Abstract
The capability of endospores of Bacillus subtilis to withstand extreme environmental conditions is secured by several attributes. One of them, the protein shell that encases the spore and is known as the coat, provides the spore with its characteristic resistance to toxic chemicals, lytic enzymes, and predation by unicellular and multicellular eukaryotes. Despite most of the components of the spore coat having been identified, we have only a vague understanding of how such a complex structure is assembled. Using the yeast two-hybrid system, we attempted to identify direct contacts among the proteins allocated to the insoluble fraction of the spore coat: CotV, CotW, CotX, CotY, and CotZ. We also examined whether they could interact with CotE, one of the most crucial morphogenetic proteins governing outer coat formation and also present in the insoluble fraction. Out of all 21 possible interactions we tested, 4 were found to be positive. Among these interactions, we confirmed the previous observation that CotE forms homo-oligomers. In addition, we observed homotypic interactions of CotY, strong interactions between CotZ and CotY, and relatively weak, yet significant, interactions between CotV and CotW. The results of this yeast two-hybrid analysis were confirmed by size exclusion chromatography of recombinant coat proteins and a pull-down assay.
Endospores formed by Bacillus subtilis are encased in a protein shell known as the coat, which is comprised of an organized, multilayered structure. Two distinct layers can be clearly distinguished by electron microscopy: a thick, electron-dense outer layer and a lightly staining inner layer composed of fine lamellae (3, 9). The role of the spore coat, whose synthesis is controlled by sporulation-specific transcription factors, is to protect the spore against lytic enzymes and toxic molecules and to provide the spores with mechanical integrity. On the other hand, the coat is also capable of allowing molecules access to the spore interior: for example, spore germinants that interact with receptors located in the inner spore membrane that is shielded by the coat. Although the spore coat traditionally has been considered as a sieving barrier, some results indicate that it has more active functions (5). In recent years, the spore coat of B. subtilis has been shown to be more complex than previously thought (12, 13). Over 50 different proteins are deposited onto the developing surface of the immature spore known as the forespore. The formation of the coat starts soon after the polar septum is formed. This asymmetrically placed septum divides the cell into two unequal compartments—the larger, mother cell compartment and the smaller forespore. The membranes gradually engulf the forespore, generating a compartmentalized forespore that will mature into the endospore that is then released from the surrounding mother cell. This forespore becomes visible by electron microscopy 4 to 5 h after the initiation of sporulation. The whole process is completed only after mother cell lysis and release of the endospore. Coat protein production is primarily controlled by two mother cell-specific RNA polymerase transcription sigma factors, σE and σK, together with three regulatory proteins, SpoIIID, GerE, and GerR. The temporal activation of all these transcription factors results in the hierarchical regulatory program that ensures that proteins are synthesized at the correct time and in the necessary amounts. Some of the existing regulatory feedback mechanisms controlling expression of particular genes allow the cell to respond directly to the changing conditions of the external environment (3). At the early stages of coat development, the synthesis of spore coat proteins is governed by the sigma factor σE, which directs transcription of several genes, among them spoIVA and cotE. Examination of their function revealed that they play a key role in spore coat morphogenesis, guiding the assembly of other coat protein components (4, 19, 29). SpoIVA, together with a small peptide referred to as SpoVM, creates a base layer of the coat on the outer forespore membrane at the very beginning of coat formation (18). A second morphogenetic protein, CotE, is localized in a SpoIVA-dependent manner and forms a ring around the forespore at a distance of approximately 75 nm from SpoIVA (4). The gap defined by the SpoIVA and CotE rings is referred to as the matrix, or precoat. Although the composition of the matrix is still unknown, it is assumed that proteins of the cotJ operon could participate in formation of this structure (3, 8, 9, 21). Later, following activation of the second mother cell-specific sigma factor, σK, which coordinates expression of the majority of coat protein genes, the matrix is transformed into the inner coat, having a typical lamellar appearance. At the same time, the outer coat proteins are assembled around the CotE ring in a CotE-dependent manner. In addition, several other proteins, such as SpoVID, SafA, and CotH, control the deposition of the rest of coat components (4, 22, 30). As has been mentioned above, the list of proteins participating in coat formation is relatively long and most of these proteins have been already shown to localize either into the inner layer or the outer layer (9), but the mechanism of their assembly is still not clear.
In this paper, we focused on a group of proteins (CotV, CotW, CotX, CotY, and CotZ) that were identified in the insoluble fraction of the spore coat. We have also studied their interaction with CotE, one of the crucial morphogenetic proteins. CotE, although abundant in the spore coat soluble extract, was also found in the insoluble fraction (1). CotY and CotZ are cysteine-rich proteins, CotY contains 15 cysteines (out of 161 residues), and CotZ contains 10 cysteines (out of 147 residues). However, minor portions of both proteins were also identified in the soluble fraction (12, 27). In addition, it was shown that CotY was also present in multimeric forms in this fraction (27). Another Cot protein, CotX, has a similarly high content of cysteines (7 out of 172 amino acid residues), and the dimeric forms of the protein were observed in spore extracts. However, in this case, it was assumed that oligomers of the protein probably contain different types of cross-links, since they could not be solubilized by reducing agents (27). After disruption of cotXYZ genes, the outer coat was significantly reduced and the surface characteristics of the spore were changed (27). Upstream of the cotXYZ operon, genes coding for CotV and CotW were identified, and because they are grouped in one gene cluster, it was assumed that they could be functionally related (27).
In this paper, we examined by the yeast two-hybrid system potential protein-protein interactions among insoluble coat proteins. The identified interactions were confirmed by additional techniques, including pull-down assays and size exclusion chromatography. The results reached within the framework of this study allowed us to propose a new scheme of insoluble coat protein assembly.
MATERIALS AND METHODS
Bacterial and yeast strains and growth conditions.
Escherichia coli strain MM294 (endA1 hsdR17 supE44 thi-1 recA1) (2) was chosen as a host for construction and maintenance of plasmids used in this study and was cultivated in Luria-Bertani (LB) medium supplemented with appropriate antibiotics. Recombinant spore coat proteins were expressed in E. coli BL21(DE3). B. subtilis PY79 (26) genomic DNA was used as a source of spore coat protein genes. Saccharomyces cerevisiae strain MaV203 (MATα leu2-3,112 trp1-901 his3Δ200 ade2-101 gal4Δ gal80Δ SPAL10::URA3 GAL1::lacZ HIS3UAS GAL1::HIS3@LYS2, can1R cyh2R) (Invitrogen) was used for the yeast two-hybrid assay. Yeast growth media were prepared as recommended by the manufacturer. Rich YPAD medium (1% yeast extract, 2% peptone, 2% dextrose, and 0.01% adenine hemisulfate) was used for routine growth of yeast, and synthetic complete medium omitting specific amino acids according to the selection requirements was used to examine protein-protein interactions.
Construction of expression plasmids.
All DNA manipulations were performed by standard methods (20). The CotY and CotW expression plasmids were constructed based on the pET28a vector system (Novagen). Full-length genes for both proteins were amplified by PCR with sense primers CotY5′ and CotW5′, respectively, carrying the NdeI restriction site and antisense primers CotY3′ and CotW3′, respectively, with the EcoRI restriction site (Table 1). The PCR fragments were digested with NdeI and EcoRI and ligated into the vector digested with the same restriction endonucleases to yield expression plasmids pETcotY and pETcotW, respectively. The expression plasmid for CotE protein pETcotE was prepared in a similar manner, using primers CotE5′ and CotE3′ respectively (Table 1). For pull-down experiments, proteins were coexpressed in the pETDuet-1 vector (Novagen). The cotZ and cotV genes were tagged with hexahistidine. The coding sequence of CotZ was amplified from B. subtilis PY79 genomic DNA with primers CotZ5′Duet and CotZ3′Duet containing EcoRI and SalI restriction sites, respectively. BamHI and PstI sites were introduced into the sense and antisense primers CotV5′Duet and CotV3′Duet, respectively, to clone the cotV gene (Table 1). The amplified fragments were digested with particular restriction enzymes and cloned into vectors (with complementary restriction sites, giving rise to the plasmids pETDuetcotY and pETDuetcotZ). In the next step, the cotY and cotW genes were cloned using NdeI and XhoI restriction sites which were added to the 5′ and 3′ ends of the coding sequences using oligonucleotides CotY5′Duet, CotY3′Duet, CotW5′ and CotW3′Duet, respectively (Table 1). PCR-amplified cotY was digested with NdeI and XhoI restriction enzymes and ligated into the plasmids pETDuet-1 and pETDuetCotZ to yield expression plasmids pETDuetcotZcotY and pETDuetcotY. cotW was cloned in the same manner into plasmids pETDuetcotV and pETDuet-1, resulting in plasmids pETDuetcotVcotW and pETDuetcotW, respectively.
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequencea |
|---|---|
| CotE5′ | AGGAGGAGGCATATGTCTGAATACAGGGAAATTAT |
| CotE3′ | TGGTGGTGGGAATTCTTATTCTTCAGGATCTCCC |
| CotY5′ | AGGAGGAGGCATATGAGCTGCGGAAAAACCC |
| CotY3′ | AGGAGGAGGGAATTCTTATCCATTGTGATGATGCTTTTTATC |
| CotW5′ | AGGAGGAGGCATATGTCAGATAACGATAAATTCAAAG |
| CotW3′ | GGAGGAGGAGAATTCCTAATTGTTATCGTTTTTCCGTC |
| CotY5′Duet | TGGTGGTGGCATATGAGCTGCGGAAAAACCCATG |
| CotY3′Duet | TGGTGGTGGCTCGAGTTATCCATTGTGATGATGCTTTTTATC |
| CotZ5′Duet | TGGTGGTGGGAATTCAAGCCAGAAAACATCAAGCTG |
| CotZ3′Duet | TGGTGGTGGGTCGACTTAATGATGATGTGTACGATTG |
| CotV5′Duet | GATGATGATGGATCCTTCATTTGAAGAAAAAGTCGAATCC |
| CotV3′Duet | ATGATGATGCTGCAGTTAAAGGACGTCAAGTTCACTAAG |
| CotW3′Duet | GGAGGAGGACTCGAGCTAATTGTTATCGTTTTTCCGTC |
| Y2HcotE5′ | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTCTGAATACAGGGAAATTATTACGAAGG |
| Y2HcotE3b′ | GGGGACCACTTTGTACAAGAAAGCTGGGTCTCATTCTTCAGGATCTCCCACTAAAAAC |
| Y2HcotX5′ | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAAGCAGACCATATTCTTGGG |
| Y2HcotX3′ | GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAGAGGACAAGAGTGATAACTAGG |
| Y2HcotY5′ | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAGCTGCGGAAAAACCCATGG |
| Y2HcotY3′ | GGGGACCACTTTGTACAAGAAAGCTGGGTCTCATCCATTGTGATGATGCTTTTTATCTTTG |
| Y2HcotZ5′ | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCAGCCAGAAAACATCAAGCTGCGTG |
| Y2HcotZ3′ | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAATGATGATGTGTACGATTGATTAATCGAG |
| Y2HcotV5′ | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTCATTTGAAGAAAAAGTCGAATCCCTG |
| Y2HcotV3′ | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAAAGGACGTCAAGTTCACTAAGAATG |
| Y2HcotW5′ | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTCAGATAACGATAAATTCAAAGAAGAGC |
| Y2HcotW3′ | GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAATTGTTATCGTTTTTCCGTCTTTGTC |
Restriction sites and attB1/attB2 sequences used for Gateway system cloning are underlined.
Overexpression of recombinant His-tagged proteins and antibody production.
E. coli BL21(DE3) was transformed with expression plasmids and grown in LB medium at 37°C. When the optical density at 600 nm (OD600) of the culture reached 0.5, the expression of recombinant proteins was induced by addition of 1 mM isopropyl-β-galactoside (IPTG). Cells were harvested 3 h after induction, centrifuged, and resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl). Bacteria were lysed by sonication (total time of 2.5 min, with 10-s bursts with 30-s cooling periods) and then centrifuged for 25 min at 30,000 rpm in a Beckman L7 ultracentrifuge, after which proteins were further purified by metal affinity chromatography. The supernatant was then applied to a 1-ml Ni Sepharose HP column (Amersham Biosciences). Proteins were eluted with a 4-ml step gradient of 40 mM to 1 M imidazole. Recombinant proteins that were more than 90% pure (according sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) were then directly used for antibody production in mice.
Production of recombinant proteins using the pETDuet construct were performed as described above for purification of His-tagged proteins. Copurified proteins were identified by Western blot analysis using specific polyclonal antibodies or the monoclonal antibody against the hexahistidine tag.
Polyclonal antibodies against CotE, CotY, and CotW.
Polyclonal antibodies were raised against CotE, CotY, and CotW recombinant proteins. All proteins were expressed in E. coli and carried the polyhistidine tag at the N terminus of the protein. Most of CotE and CotY was produced as an insoluble protein, but for antibody production, the proteins were purified from soluble fractions as described above. CotW, CotE, and CotY were produced as soluble proteins. Purified proteins were used to raise mouse polyclonal antibodies according to standard procedures.
Western blot analysis.
Protein samples were mixed with an equal volume of 2× SDS sample buffer and boiled for 10 min. Proteins were fractionated by 12% SDS-PAGE and transferred to a nitrocellulose membrane (Hybond ECL; Amersham Bioscience) by standard procedures. The membrane was then treated with 5% nonfat milk in Tris-buffered saline, containing 0.05% Tween 20 to prevent nonspecific protein binding. Bound proteins were probed with specific monoclonal or polyclonal antibodies and detected using antimouse horseradish peroxidase-conjugated secondary antibodies.
Analysis of proteins by size exclusion chromatography.
The Superose 12 column 10/300 GL (Amersham Pharmacia) was calibrated with low-molecular-weight calibration proteins (Amersham Pharmacia) in 50 mM Tris-HCl buffer-150 mM NaCl (pH 8.0). Purified recombinant proteins were loaded in a total volume of 0.5 ml. Chromatography was performed at a flow rate of 0.5 ml per minute. The eluted proteins were detected with UV light at 280 nm and confirmed by Western blot analysis.
Construction of yeast plasmids.
A Gal4-based system with Gateway Technology Proquest (Invitrogen) was used for yeast two-hybrid analysis. The genes cotE, cotV, cotW, cotX, cotY, and cotZ were amplified by PCR using B. subtilis PY79 genomic DNA as a template. The PCR primers (Table 1) were flanked with attB recombination sites required by the Gateway cloning system. PCR products were purified using the QIAquick gel extraction kit (Qiagen) and initially cloned into the pDONR221 vector by the BP recombination reaction to create the entry clones. Subsequently, the target genes were transferred into yeast destination vectors pDEST22 and pDEST32 via the LR recombination reaction. pDEST22 encodes the GAL4 transcription activation domain and contains selection marker (TRP1) for tryptophan auxotrophy, pDEST32 DNA encodes the DNA binding domain of Gal4 and for auxotrophic selection carries the gene LEU2. All clones were checked by restriction analysis and DNA sequencing.
Yeast two-hybrid assay of spore coat protein interactions.
Both bait and target plasmids were cotransformed simultaneously into MaV203 yeast strains by the lithium acetate method (7). Yeast transformants were plated on synthetic complete medium (SC) lacking leucine (−Leu) and tryptophan (−Trp). After 72 h, large colonies were picked up and suspended in 80 μl of sterile water. Two-microliter drops of cell suspension were applied onto selection plates for screening expression of three reporter genes (HIS3, URA3, and lacZ). Interactions were assessed by growth on SC −Leu −Trp −His plus 100 mM 3-amino-1,2,4-triazole (3AT) and SC −Leu −Trp −Ura, by no growth on SC −Leu −His plus 0.5% 5-fluoroorotic acid (5FOA), and by the change of color to blue in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). All possible bait-target plasmid combinations were analyzed. To check autoactivation of reporter genes, all bait plasmids were also combined with the pEXP-AD502 vector and target plasmids were combined with the pDBLeu vector.
β-Galactosidase assay in liquid culture.
β-Galactosidase activity was measured using o-nitrophenol-β-d-galactopyranoside (ONPG) as a substrate (17). β-Galactosidase activity was expressed in Miller units: 1 Miller unit = 1,000 × OD420/(t × V × OD600), where t is incubation time in minutes, V is volume of culture used in the assay, OD420 is absorbance of o-nitrophenol, and OD600 represents the cell density of the culture.
Reassessment of interactions: retransformation assay.
The yeast plasmids from the candidate clones were prepared by being cultured in 10 ml of SC −Leu −Trp for 24 h at 30°C. After mechanical disruption of cells with autoclaved acid-washed glass beads by vortexing for 2 min, the QIAprep spin miniprep kit (Qiagen) was used for isolation of plasmids that were transformed into E. coli XL1 Blue. Transformants were selected on LB plates with 100 μg/ml ampicillin to selectively isolate pDEST22cot plasmids and 10 μg/ml gentamicin to selectively isolate pDEST32cot plasmids. All plasmids were analyzed by restriction analysis and reintroduced into MaV203. Transformants were examined on selection plates for appearance of the original phenotype.
RESULTS
Protein-protein interactions of Cot proteins revealed by yeast two-hybrid analysis.
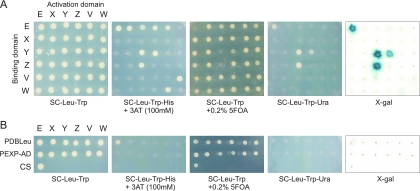
The proteins encoded by the cotVWXYZ gene cluster are required for the proper formation of the outer coat, and we focused on addressing three principal questions. First, can these proteins assemble spontaneously? Second, which of these proteins are in contact with others? Finally, what is the possible role of the CotE morphogenetic protein in the assembly of these coat proteins? As a first step toward a better understanding of these questions, the yeast two-hybrid analysis was employed. Full-length structural genes encoding CotE, CotV, CotW, CotX, CotY, and CotZ were fused with both the DNA binding domain and transcription activation domains of the Gal4 transcription factor. All possible combinations of interacting pairs were tested, and every potential protein-protein interaction was examined using reciprocal bait/target setups to avoid potential folding or steric constraints. The direct interactions of fusion partners were investigated by expression of all three reporter genes (HIS3, URA3, and lacZ) as described in Materials and Methods. We identified well-growing yeast cells: HIS3 prototrophs on plates lacking Leu, Trp, and His and containing 100 mM 3AT in 6 out of 36 possible combinations (Fig. 1). The homotypic interactions were detected for CotE and CotY proteins. We also identified two heterotypic interactions between CotY-CotZ and CotV-CotW in both combinations of prey and target plasmids (Table 2). Although the presence of 3AT as a competitive inhibitor of the HIS3 reporter should have helped to discriminate strong and weak interactions, we did not observe any differences in the amount of growth of positive candidate clones even at 100 mM concentrations of 3AT. However, we were able to distinguish between the strong and weak interactions by examination of the expression level of the lacZ gene, which was monitored by a filter assay using X-Gal as the substrate. The blue color was developed within 1 h in the yeast colonies carrying bait/target plasmids with CotE-CotE, CotY-CotY, and CotY-CotZ fusions, indicating strong interactions. Significantly weaker interactions were observed between CotV and CotW, since yeast cells turned blue in an X-Gal assay only after overnight incubation (Table 2). Expression of a third reporter gene, URA3, entailed the examination of cell growth on two different plates: plates lacking uracil, where cells containing interacting proteins grow, and plates containing 5FOA, where cells do not grow due to the toxicity of 5-fluorouracil originating from conversion of 5FOA after URA3 induction. In the case of CotE-CotE, CotY-CotY, and CotY-CotZ interactions, the phenotype of the stronger interactors was confirmed since yeast cells grew well on SC −Leu −Trp −Ura plates and they did not grow on plates containing 5FOA. As mentioned above, CotV-CotW interaction was identified as weak according to the X-Gal assay. The yeast cells expressing CotV and CotW fusion proteins were not able to grow on plates lacking uracil, and inhibition by 5FOA was only partial (Table 2). These results indicate that growth on uracil and 5FOA selection plates was not sufficiently sensitive for such weak protein-protein interactions and only strong interactions can sufficiently induce the URA3 promoter, as described previously (25).
FIG. 1.
Protein-protein interactions among the spore coat proteins. The images represent the summary of results obtained from the yeast two-hybrid analysis. Panel A shows yeast MaV203 cells cotransformed with multiple bait-target combinations of plasmids as specified and analyzed for gene reporter activity by growth on selection media as described in Materials and Methods. E, CotE; V, CotV; W, CotW; X, CotX; Y, CotY; Z, CotZ. Panel B shows the self-activation test. In the first line, the pDBleu plasmid was cotransformed with the yeast target plasmids, containing the indicated Cot protein fused with the Gal4 activation domain. Though the CotE bait is a weak self-activator, when coexpressed with the CotE target, the activation of reporter genes is significantly enhanced. In the second line, the pEXP-AD502 plasmid was cotransformed with bait plasmids containing the Cot protein fused with the Gal4 binding domain. The control strain carries the pDBleu and pEXP-AD502 plasmids. CS, negative control strain.
TABLE 2.
Interaction of spore coat proteins in the yeast two-hybrid system
| Activation domain | Protein activity in binding domaina
|
|||||
|---|---|---|---|---|---|---|
| CotE | CotV | CotW | CotX | CotY | CotZ | |
| CotE | +++ | − | − | − | − | − |
| CotV | − | − | + | − | − | − |
| CotW | − | + | − | − | − | − |
| CotX | − | − | − | − | − | − |
| CotY | − | − | − | − | +++ | +++ |
| CotZ | − | − | − | − | ++ | − |
Quantitative estimate of interactions was made based on the growth profiles on selection plates and the X-Gal assay. +++, strong interactions when the yeast colonies grow well on plates lacking histidine and uracil, do not grow in the presence of 5FOA, and an intensive blue color in the filter assay develops in <1 h; ++, moderately strong interactions when the yeast colonies grow well on plates lacking histidine and slightly less on plates lacking uracil, do not grow in the presence of 5FOA, and an intensive blue color in filter assay develops in <6 h; +, weak interactions when the yeast colonies grow slightly less than the control representing the strong interaction (CotE/CotE) on plates lacking histidine, do not grow on plates lacking uracil, grow slightly slower in the presence of 5FOA, and a light blue color in the filter assay develops overnight.
The relative strength of all identified protein-protein interactions was examined further by quantitative assay of β-galactosidase activity in liquid culture (see Materials and Methods). The strong interactions revealed by the filter assay were confirmed for the pairs CotE-CotE and CotY-CotY with β-galactosidase activity of about 30 Miller units (Table 3). In the case of CotY-CotZ, we observed that this interaction was sensitive to the direction in which it was tested. β-Galactosidase activity was more than 1 order higher when the cotZ gene was fused with the GAL4 DNA binding domain (38 Miller units) versus when fused with the transcription activation domain (2.2 Miller units) (Table 3). The β-galactosidase activity, in principle, did not differ significantly in both directions for the CotV-CotW pair, but only reached approximately 1 Miller unit (Table 3). Although this value is rather low, there was an obvious difference in the intensity of the yellow color of the reaction mixture containing the lysate of CotV-CotW and that of the negative control containing the empty vectors. Clearly, after the overnight incubation, the reaction mixture with CotV-CotW developed a yellow color in comparison with the completely colorless negative control. We also examined the β-galactosidase activity of three control strains, obtained from the manufacturer (Table 3), where the strength of interactions is known. After comparing all results, we can consider CotE-CotE, CotY-CotY, and CotZ-CotY to be the strong interactions, while CotW-CotV could be regarded as a weak interaction.
TABLE 3.
Level of lacZ expression in yeast
| Fusion protein binding domain/ activation domaina | β-Galactosidase activity (Miller units)b |
|---|---|
| CotE/CotE | 48.2 ± 8.6 |
| CotY/CotY | 45.4 ± 5.1 |
| CotZ/CotY | 58.2 ± 2.8 |
| CotY/CotZ | 3.4 ± 0.5 |
| CotW/CotV | 1.6 ± 0.1 |
| CotV/CotW | 0.8 ± 0.09 |
| Human RB/human E2F1c | 0.05 ± 0.00 |
| Drosophila DP/Drosophila E2Fd | 6.7 ± 0.2 |
| Gal4/no inserte | 212.7 ± 23.0 |
Shown are the different pairs of Cot protein fusions and control protein pairs.
β-Galactosidase activity was measured in Miller units, with ONPG as the substrate. The values shown represent the average of at least five independently isolated colonies, and every sample was measured in triplicate. Control strains were obtained from Invitrogen Life Technologies, and more details can be found in the ProQuest two-hybrid system with Gateway Technology instruction manual.
Strain shows weak interaction. Human RB, accession no. M28419, amino acids 302 to 928; human E2F1, accession no. M96577, amino acids 324 to 437.
Strain shows moderately strong interaction. Drosophila DP, accession no. U10184, amino acids 1 to 377; Drosophila E2F, accession no. X12761, amino acids 250 to 325.
Strain shows very strong interaction. Gal4, accession no. K10486, amino acids 1 to 881.
To reassess the positive interactions which we have revealed, the plasmids were isolated from yeast and evaluated by restriction analysis. After retransformation into yeast and examination of their phenotype on selection plates, all previously identified interactions were confirmed. Fusion proteins after interacting with their partners retained the ability to induce reporter genes.
Coexpression of spore coat proteins and pull-down assay.
Although yeast two-hybrid analysis is a useful tool for the identification of protein-protein interactions, it is crucial to confirm them by other independent biochemical assays. In the case of Cot proteins, the selection of another method was problematic due to the difficulties with their solubility when produced in E. coli, which varies from highly soluble CotW, via partially soluble CotE and CotY, up to almost completely insoluble CotV and CotZ. Moreover, as mentioned above, Cot proteins often form large multimeric structures that in some cases limit their capability to bind onto the Ni Sepharose. Similarly, it is possible that such large aggregates could not interact in vitro. Therefore, we selected the pETDuet system to coexpress two proteins, allowing possible interaction inside the cells and consequently purifying the protein complex. In this case, one protein carries the polyhistidine tag and could be affinity purified on a Ni column and the second was produced as a native protein that could be pulled down. Immunoblot analysis of bacterial extracts using antibodies to His tag or specific Cot protein antibodies confirmed that all proteins were produced in E. coli, and at least a portion of the protein could be detected in a soluble fraction. To identify potential interactions, the proteins after purification on Ni Sepharose were analyzed by Western blotting (Fig. 2). The obtained results showed that native untagged CotY protein was not detected in elution fractions when produced alone (Fig. 2, lane 1) but was pulled down with His-tagged CotZ after coexpression of both proteins (Fig. 2, lane 2). Similarly, CotW was pulled down with His-tagged CotV (Fig. 2, lane 8) and was not binding onto the affinity column when expressed by itself (Fig. 2, lane 7). Both results suggest that untagged proteins made direct contacts with their protein partners. We also observed that the solubility of CotV when produced together with CotW was increased significantly. The level of purified soluble protein could barely be seen by immunoblotting when using the monoclonal anti-His antibody. However, when coexpressed with CotW it could be simply detected on SDS-PAGE gels stained with Coomassie brilliant blue (data not shown).
FIG. 2.

The pull-down assay. Proteins were expressed in E. coli BL21(DE3) cells, and bacterial extracts were applied onto a Ni Sepharose HP column. Eluted proteins were probed with anti-CotY (A) or anti-CotW (C) polyclonal antibodies or anti-His monoclonal antibody (B and D) by Western blot analysis. Panels A and B show the results of the CotY/CotZ pull-down assay. Lanes 1 and 4 represent the negative control, where untagged CotY is not binding on the affinity column. Lanes 3 and 6 contain His-tagged CotZ. Lanes 2 and 5 show CotY and His-tagged CotZ proteins copurified from the Ni column. Panels C and D show the results of CotV/CotW pull-down. Lanes 7 and 10 represent the negative control, when untagged CotW is not binding on the Ni column. Lanes 9 and 12 contain His-tagged CotV protein, which is not detectable by immunoblotting in this arrangement because of a large difference in the levels of expression of CotV when expressed alone and in the presence of CotW. Lanes 8 and 11 show CotW copurified with His-tagged CotV.
Size exclusion chromatography indicates multimeric structure formation by CotE, CotY, and CotZ.
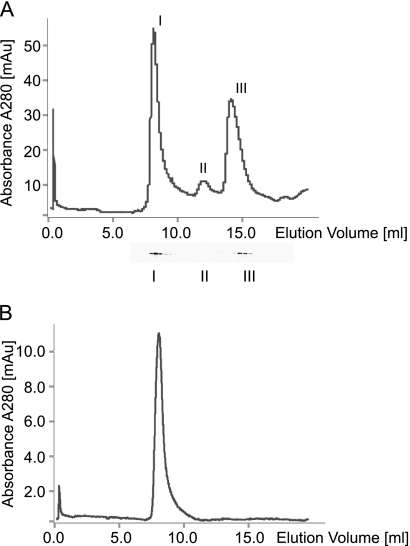
Self-assembly of the CotE, CotY, and CotZ proteins has been previously reported, and several multimeric forms have been detected in spore coat protein extracts (14, 27). To analyze the size of possible multimeric forms of recombinant variants of these proteins, we employed a size exclusion chromatography procedure. We observed the propensity to form high-molecular-weight species in the case of recombinant proteins. We found out that soluble native CotE protein was binding onto the Ni Sepharose column very poorly, whereas after solubilization in 8 M urea, most of the protein was binding efficiently (data not shown). After analysis of the native protein on calibrated Superose 12, we found that the protein is eluted in a void volume of the column, suggesting that CotE was self-assembling into large multimeric structures. The monomeric or dimeric forms of CotE could be observed only if the native protein, eluted from the Ni column, was immediately analyzed by gel filtration (Fig. 3A). Moreover, the overnight incubation of the sample at 4°C led to the state when the entire protein was assembled into high-molecular-weight oligomers, as seen in Fig. 3B. The identity of CotE in all fractions was ensured by immunoblotting using CotE-specific antibody (see Materials and Methods). In the case of CotY and CotZ, only the multimeric forms of proteins were eluted in the void volume of a Superose 12 column; the smaller forms were not detected (data not shown). Since reducing agents have to be avoided in all buffers for Ni Sepharose column, 2 mM dithiothreitol (DTT) was normally added after the proteins were eluted. Obviously, such a concentration of DTT was not sufficient to disrupt the extensive cross-linking of CotY or CotZ, which could result from various intermolecular disulfide bonds. Interestingly, when analyzing proteins eluted from Ni Sepharose even in the presence of urea, we also observed high-molecular-weight species on the top of the 12% SDS-PAGE gel. Apparently, once these proteins are cross-linked, the ability to solubilize them is quite limited.
FIG. 3.
Size exclusion chromatography of CotE protein. Chromatography was performed using a Superose 12 column. (A) Native CotE protein analyzed by gel filtration directly after elution from the Ni column. Values are expressed in milliabsorbance units. (B) Analysis of the multimeric state of CotE approximately 24 h after elution from the Ni column. Proteins eluted from the column were identified using specific CotE antibodies, and the different forms detected were the multimer (I), dimer (II), and the monomer (III).
DISCUSSION
Protein assembly is a crucial process in enabling complex macromolecular structures important for cell function to be built (10, 15, 23, 24). These structures usually differ significantly in function, morphology, and complexity. The spore coats of B. subtilis endospores represent a unique and compact system in which proteins arrange themselves into a three-dimensional structure. Though probably most of the components have been identified, we have only an incomplete understanding of how this specific protein assembly process has evolved. In this work, we focused on the group of the proteins identified as insoluble fractions of spore coat proteins which are likely highly cross-linked, as was proposed in earlier studies (27). In this respect, to study these protein-protein interactions is technically challenging. As an initial approach, we probed their interactions using the yeast two-hybrid system. This method enabled detection of interactions between CotY and CotZ, CotV and CotW, and two homotypic interactions of CotE and CotY. As mentioned earlier, the CotY and CotZ proteins are homologous, tyrosine-rich components of the coat, and their direct contact had been predicted assuming that the binding is accomplished via disulfide bonds. According to the high level of β-galactosidase activity measured in yeast, implying strong interactions, we could speculate that there is more than one disulfide bond and that both proteins are in fact highly cross-linked. Similarly the presence of potential disulfides is plausible within individual CotY and CotZ molecules. Although we did not identify the homotypic interaction of CotZ by the yeast two-hybrid assay, we have seen multimeric forms of this protein by gel filtration. These high-molecular-weight CotZ species were also detected by SDS-PAGE and could not be reduced by a large excess of dithiothreitol or β-mercaptoethanol. It is reasonable to assume that the failure to detect the self-interaction of CotZ by two-hybrid techniques could be the effect of propensity of the CotZ to form large aggregates, which precludes the reconstitution of a functional Gal4 yeast transcription factor. All of our experiments showed that there are three different functional complexes present in the spore coat (CotY-CotY, CotZ-CotZ, and CotZ-CotY), but it is still unclear how exactly these proteins are arrayed in the coat. The specific interactions between CotY/CotZ and CotX were proposed by Zhang (27) since in a cotX-null mutant an excess of the CotY and CotZ proteins was observed in the soluble coat fraction in comparison to the wild-type spores. However, our data did not show direct contact. We believe that the interactions of CotX, which is rich in hydrophobic regions in its C terminus, could be restricted by incorrect folding or inefficient targeting of hybrid protein into the yeast nucleus. Other explanations include instability of the protein or steric hindrance arising from the adjacent Gal4 domain in the hybrid protein so that transcriptional activation of reporter genes is prevented.
In spite of numerous lines of evidence that CotE controls the assembly of the outer coat, until now, no direct interaction of CotE with another coat component has been reported. It has been shown that assembly of CotX, CotZ, and CotW is CotE dependent (11). Moreover, as shown by Eichenberger et al. (6), CotYZ proteins are synthesized earlier than has been suggested previously (28), since their expression is controlled not only by σK, but also by the σE transcription factor. σE directs spore coat formation during the initial stages of development and transcribes the genes of the most important morphogenetic proteins, including CotE. Taking into account these results, together with the fact that the CotYZ and CotE proteins were found in the insoluble coat fraction, we hypothesized that these proteins together with some other insoluble fraction components form an assembly platform for other outer coat proteins. However, our results from the yeast two-hybrid screening did not reveal any direct interaction involving CotE and CotVWXYZ. Nevertheless we cannot exclude that some interactions were missed because of the inherent limits of the yeast two-hybrid method arising from the specific properties of hybrid coat proteins. For example, some of our in vitro experiments suggest that there is some affinity between CotE and CotZ. We observed that the recombinant CotZ protein did not precipitate when dialyzed together with CotE, but was heavily precipitating when dialyzed alone (data not shown). Another indication of their interaction was obtained from a pull-down assay in which CotE was significantly retained by CotZ (data not shown). Although the untagged CotE was partially binding nonspecifically on a Ni Sepharose HP column, the pattern of CotE elution when coexpressed with CotZ was distinct. It appears that CotE/CotZ forms a low-affinity interaction which could lie below the detection limits of the yeast two-hybrid method, which usually represents an equilibrium dissociation constant (KD) ranging around 20 to 50 μM.
We also found that CotV associates with CotW. The ability of these bait/target hybrid proteins to activate transcription of reporter genes in the yeast two-hybrid system was relatively lower than those in the other positive cases that were identified. We could speculate that similarly, as in the case of CotX, this could also be caused by insufficient nuclear targeting or misfolding of CotV, which has, like CotX a highly hydrophobic region in the C terminus. Although CotV/CotW appears to be a weak interaction in the yeast two-hybrid assay, the result of pull-down experiments was well evident. The CotW protein was pulled-down with CotV, and additionally, the solubility of CotV when produced with CotW was remarkably increased. Obviously coexpression of both proteins facilitates the formation of a functional complex. These results clearly show that CotV is bona fide coat protein as originally presumed (27), although it has not been detected in spore coat extract (12, 13).
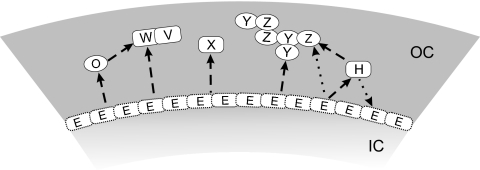
In conclusion, our study provides the first view in which direct interactions could be expected among the insoluble spore coat proteins (Fig. 4). As we mentioned above, we cannot truly rule out the possibility that we were not able to detect all existing contacts among the investigated proteins, likely resulting from their insoluble nature. It is also possible that formation of functional complexes requires the presence of more than two proteins, which was not investigated within the scope of this work. Nevertheless, our results suggest that the insoluble fraction of the spore coat is formed by self-assembly of at least two pairs of its components, CotY and CotZ and CotV and CotW. It is possible to speculate that CotX, containing seven cysteines and a hydrophobic C-terminal region, could be the third potential binding partner for both protein couples. These contacts could be mediated by cross-linking via disulfide bonds with CotY/CotZ or by a contact with very hydrophobic CotV (27). We still have minimal knowledge of how these proteins are guided into the proper position. Although we observed some affinity between CotE and CotZ, we do not have enough data to support the idea that CotE is responsible for directing insoluble Cot proteins onto the spore surface. It is still possible that the CotE protein partner/s among the insoluble Cot proteins will be uncovered by using more advanced techniques. Alternatively, other Cot protein or proteins could act as intermediates in this process, such as CotO or CotH. Namely, CotO is a good candidate since it was shown previously that the assembly of CotW is CotO dependent (16).
FIG. 4.
Model of interactions among the insoluble spore coat proteins. Only proteins localized in the outer coat (OC) are indicated. Capital letters refer to appropriate Cot proteins. Schematic sections of the outer coat and inner coat (IC) are shaded. The direct contacts between proteins revealed by yeast two-hybrid assay are indicated by the direct contacts of the blocks representing individual Cot proteins. Dashed arrows show the assembly dependencies of one protein on another. In the case of CotE/CotZ, the dotted arrow shows some affinity between these proteins observed in the pull-down assay. In the case of CotE/CotH, the dotted arrow refers to an effect of CotH on CotE deposition (11).
Taken together, the obtained results clearly show that proteins of insoluble spore coat fractions participate in specific protein-protein interactions. These interactions, together with the contribution of cross-linking enzymes, determine the resilient and robust nature of the spore coat.
Acknowledgments
The work in our laboratory is supported by grant NMP4-CT-2004-013523 from the EU 6th FP to S.M.C. and I.B., by grant 2/7007/27 to I.B. from the Slovak Academy of Sciences, and by grant APVT-51-027804 from the Ministry of Education of the Slovak Republic to I.B.
We thank Milka Chovancová for technical assistance.
Footnotes
Published ahead of print on 20 March 2009.
REFERENCES
- 1.Aronson, A. I., L. Ekanayake, and P. C. Fitz-James. 1992. Protein filaments may initiate the assembly of the Bacillus subtilis spore coat. Biochimie 74661-667. [DOI] [PubMed] [Google Scholar]
- 2.Backman, K., M. Ptashne, and A. W. Gilbert. 1976. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 734174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 631-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driks, A., S. Roels, B. Beall, C. P. Moran, and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8234-244. [DOI] [PubMed] [Google Scholar]
- 5.Driks, A. 2003. The dynamic spore. Proc. Natl. Acad. Sci. USA 1003007-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. González-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327945-972. [DOI] [PubMed] [Google Scholar]
- 7.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by the lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 35087-96. [DOI] [PubMed] [Google Scholar]
- 8.Henriques, A. O., and C. P. Moran, Jr. 2000. Structure and assembly of the bacterial endospore coat. Methods 2095-110. [DOI] [PubMed] [Google Scholar]
- 9.Henriques, A. O., and C. P. Moran, Jr. 2007. Structure, assembly and function of the spore surface layers. Annu. Rev. Microbiol. 61555-588. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. E., and W. Chiu. 2007. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 17237-243. [DOI] [PubMed] [Google Scholar]
- 11.Kim, H., M. Hahn, P. Grabowski, D. C. McPherson, M. M. Otte, R. Wang, C. C. Ferguson, P. Eichenberger, and A. Driks. 2006. The Bacillus subtilis spore coat protein interaction network. Mol. Microbiol. 59487-502. [DOI] [PubMed] [Google Scholar]
- 12.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 1483971-3982. [DOI] [PubMed] [Google Scholar]
- 13.Lai, E.-M., N. D. Phadke, M. T. Kachman, R. Giorno, S. Vazquez, J. A. Vazquez, J. R. Maddock, and A. Driks. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 1851443-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little, S., and A. Driks. 2001. Functional analysis of the Bacillus subtilis morphogenetic spore coat protein CotE. Mol. Microbiol. 421107-1120. [DOI] [PubMed] [Google Scholar]
- 15.Macnab, R. M. 2004. F type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694207-217. [DOI] [PubMed] [Google Scholar]
- 16.McPherson, D. C., H. Kim, M. Hahn, R. Wang, P. Grabowski, P. Eichenberger, and A. Driks. 2005. Characterization of the Bacillus subtilis spore morphogenetic coat protein CotO. J. Bacteriol. 1878278-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Ramamurthi, K. S., K. R. Clapham, and R. Losick. 2006. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol. Microbiol. 621547-1557. [DOI] [PubMed] [Google Scholar]
- 19.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Seyler, R. W., Jr., A. O. Henriques, A. J. Ozin, and C. P. Moran, Jr. 1997. Assembly and interactions of cotJ-encoded proteins, constituents of the inner layers of the Bacillus subtilis spore coat. Mol. Microbiol. 25955-966. [DOI] [PubMed] [Google Scholar]
- 22.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. 1999. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol. 1814986-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thanassi, D. G., and S. J. Hultgren. 2000. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods 20111-126. [DOI] [PubMed] [Google Scholar]
- 24.Vicente, M., A. I. Rico, R. Martínez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 18819-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidal, M. 1997. The reverse two-hybrid system, p. 109-147. In p. Bartels and S. Fields (ed.), The two-hybrid system. Oxford University Press, New York, NY.
- 26.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 121-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, J., P. C. Fitz-James, and A. I. Aronson. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 1753757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, J., H. Ichikawa, R. Halberg, L. Kroos, and A. I. Aronson. 1994. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J. Mol. Biol. 240405-415. [DOI] [PubMed] [Google Scholar]
- 29.Zheng, L. B., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 21047-1054. [DOI] [PubMed] [Google Scholar]
- 30.Zilhão, R., M. Serrano, R. Isticato, E. Ricca, C. P. Moran, Jr., and A. O. Henriques. 1994. Interactions among CotB, CotG, and CotH during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 1861110-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]