there were enough cyanate in the medium to saturate the transporter.
In previous studies on cyanate metabolism in S. elongatus cells, the rate of cyanate decomposition by nitrate-grown, nitrogen-replete cells was determined to be 80 μmol mg Chl−1 h−1 by measuring cyanate-dependent ammonium excretion from the cells (15, 31). Even higher cyanate decomposition activity was observed by measuring cyanate-dependent O2 evolution (11, 31). The Vmax and K0.5 (the substrate concentration that yields one-half of the maximum rate) of cyanate-dependent O2 evolution were reported to be 188 μmol mg Chl−1 h−1 and 450 μM, respectively (31). These kinetic parameters cannot be accounted for by the properties of the ABC-type cyanate transporter. It should be noted that the previous studies were performed at high extracellular cyanate concentrations (1 to 2 mM) and at pH values of 8.0 to 8.3. Under these conditions, passive permeation of HNCO into the cells would be fast, given that HNCO is a weak acid (pKa = 3.48), as HNO2 is (pKa = 3.15). In the case of nitrite, a roughly linear relationship was observed between the uptake rate and the substrate concentration up to 150 μM in the medium at pH 7.2 (27). The uptake rate at 100 μM nitrite was 45 μmol mg Chl−1 h−1, from which the rate of passive HNO2 entrance into the cells at 100 μM nitrite and pH 7.2 was calculated to be 34 μmol mg Chl−1 h−1 by subtracting the rate of active nitrite transport determined at pH 9.6 (11 μmol mg Chl−1 h−1). Assuming that the diffusion coefficients of HNCO and HNO2 are the same and that the rate of passive permeation into the cells is proportional to the concentrations of HNCO and HNO2 in the medium, calculations with the Henderson-Hasselbach equation predict that the rate of HNCO permeation is 220 μmol mg Chl−1 h−1 at a cyanate concentration of 2 mM and pH 8.0, which is high enough to account for the rates of cyanate decomposition reported for S. elongatus cells. Nitrate-grown cells of Synechocystis sp. strain PCC6803, which lacks the cynABD genes, showed a cyanate decomposition rate of 54 μmol mg Chl−1 h−1 at a 2 mM extracellular cyanate concentration and pH 8.0 (15), showing that passive HNCO permeation into the cells was not slower than 54 μmol mg Chl−1 h−1. It should also be mentioned that Synechococcus sp. strain WH7803, which also lacks the cynABD genes, grew in medium containing 0.8 mM cyanate as the sole nitrogen source, indicating that the passive permeation of HCNO is rapid enough to satisfy the nitrogen requirements of the cells (18). These considerations led to the conclusion that passive HNCO permeation plays a far more important role in cellular cyanate uptake than does active cyanate transport in the presence of millimolar concentrations of cyanate in the medium. It was reasonable that Espie et al. (11) failed to detect inhibitory effects of nitrite on cyanate uptake in the presence of 1 mM cyanate in the medium. The Km (NCO−) of cyanase has been estimated to be 600 μM in E. coli (3) and 70 μM in Methylobacterium thiocyanatum (51). The K0.5 of cyanate-dependent O2 evolution by S. elongatus cells thus seems to represent the kinetic property of cyanase. This high cyanase activity seems to be required to compensate for the low affinity of the enzyme for the substrate, compared to the extremely high affinity of the cyanate transporter.
The cynS gene for cyanase is found in 17 out of the 33 cyanobacterial genome sequences available in GenBank (as of November 2008). The cynABD genes are present in six cynS-carrying strains, i.e., S. elongatus strains PCC7942 and PCC6301, Synechococcus sp. strains PCC7002 and WH8102, P. marinus strain MED4, and Acaryochloris marina, but absent in the rest of the cynS-carrying strains or in strains that lack cynS. In five of the six strains, the genes form a putative cynABDS operon, and in the other strain (Synechococcus sp. strain WH8102), they are separated from cynS by only two genes, suggesting that the general role of cynABD is to transport cyanate. Because of the high activity of ABC-type NRT (∼40 μmol mg Chl−1 h−1), the contribution of cynABD to nitrite transport seems to be marginal in S. elongatus strains PCC7942 and PCC6301. Since P. marinus strain MED4 lacks NR and NiR, there is no physiological significance of cynABD in nitrite transport. In Synechococcus sp. strain WH8102 and A. marina, on the other hand, CynABD may have physiological relevance to nitrite assimilation. These two strains have NR and NiR, indicating that they assimilate nitrate, and have NrtP instead of NrtABCD for active uptake of nitrate and nitrite. Characterization of NrtP from N. punctiforme has shown that its affinity for nitrate is as high as that of the ABC-type NRT, but that for nitrite is low; the Km (NO2−) seems to be larger than 50 μM (2). Since Synechococcus sp. strain WH8102 and A. marina have no other potential nitrite transporter genes, CynABD may play a significant role in nitrite assimilation by enhancing the nitrite transport activity of the cell. The last of the cynABD-carrying cyanobacteria known to date is Synechococcus sp. strain PCC7002, which has NrtP and a FocA-like transporter that presumably transport nitrite (41), and the significance of cynABD in nitrite transport is unclear. FocA-like proteins involved in nitrite transport have been identified in C. reinhardtii (40), Aspergillus nidulans (49), and E. coli (8), and the Ks (NO2−) or Km (NO2−) of the transporter has been estimated to be 5 μM in C. reinhardtii (40) and 4 μM in A. nidulans (49). Their homologs are found in many marine cyanobacterial strains, forming a putative operon with the NiR structural gene nirA, except in Synechococcus sp. strain PCC7002, strongly suggesting that it has a role in nitrite transport. It is intriguing that many of the nrtP-containing marine cyanobacteria have the focA-like gene, whereas none of the nrtABCD-containing freshwater strains has it. Although the role of the cyanobacterial focA-like genes in nitrite transport needs to be experimentally verified, the distribution of the gene seems to support our notion that enhancement of nitrite transport activity is important in NrtP-carrying strains.
Acknowledgments
This work was supported by the 21st Century COE Program and in part by a Grant-in-Aid for Scientific Research (18570039) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Aichi, M., N. Takatani, and T. Omata. 2001. Role of NtcB in activation of nitrate assimilation genes in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 1835840-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aichi, M., S. Yoshihara, M. Yamashita, S. Maeda, K. Nagai, and T. Omata. 2006. Characterization of the nitrate-nitrite transporter of the major facilitator superfamily (the nrtP gene product) from the cyanobacterium Nostoc punctiforme strain ATCC 29133. Biosci. Biotechnol. Biochem. 702682-2689. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, P. M. 1980. Purification and properties of the inducible enzyme cyanase. Biochemistry 192882-2888. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, P. M., and R. M. Little. 1986. Kinetic properties of cyanase. Biochemistry 251621-1626. [DOI] [PubMed] [Google Scholar]
- 5.Andriesse, X., H. Bakker, and P. Weisbeek. 1990. Analysis of nitrate reduction genes in cyanobacteria, p. 303-307. In W. R. Ullrich, C. Rigano, A. Fuggi, and P. J. Aparicio (ed.), Inorganic nitrogen in plants and microorganisms. Springer-Verlag, Berlin, Germany.
- 6.Bird, C., and M. Wyman. 2003. Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression. Appl. Environ. Microbiol. 697009-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, Y., and C. P. Wolk. 1997. Nitrogen deprivation of Anabaena sp. strain PCC 7120 elicits rapid activation of a gene cluster that is essential for uptake and utilization of nitrate. J. Bacteriol. 179258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clegg, S., F. Yu, L. Griffiths, and J. A. Cole. 2002. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Mol. Microbiol. 44143-155. [DOI] [PubMed] [Google Scholar]
- 9.Dufresne, A., M. Salanoubat, F. Partensky, F. Artiguenave, I. M. Axmann, V. Barbe, S. Duprat, M. Y. Galperin, E. V. Koonin, F. Le Gall, K. S. Makarova, M. Ostrowski, S. Oztas, C. Robert, I. B. Rogozin, D. J. Scanlan, N. Tandeau de Marsac, J. Weissenbach, P. Wincker, Y. I. Wolf, and W. R. Hess. 2003. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA 10010020-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dzelzkalns, V. A., G. C. Owens, and L. Bogorad. 1984. Chloroplast promoter driven expression of the chloramphenicol acetyl transferase gene in a cyanobacterium. Nucleic Acids Res. 128917-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espie, G. S., F. Jalali, T. Tong, N. J. Zacal, and A. K. So. 2007. Involvement of the cynABDS operon and the CO2-concentrating mechanism in the light-dependent transport and metabolism of cyanate by cyanobacteria. J. Bacteriol. 1891013-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores, E., A. Herrero, and M. G. Guerrero. 1987. Nitrite uptake and its regulation in the cyanobacterium Anacystis nidulans. Biochim. Biophys. Acta 896103-108. [Google Scholar]
- 13.Fogg, G. E., W. D. P. Stewart, P. Fay, and A. E. Walsby. 1973. The blue-green algae. Academic Press, Inc., New York, NY.
- 14.García-Fernández, J. M., N. Tandeau de Marsac, and J. Diez. 2004. Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments. Microbiol. Mol. Biol. Rev. 68630-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harano, Y., I. Suzuki, S. Maeda, T. Kaneko, S. Tabata, and T. Omata. 1997. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J. Bacteriol. 1795744-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero, A., E. Flores, and M. G. Guerrero. 1981. Regulation of nitrate reductase levels in the cyanobacteria Anacystis nidulans, Anabaena sp. strain 7119, and Nostoc sp. strain 6719. J. Bacteriol. 145175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrero, A., and M. G. Guerrero. 1986. Regulation of nitrite reductase in the cyanobacterium Anacystis nidulans. J. Gen. Microbiol. 1322463-2468. [Google Scholar]
- 18.Kamennaya, N. A., M. Chernihovsky, and A. F. Post. 2008. The cyanate utilization capacity of marine unicellular cyanobacteria. Limnol. Oceanogr. 532485-2494. [Google Scholar]
- 19.Koropatkin, N. M., D. W. Koppenaal, H. B. Pakrasi, and T. J. Smith. 2007. The structure of a cyanobacterial bicarbonate transport protein, CmpA. J. Biol. Chem. 2822606-2614. [DOI] [PubMed] [Google Scholar]
- 20.Koropatkin, N. M., H. B. Pakrasi, and T. J. Smith. 2006. Atomic structure of a nitrate-binding protein crucial for photosynthetic productivity. Proc. Natl. Acad. Sci. USA 1039820-9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhlemeier, C. J., A. A. Thomas, A. van der Ende, R. W. van Leen, W. E. Biorrias, C. A. van den Hondel, and G. A. van Arkel. 1983. A host-vector system for gene cloning in the cyanobacterium Anacystis nidulans R2. Plasmid 10156-163. [DOI] [PubMed] [Google Scholar]
- 22.Lawrie, A. C. 1979. Effect of carbamoyl phosphate on nitrogenase in Anabaena cylindrica Lemm. J. Bacteriol. 139115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luque, I., E. Flores, and A. Herrero. 1993. Nitrite reductase gene from Synechococcus sp. PCC 7942: homology between cyanobacterial and higher-plant nitrite reductases. Plant Mol. Biol. 211201-1205. [DOI] [PubMed] [Google Scholar]
- 24.Mackinney, G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140315-322. [Google Scholar]
- 25.Maeda, S., M. R. Badger, and G. D. Price. 2002. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol. Microbiol. 43425-435. [DOI] [PubMed] [Google Scholar]
- 26.Maeda, S., Y. Kawaguchi, T. Ohe, and T. Omata. 1998. cis-acting sequences required for NtcB-dependent, nitrite-responsive positive regulation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1804080-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda, S., M. Okamura, M. Kobayashi, and T. Omata. 1998. Nitrite-specific active transport system of the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1806761-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda, S., and T. Omata. 1997. Substrate-binding lipoprotein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in the transport of nitrate and nitrite. J. Biol. Chem. 2723036-3041. [DOI] [PubMed] [Google Scholar]
- 29.Maeda, S., G. D. Price, M. R. Badger, C. Enomoto, and T. Omata. 2000. Bicarbonate binding activity of the CmpA protein of the cyanobacterium Synechococcus sp. strain PCC 7942 involved in active transport of bicarbonate. J. Biol. Chem. 27520551-20555. [DOI] [PubMed] [Google Scholar]
- 30.Merchán, F., K. L. Kindle, M. J. Llama, J. L. Serra, and E. Fernández. 1995. Cloning and sequencing of the nitrate transport system from the thermophilic, filamentous cyanobacterium Phormidium laminosum: comparative analysis with the homologous system from Synechococcus sp. PCC 7942. Plant Mol. Biol. 28759-766. [DOI] [PubMed] [Google Scholar]
- 31.Miller, A. G., and G. S. Espie. 1994. Photosynthetic metabolism of cyanate by the cyanobacterium Synechococcus UTEX 625. Arch. Microbiol. 162151-157. [Google Scholar]
- 32.Miller, S. R., and R. W. Castenholz. 2001. Ecological physiology of Synechococcus sp. strain SH-94-5, a naturally occurring cyanobacterium deficient in nitrate assimilation. Appl. Environ. Microbiol. 673002-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47989-996. [Google Scholar]
- 34.Nagore, D., B. Sanz, J. Soria, M. Llarena, M. J. Llama, J. J. Calvete, and J. L. Serra. 2006. The nitrate/nitrite ABC transporter of Phormidium laminosum: phosphorylation state of NrtA is not involved in its substrate binding activity. Biochim. Biophys. Acta 1760172-181. [DOI] [PubMed] [Google Scholar]
- 35.Omata, T. 1991. Cloning and characterization of the nrtA gene that encodes a 45-kDa protein involved in nitrate transport in the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 32151-157. [Google Scholar]
- 36.Omata, T., X. Andriesse, and A. Hirano. 1993. Identification and characterization of a gene cluster involved in nitrate transport in the cyanobacterium Synechococcus sp. PCC 7942. Mol. Gen. Genet. 236193-202. [DOI] [PubMed] [Google Scholar]
- 37.Omata, T., T. J. Carlson, T. Ogawa, and J. Pierce. 1990. Sequencing and modification of the gene encoding the 42-kilodalton protein in the cytoplasmic membrane of Synechococcus PCC 7942. Plant Physiol. (Rockville) 93305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omata, T., M. Ohmori, N. Arai, and T. Ogawa. 1989. Genetically engineered mutant of the cyanobacterium Synechococcus PCC 7942 defective in nitrate transport. Proc. Natl. Acad. Sci. USA 866612-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palinska, K. A., W. Laloui, S. Bédu, S. Loiseaux-de Goër, A. M. Castets, R. Rippka, and N. Tandeau de Marsac. 2002. The signal transducer P(II) and bicarbonate acquisition in Prochlorococcus marinus PCC 9511, a marine cyanobacterium naturally deficient in nitrate and nitrite assimilation. Microbiology 1482405-2412. [DOI] [PubMed] [Google Scholar]
- 40.Rexach, J., E. Fernández, and A. Galván. 2000. The Chlamydomonas reinhardtii Nar1 gene encodes a chloroplast membrane protein involved in nitrite transport. Plant Cell 121441-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocap, G., F. W. Larimer, J. Lamerdin, S. Malfatti, P. Chain, N. A. Ahlgren, A. Arellano, M. Coleman, L. Hauser, W. R. Hess, Z. I. Johnson, M. Land, D. Lindell, A. F. Post, W. Regala, M. Shah, S. L. Shaw, C. Steglich, M. B. Sullivan, C. S. Ting, A. Tolonen, E. A. Webb, E. R. Zinser, and S. W. Chisholm. 2003. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 4241042-1047. [DOI] [PubMed] [Google Scholar]
- 42.Sakamoto, T., K. Inoue-Sakamoto, and D. A. Bryant. 1999. A novel nitrate/nitrite permease in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 1817363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snell, F. D., and C. T. Snell. 1949. Colorimetric methods of analysis, vol. 2., p.802-807. Van Nostrand, New York, NY. [Google Scholar]
- 44.Stanier, R. Y., R. Kunisawa, M. Mandel, and G. Cohen-Bazire. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35171-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki, I., T. Sugiyama, and T. Omata. 1993. Primary structure and transcriptional regulation of the gene for nitrite reductase from the cyanobacterium Synechococcus PCC 7942. Plant Cell Physiol. 341311-1320. [Google Scholar]
- 46.Suzuki, I., T. Sugiyama, and T. Omata. 1996. Regulation by cyanate of the genes involved in carbon and nitrogen assimilation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1782688-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vega-Palas, M. A., E. Flores, and A. Herrero. 1992. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 61853-1859. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Q., H. Li, and A. F. Post. 2000. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601. J. Bacteriol. 1821764-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Y., W. Li, Y. Siddiqi, V. F. Symington, J. R. Kinghorn, S. E. Unkles, and A. D. Glass. 2008. Nitrite transport is mediated by the nitrite-specific high-affinity NitA transporter and by nitrate transporters NrtA, NrtB in Aspergillus nidulans. Fungal Genet. Biol. 4594-102. [DOI] [PubMed] [Google Scholar]
- 50.Williams, J. G. 1988. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167766-778. [Google Scholar]
- 51.Wood, A. P., D. P. Kelly, I. R. McDonald, S. L. Jordan, T. D. Morgan, S. Khan, J. C. Murrell, and E. Borodina. 1998. A novel pink-pigmented facultative methylotroph, Methylobacterium thiocyanatum sp. nov., capable of growth on thiocyanate or cyanate as sole nitrogen sources. Arch. Microbiol. 169148-158. [DOI] [PubMed] [Google Scholar]