Abstract
Using comparative genome sequencing analysis, we identified a novel mutation in Bacillus subtilis that confers a low level of resistance to fusidic acid. This mutation was located in the mdtR (formerly yusO) gene, which encodes a MarR-type transcriptional regulator, and conferred a low level of resistance to several antibiotics, including novobiocin, streptomycin, and actinomycin D. Transformation experiments showed that this mdtR mutation was responsible for multidrug resistance. Northern blot analysis revealed that the downstream gene mdtP (formerly yusP), which encodes a multidrug efflux transporter, is cotranscribed with mdtR as an operon. Disruption of the mdtP gene completely abolished the multidrug resistance phenotype observed in the mdtR mutant. DNase I footprinting and primer extension analyses demonstrated that the MdtR protein binds directly to the mdtRP promoter, thus leading to repression of its transcription. Moreover, gel mobility shift analysis indicated that an Arg83 → Lys or Ala67 → Thr substitution in MdtR significantly reduces binding affinity to DNA, resulting in derepression of mdtRP transcription. Low concentrations of fusidic acid induced the expression of mdtP, although the level of mdtP expression was much lower than that in the mdtR disruptant. These findings indicate that the MdtR protein is a repressor of the mdtRP operon and that the MdtP protein functions as a multidrug efflux transporter in B. subtilis.
The bacterial ribosome is a major target for antibiotics. For example, the steroid-like antibiotic fusidic acid is characterized by its ability to interfere with the translation factor elongation factor G (EF-G), thus inhibiting protein synthesis (5, 21). Bacterial cells often mutate spontaneously, producing cells that are resistant to various antibiotics. The mechanisms of antibiotic resistance can be classified into four major classes: (i) alteration or modification of the target of the antibiotic, leading to a loss or reduction of the interaction of the target with the antibiotic; (ii) acquisition of impermeability or increased efflux of antibiotic, decreasing its intracellular concentration; (iii) enzymatic detoxification of antibiotic; and (iv) target bypass (6, 35). High-level resistance to fusidic acid is often due to alterations in its target molecule, EF-G, a protein encoded by fusA (4, 18). In contrast, mutations that confer a low-level resistance to antibiotics, including fusidic acid, have been largely overlooked due to their reduced significance in antibiotic chemotherapy. However, the importance of studying low-level resistance to antibiotics has increased in both medical and industrial microbiology. For example, we recently found that low-level resistance to streptomycin was due to a mutation in the rsmG gene, which encodes a 16S rRNA methyltransferase, and that low-level resistance to kasugamycin was due to a mutation in the speD gene, which encodes S-adenosylmethionine decarboxylase (25, 26, 29). These findings provided clues to study the mechanism of high-frequency appearance of high-level streptomycin resistance in Mycobacterium tuberculosis (30) and the overproduction of antibiotics by Streptomyces coelicolor (25). Similarly, our laboratory has focused on strain improvement for antibiotic overproduction and has developed a new method to activate or enhance antibiotic production in bacteria. In this method, a specific mutation is introduced, by isolating spontaneously developed drug-resistant mutants, into the rpsL gene (encoding the ribosomal protein S12) or the rpoB gene (encoding the RNA polymerase β subunit), conferring resistance to a drug such as streptomycin, gentamicin, or rifampin (rifampicin). These findings indicated that bacterial gene expression can be altered dramatically by modulating ribosomal proteins and/or RNA polymerase (12, 16). The rpsL mutant ribosomes carrying an amino acid substitution in S12, which confers a high level of resistance to streptomycin, are more stable than those of wild-type controls at low magnesium concentrations, indicating that this increase in stability could enhance protein synthesis at the late growth phase (11, 20, 31, 33). We later found that increased expression of the translation factor ribosome recycling factor also contributes to the enhanced protein synthesis observed during the late growth phase in the rpsL K88E mutant of S. coelicolor. This led us to conclude that both the greater stability of the 70S ribosomes and the elevated levels of ribosome recycling factor caused by the rpsL K88E mutation are responsible for the enhanced protein synthesis seen during the late growth phase and that this underlies the observed antibiotic overproduction in the rpsL K88E mutant (12). Likewise, certain mutations that confer low-level resistance to fusidic acid or thiostrepton result in enhanced antibiotic production in S. coelicolor and Streptomyces lividans (27, 36). Although the mutated genes have not yet been identified, these findings indicate the importance of low-level resistance mutations, especially in industrial microbiology. In the case of the rpsL K88E mutant of S. coelicolor, a high level of protein synthesis during the late growth phase underlies the enhanced antibiotic production of this mutant (12). Thus, our ultimate aim was to construct “ribosome engineering” (27, 28) as a rational approach to elicit bacterial capabilities fully within an industrial application. Working with the gram-positive model bacterium Bacillus subtilis and a new mutation search technique (1, 25), an alternative site for mutation (mdtR) to fusidic acid resistance, instead of a ribosome alteration, was found. The results are reported here, along with physiological and molecular analyses of the resistance phenotype.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All spontaneous fusidic acid-resistant mutants (HF01 to HF03 and LF01 to LF06) were derived from B. subtilis strain 168. Strain KJ01 (yusO1) was obtained by congression using B. subtilis strain YO-005 (hisC101) (13) as a recipient (see Results). Strain YUSOd was constructed by integration of the plasmid pMUTIN2 (34) into the yusO gene (19). To disrupt the yusP gene, the DNA fragment containing a partial yusP gene was amplified using the primers yusPH-F and yusPB-R (Table 2), digested with HindIII and BamHI, and cloned into the corresponding sites of pMutinT3 (24). The resulting plasmid, pMutinT3-yusP, was used for transformation of strains 168 and KJ01 (yusO1), generating strains KJ02 and KJ03, respectively. Erythromycin (0.5 μg/ml) was used for selection of transformants. B. subtilis strains were grown in L medium (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) at 37°C with vigorous shaking. Ampicillin (100 μg/ml) was used for selection of Escherichia coli transformants. MICs were determined as follows. Cells were grown in L medium for 18 h and diluted 200-fold (approximately 106 cells/ml). Then, 5 μl of cell suspension was spotted onto an L agar plate containing various concentrations of a drug, followed by incubation for 18 h at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| B. subtilis strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| 168 | trpC2 | Laboratory stock |
| YO-005 | hisC101 | 13 |
| HF01 | trpC2 fusA1 | This study |
| HF02 | trpC2 fusA2 | This study |
| HF03 | trpC2 fusA3 | This study |
| LF01 | trpC2 yusO1 | This study |
| LF02 | trpC2 yusO2 | This study |
| LF03 | trpC2 yusO3 | This study |
| LF04 | trpC2 yusO4 | This study |
| LF05 | trpC2 yusO5 | This study |
| LF06 | trpC2 yusO6 | This study |
| KJ01 | trpC2 yusO1 | This study |
| KJ02 | trpC2 yusP::pMutinT3 | This study |
| KJ03 | trpC2 yusO1 yusP::pMutinT3 | This study |
| YUSOd | trpC2 yusO::pMUTIN2 | This study |
| Plasmids | ||
| pCR2.1 | Cloning vector for PCR product | Invitrogen |
| pCR2.1-yusO | pCR2.1 containing yusO gene | This study |
| pCR2.1-yusP | pCR2.1 containing yusP gene | This study |
| pMutinT3 | Integration vector | 24 |
| pMutinT3-yusP | pMutinT3 containing 5′ region of yusP gene | This study |
| pMUTIN2 | Integration vector | 34 |
| pMUTIN2-yusO | pMUTIN2 containing 5′ region of yusO gene | This study |
| pET19b | Expression vector | Novagen |
| pET19b-yusO(wild type) | pET19b containing yusO gene (wild-type) | This study |
| pET19b-yusO(R83K) | pET19b containing yusO gene (R83K) | This study |
| pET19b-yusO(A67T) | pET19b containing yusO gene (A67T) | This study |
| pET22b(+) | Expression vector | Novagen |
| pET22b(+)-yusO | pET22(+) containing yusO gene | This study |
TABLE 2.
Oligonucleotides used in this study
| Primer | Oligonucleotide sequence (5′ → 3′)a |
|---|---|
| fusA1-F-seq | GGCTGAAGCAAACAAAGC |
| fusA1-R-seq | GATATCCCCTGCGTATACAG |
| fusA2-F-seq | GTTCTTCCGTGTGTACTCTG |
| fusA2-R-seq | TACCATGGTCAACGTGTC |
| yusO-F-seq | GATGTTCTTTCGGGCATC |
| yusO-R-seq | GTTGTCAGCCAGGTCATC |
| NyusO-F | AGCCTGAAATGCTGGAAAGC |
| NyusO-R | TCATCCGTT TCTGCTGCTTG |
| yusPH-F | CCCAAGCTTAGCCCTTGACGGTACC |
| yusPB-R | CGGGATCCCCGCAATAATTCCGACTG |
| PyusO-del-F | GTAAAATCAGTTGACTTTTCACTATTTTGTCATTAAAATATATATAC |
| PyusO-del-R | GTATATATATTTTAATGACAAAATAGTGAAAAGTCAACTGATTTTAC |
| PyusO-F1 | TACGATGTTCTTTCGGGCAT |
| PyusO-R1 | CACTCTTCATATTCATTTCCC |
| PyusO-F2 | TCTGTTGAAGCGGAGGATTC |
| PyusO-R2 | CTTGTTTCTCCATGCTTTCCAGC |
| yusON-F1 | CATATGAAGAGTGCGGATCAGTTAATG |
| yusOB-R1 | GGATCCTTATCCGTTTCCTCTTTTCATG |
| yusON-F2 | CATCATATGAAGAGTGCGGATCAGTT |
| yusOB-R2 | CGCGGATCCAATGGTACCGTCAAGGGCTG |
| RT-rrn16S-F | ATCTTCCGCAATGGACGA |
| RT-rrn16S-R | GCCGTGGCTTTCTGGTTA |
| RT-yusPF | TTCAGCCCTTGACGGTAC |
| RT-yusPR | TCCGCACAAAGCAGAAGC |
Underlining indicates sequences cleaved by the restriction enzymes HindIII, NdeI, and BamHI.
Northern blot analysis.
Northern blot analysis was performed as described previously (15). Cells were harvested at 2 h (exponential growth phase) and 5 h (early stationary phase) after inoculation, and total RNA was prepared as described previously (17). RNA samples (10 μg) were separated by electrophoresis and transferred to nylon membranes (Hybond-N+; GE Healthcare Bioscience), and the membranes were incubated with a digoxigenin (DIG)-labeled RNA probe. To prepare the yusO RNA probe, the DNA fragment containing the partial yusO gene was amplified by PCR using the primers NyusOF and NyusOR and cloned into pCR2.1 vector (Invitrogen). The resulting plasmid, pCR2.1-yusO, was used as a template for in vitro transcription from a T7 promoter. The RNA labeling for hybridization was performed using a DIG RNA labeling kit (SP6/T7; Roche Applied Science).
RT-qPCR.
Cells were grown in L medium until the optical density at 650 nm (OD650) reached 0.4, and antibiotics were added to the culture so that their final concentrations were 1 μg/ml for fusidic acid, 2 μg/ml for novobiocin, 10 μg/ml for streptomycin, and 0.2 μg/ml for actinomycin D. After further incubation for 30 min, the cells were harvested and total RNAs were prepared as described previously (17). Real-time quantitative PCR (RT-qPCR) was performed as described previously, using the primers shown in Table 2 (17). Amplification of the 16S rRNA gene was used as an internal control. The oligonucleotides used for PCR amplification (RT-rrn16S-F and RT-rrn16S-R for 16S rRNA and RT-yusPF and RT-yusPR for yusP) are listed in Table 2.
Primer extension analysis.
Primer extension analysis to determine the transcription start site of the yusOP operon was performed as described previously (32). Total RNA was extracted from the cells (37), and 45-μg aliquots were each annealed to 1 pmol of PyusO-R1 primer (Table 2), which had been 5′ end labeled with [γ-32P]ATP (MP Biomedicals) using a Megalabel kit (Takara Bio). The primer extension reaction and dideoxy sequencing reactions were performed using ThermoScript reverse transcriptase (Invitrogen). The synthesized cDNA and sequencing ladders were subjected to urea-polyacrylamide gel electrophoresis (PAGE) and quantified using a Typhoon 9400 variable image analyzer (GE Healthcare Bioscience).
DNase I footprinting analysis.
For footprinting analysis, the complete yusO gene was amplified by PCR using the primers yusON-F2 and yusOB-R2 (Table 2), digested with NdeI and BamHI, and cloned into the expression vector pET22b(+) (Novagen), generating pET22b(+)-yusO. E. coli BL21(DE3) harboring pET22b(+)-yusO produced YusO protein at 23% of total soluble proteins, as determined by sodium dodecyl sulfate-PAGE. Using this lysate without further purification, DNase I footprinting analysis was performed as described previously (7, 10). The yusOP probe was prepared by PCR using the primer pair PyusO-F1 and PyusO-R1. Prior to PCR amplification, only the 5′ terminus of one of the primer pairs had been labeled with [γ-32P]ATP, using a Megalabel kit (Takara Bio). This DNA probe (0.04 pmol) was mixed with the YusO-containing crude extract to form the DNA-protein complex and then partially digested with DNase I (Takara Bio) in a 50-μl reaction mixture, followed by urea-PAGE.
Purification of His10-tagged YusO protein.
For expression of His10-tagged wild-type and mutant YusO, the DNA fragment that had been synthesized by PCR using the primers yusON-F1 and yusON-R1 was cloned into the plasmid pCR2.1 and fully sequenced. Genomic DNA of strain 168, KJ01 (Arg83 → Lys), or LF05 (Ala67 → Thr) was used as a template. An NdeI-BamHI fragment containing a full-length yusO coding region was inserted into the expression vector pET19b (Novagen), generating pET19b-yusO(wild type), -yusO(R83K), and -yusO(A67T), respectively. The His10-tagged YusO protein thus designed has the sequence Met-Gly-His10-Ser-Ser-Gly-His-Ile-Asp-Asp-Asp-Asp-Lys-His at the N terminus.
To overexpress YusO, E. coli BL21(DE3) harboring the appropriate expression plasmid pET19b-yusO(wild-type), -yusO(R83K), or -yusO(A67T) was grown in L medium supplemented with 1% glucose until the OD650 reached 1.0, followed by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 2 mM. After further incubation for 3 h, the cells were harvested by centrifugation and disrupted by sonication. The cell lysate was centrifuged (8,000 × g for 10 min) to remove insoluble material, and the crude extract was fractionated using 30% to 60% saturated ammonium sulfate. The His10-tagged proteins were purified using a HisTrap HP column (GE Healthcare Bioscience) according to the manufacturer's manual. The purified protein (95% purity) was stored in storage buffer (50 mM Tris-HCl buffer [pH 8.0] and 50% glycerol).
Gel mobility shift analysis.
A DIG gel shift kit (Roche Diagnostics) was used for gel mobility shift assay (16). The His10-tagged YusO protein and its mutants (R83K and A67T) were prepared as described above. To prepare probes A and B, the DNA fragments were amplified by PCR with the primer pairs PyusO-F2 and PyusO-R2 for probe A and PyusO-F1 and PyusO-R1 for probe B. To prepare the deletion probe C, two PCR fragments, synthesized with PyusO-F1 and PyusO-del-R or PyusO-del-F and PyusO-R1, were annealed to each other, incubated with Taq DNA polymerase without primer, and used as a template for a PCR using the primers PyusO-F1 and PyusO-R1. This amplified DNA fragment was cloned into pCR2.1 and fully sequenced to confirm the 31-bp deletion. To determine the effect of the R83K and A67T substitutions on the binding ability of YusO, DIG-labeled probe A and various amounts of His10-tagged YusO (wild type, R83K, or A67T) were mixed into 15 μl of binding buffer [20 mM HEPES (pH 7.6), 30 mM KCl, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM dithiothreitol, 0.2% Tween 20, and 1 μg poly(dA-dT)] and incubated at 25°C for 15 min. The YusO binding site in the DNA was determined by incubating DIG-labeled probe B or C with various amounts of His10-tagged YusO protein (wild type). DNA and DNA-protein complexes were separated by 5% nondenaturing PAGE, transferred onto a Hybond-N+ membrane, and detected according to the manufacturer's instructions.
Assay of β-galactosidase.
Strains were grown in L medium, and an appropriate volume of cell suspension was withdrawn and centrifuged. Each pellet was resuspended in 0.5 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, and 50 mM 2-mercaptoethanol), to which 3 drops of toluene (30 μl) was added. The suspensions were vortexed for 5 s and incubated at 28°C for 1 min, followed by addition of 0.2 ml of o-nitrophenol-β-d-galactopyranoside solution (4 mg/ml in Z buffer without 2-mercaptoethanol) and further incubation at 28°C. When a sufficient amount of yellow color had developed, the reaction was stopped by adding 0.5 ml of 1 M Na2CO3 solution, followed by centrifugation for 3 min, and the absorbance of the supernatant at 420 nm (A420) was measured. Specific activity was expressed as A420·t−1·V−1·OD650−1 × 1,000, where t and V indicate the time (min) of reaction and the volume (ml) of culture used in the assay, respectively.
RESULTS
Isolation of mutants resistant to low levels of fusidic acid.
High-level resistance to fusidic acid is often due to mutations in fusA, which encodes EF-G (4, 18). To investigate the mechanism underlying low-level resistance to fusidic acid in B. subtilis, we isolated 14 resistance mutants that had developed spontaneously on L agar plates containing various concentrations (0.5 to 2 μg/ml) of fusidic acid. Of these 14 mutants, 6 (LF01 to LF06) had no mutations in the fusA gene, whereas the other 8 carried mutations in fusA (several representatives, HF01 to HF03, are listed in Table 3). The former group (LF01 to LF06) grew normally in L medium and sporulated well in sporulation medium (data not shown). Although the parent strain, 168, produces the antibiotic bacilysin (14, 15), bacilysin production was not altered in these mutant strains (data not shown). To further characterize these mutants, we determined the MICs of various antibiotics, including actinomycin, chloramphenicol, erythromycin, fusidic acid, gentamicin, kasugamycin, lincomycin, nalidixic acid, novobiocin, penicillin, rifampin, spectinomycin, streptomycin, tetracycline, and thiostrepton. Interestingly, all six mutants with low-level fusidic acid resistance exhibited cross-resistance to novobiocin, streptomycin, and actinomycin but not to the other antibiotics (Table 3), suggesting that each of these mutants has a multidrug resistance mutation.
TABLE 3.
Antibiotic susceptibilities of isolated fusidic acid-resistant mutants
| B. subtilis strain | Identified mutationa in:
|
MIC (μg/ml)b
|
||||
|---|---|---|---|---|---|---|
| fusA | yusO | FA | NB | SM | ACT | |
| 168 (wild type) | 0.4 | 0.8 | 8 | 0.04 | ||
| HF01 | 2001C → G (Phe667 → Leu) | 2 | 0.8 | 8 | 0.04 | |
| HF02 | 1352G → C (Gly451 → Ala) | 2 | 0.8 | 8 | 0.04 | |
| HF03 | 1999T → C (Phe667 → Leu) | 2 | 0.8 | 8 | 0.04 | |
| LF01 | 248G → A (Arg83 → Lys) | 1.6 | 1.6 | 12 | 0.08 | |
| LF02 | I(398ggcgc402)c (frameshift from His133, codon 152 → stop codon) | 1.6 | 1.6 | 12 | 0.08 | |
| LF03 | 232A → G (Lys78 → Glu) | 0.8 | 1.6 | 8 | 0.08 | |
| LF04 | 200C → T (Ala67 → Val) | 1.6 | 1.6 | 12 | 0.08 | |
| LF05 | 199G → A (Ala67 → Thr) | 0.8 | 1.6 | 12 | 0.08 | |
| LF06 | Δ(249aacacac255)d (frameshift from His85, codon 90 → stop codon) | 1.6 | 1.6 | 12 | 0.08 | |
Numbering is from the first nucleotide of the start codon.
MICs were determined after 18 h of incubation at 37°C on L agar plates. FA, fusidic acid; NB, novobiocin; SM, streptomycin; ACT, actinomycin D.
I, 5-base insertion at position 398.
Δ, 7-base deletion at positions 249 to 255.
Identification of mutations conferring multidrug resistance.
To identify the mutation conferring fusidic acid resistance, we performed comparative genome sequencing analysis using the genomic DNAs of the LF01 mutant and its parent strain 168. We observed a single-base substitution (248G → A) within the yusO gene (Fig. 1), which encodes a MarR family transcriptional regulator, YusO. Furthermore, the five other mutants (LF02 to LF06) that were resistant to low levels of fusidic acid were all found to carry a mutation within the yusO gene as determined by DNA sequencing (Table 3), suggesting that mutations in this gene cause resistance to low levels of fusidic acid. A causal relationship between yusO mutations and multidrug resistance was confirmed by transformation as follows. Since the hisC and trpC genes are near each other in the B. subtilis genome, the two genes are cotransformed at high frequency (approximately 70%). Using these selectable markers, a histidine auxotrophic strain, YO-005 (hisC yusO+), was transformed with the genomic DNA of LF01 (trpC yusO1). Of 100 His+ Trp− transformants, 4 also showed fusidic acid resistance. As expected, these fusidic acid-resistant transformants all carried the yusO1 mutation (data not shown) and displayed resistance to novobiocin, streptomycin, and actinomycin.
FIG. 1.
SignalMap (Roche NimbleGen) representation of comparative genome sequencing analysis of the fusidic acid-resistant mutant LF01. The lowest two traces show the signal intensities for wild-type strain 168 (blue) and mutant strain LF01 (green) hybridizations; the red trace above shows their ratio. The blue bar depicts a single-nucleotide polymorphism (SNP), which was confirmed by sequencing.
YusO functions as a repressor for the yusOP operon.
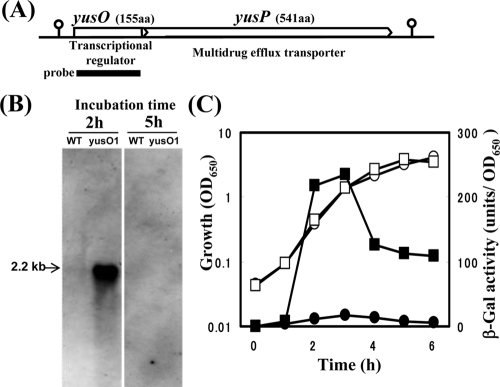
The results described above suggest that the yusO gene product, YusO, regulates the multidrug resistance gene in B. subtilis, and that the downstream gene yusP, which encodes a protein similar to a multidrug efflux transporter, forms an operon with yusO gene (Fig. 2A). To assess these hypotheses, we performed Northern blot analysis using a yusO probe. This probe hybridized with a 2.2-kb transcript, corresponding to the full length of yusOP, which was strongly induced in the exponentially growing yusO1 mutant KJ01 but not in the parent strain 168 (Fig. 2B). Similar results were obtained with the yusO disruption mutant YUSOd (data not shown). Furthermore, the 2.2-kb transcript was also detected when a specific probe for yusP was used (data not shown). RT-qPCR analysis revealed that the level of the yusP transcript in the yusO1 mutant was 70-fold higher than that in its parent strain 168 (data not shown). The yusOP transcript, however, disappeared at stationary growth phase (Fig. 2B).
FIG. 2.
Alignment of yusO and yusP and effect of yusO mutation on the expression of the yusO gene in B. subtilis. (A) Map of the genomic DNA region containing the yusO and yusP genes. The amino acid (aa) length of each product is shown in parentheses. Their deduced functions are also indicated. The thick line represents the DNA probe. The length of yusOP is about 2.2 kb. The stem-loop structure indicates the transcriptional terminator. (B) Northern analysis of the yusO transcript. Strains 168 (wild type [WT]) and KJ01 (yusO1) were grown in L medium for 2 h (exponential growth phase) or 5 h (early stationary phase). Total RNA was extracted from each strain, and 10-μg aliquots were subjected to electrophoresis, transferred to a membrane, and hybridized with the RNA probe for yusO. (C) Transcriptional fusion analysis of yusOP′-lacZ. Strains KJ02 (yusP::pMutinT3, circles) and KJ03 (yusO1 yusP::pMutinT3, squares) were grown in L medium. Culture samples were withdrawn at the indicated times, and cell densities (OD650, open symbols) and β-galactosidase (β-Gal) activities (closed symbols) were measured.
We further analyzed the function of YusO and YusP by constructing yusP disruption mutants. Insertion of the plasmid pMutinT3 into the yusP genes of strains 168 and KJ01 yielded the yusP disruption mutants KJ02 (yusP::pMutinT3) and KJ03 (yusO1 yusP::pMutinT3), which carry the transcriptional fusion yusOP′-lacZ. Consistent with the results from Northern analysis, the expression of yusOP′-lacZ in the yusO1 mutant was highly induced during exponential growth phase but then declined during stationary phase (Fig. 2C). When we tested the effect of yusP disruption on multidrug resistance, we found that disruption of the yusP gene completely abolished the multidrug resistance observed in the yusO1 mutant KJ01 (Table 4). In contrast, no substantial effect of the yusP disruption on antibiotic susceptibility was detected in the yusO+ strain. These results indicate that YusO acts as a repressor of the yusOP operon and that an elevated level of YusP renders cells resistant to fusidic acid, novobiocin, streptomycin, and actinomycin.
TABLE 4.
Effect of yusP disruption on antibiotic resistance
| B. subtilis strain | Relevant genotype | MIC (μg/ml)a
|
|||
|---|---|---|---|---|---|
| FA | NB | SM | ACT | ||
| 168 | Wild type | 0.4 | 0.8 | 8 | 0.04 |
| KJ01 | fus1 (yusO1) | 1.6 | 1.6 | 12 | 0.08 |
| KJ02 | yusP::pMutinT3 | 0.4 | 0.8 | 8 | 0.04 |
| KJ03 | yusO1 yusP::pMutinT3 | 0.4 | 0.8 | 8 | 0.04 |
See Table 3, footnote b.
Identification of cis-acting elements for transcriptional regulation of the yusOP operon.
Since Northern blot experiments had shown that the yusOP operon is transcribed from a single promoter located upstream of the yusO gene (Fig. 2), we determined the transcription start site of the yusOP operon by primer extension analysis. We found that transcription of yusOP was initiated at the G residue 60 bases upstream of the translational start codon, which was detected only in the sample of strain YUSOd and not in that of the wild-type strain 168 (Fig. 3A). The most probable −35 and −10 regions (TTGACT and TATATT with a 17-bp spacer), which are likely recognized by σA-RNA polymerase (9), were found in the region upstream of the transcriptional start site.
FIG. 3.
Identification of the YusO binding region. (A) Determination of the transcription start site of the yusOP operon. Total RNAs (45 μg) of strains 168 (wild type, lane 1) and YUSOd (yusO::pMUTIN2, lane 2) were reverse transcribed to generate runoff cDNA (bold arrow). Lanes G, A, T, and C contain the products of the dideoxy sequencing reactions with the same primer used for reverse transcription. The partial nucleotide sequence of the coding strand corresponding to the ladders is shown, where the −10 regions are underlined and the transcription start sites (+1) are boxed. (B) DNase I footprinting of YusO in the yusOP promoter region. DNA probes corresponding to the coding or noncoding strand of the yusOP promoter region were 5′ end labeled, and each was incubated at a final concentration of 0.8 nM with a crude extract from E. coli BL21(DE3) expressing yusO in the absence (lanes 1 and 5) or presence (lane 2, 2.6 μM; lane 3, 1.3 μM; lane 4, 0.65 μM as a dimer) of crude YusO protein. After partial digestion with DNase I, the resulting mixtures were subjected to urea-PAGE. Lanes G, A, T, and C contain the products of the dideoxy sequencing reactions with the corresponding 5′-labeled primer. Nucleotide sequences protected by YusO are indicated on the right of each panel. (C) Organization of the yusOP promoter region. The stop codon of the yusN gene and the −35 and −10 regions of the yusOP promoter are underlined. The transcription start site (+1) of the yusOP operon is shown by capital boldface letter. The Shine-Dalgarno (SD) sequence of yusO is boxed, and the three inverted repeat sequences, IR1, IR2, and IR3, are indicated by pairs of facing arrows with bold letters showing the matching bases. The open reading frames of yusN and yusO genes are depicted by thick lines. The protected regions in the coding and noncoding strands are indicated by gray bars.
To determine whether YusO protein binds directly to the yusOP promoter region, we performed DNase I footprinting analysis using a crude extract prepared from E. coli cells that overexpress the recombinant yusO gene. The YusO protein was found to protect the region, including the predicted −10 region of the yusOP promoter (bases −29 to +34 of the coding strand and bases −30 to +35 of the noncoding strand) (Fig. 3B). Binding of E. coli proteins to the yusOP promoter region was not detected by gel mobility shift assay, as examined with lysate prepared from E. coli cells not expressing YusO (data not shown). Importantly, this region contained two inverted repeats, IR1 (bases −24 to +7) and IR2 (bases +5 to +30), and part of another, IR3 (bases +27 to +43) (Fig. 3C).
Determination of binding affinity of YusO to the yusOP promoter region.
To determine the binding affinity, we purified His10-tagged YusO protein and two mutant proteins (R83K and A67T). The His10-tagged YusO protein, which was approximately 95% pure, formed a dimer in Tris-HCl buffer, as examined by gel filtration analysis (data not shown). Gel mobility shift analysis indicated that wild-type YusO protein bound to probe A (348 bp), which contained a yusOP promoter, with a binding dissociation constant of 81 nM (calculated as a dimer) (Fig. 4B). A similar result was obtained when nontagged YusO was used (data not shown). This result indicates that the His10 tag has no effect on the binding activity of YusO. Strikingly, both variants (R83K and A67T) had much lower binding affinity (Fig. 4B and data not shown), indicating that residues Arg83 and Ala67 play a critical role in the binding of YusO to the yusOP promoter.
FIG. 4.
Binding affinity of YusO to the yusOP promoter region. (A) Probes used for gel mobility shift analyses. The −35 and −10 regions of the yusOP operon are indicated as dashed lines. The transcription start site (+1) is indicated by a bent arrow, and the inverted repeat sequence IR1 is indicated by a pair of facing arrows. The three DNA probes (A, B, and C) used for gel mobility shift analysis are shown. The dashed line indicates the deleted region. (B) Effect of the R83K substitution in YusO on its binding affinity to the yusOP promoter. A DIG-labeled DNA fragment (probe A) was mixed with His10-tagged YusO (wild type) or its R83K variant. Purified YusO protein was added at the indicated concentrations. (C) Effect of IR1 deletion on the binding affinity of YusO. A DIG-labeled DNA fragment (probe B or C) was incubated with His10-tagged YusO at the indicated concentrations.
To identify the cis-acting element required for the binding of YusO protein, we prepared shortened DNA probes B (150 bp) and C (119 bp). Probe B contains the entire region protected from DNase I by YusO protein, whereas probe C lacks the IR1 sequence entirely (Fig. 4A). Wild-type YusO protein bound to probe B with an affinity similar to that for probe A (Fig. 4C). YusO, however, failed to bind to the IR1 deletion probe C, indicating that the IR1 sequence is required for binding.
Fusidic acid induces yusP expression in vivo.
As described above, loss of YusO function resulted in increased expression of the multidrug transporter YusP, leading to multidrug resistance. To analyze whether these antibiotics can inhibit the binding of YusO to the YusO binding site, we performed gel mobility shift assays in the presence or absence of antibiotics. We found that the binding of YusO was inhibited in the presence of fusidic acid or novobiocin (Fig. 5), with 50% inhibitory concentrations (Ki) of 1.8 to 3.7 mM and 5 to 10 mM, respectively. No significant dissociation was detected when up to 10 mM streptomycin or up to 0.1 mM actinomycin was added to the reaction mixture. To evaluate whether these antibiotics can induce yusP expression in vivo, antibiotics were added to cultures of the wild-type strain 168 at concentrations that fully inhibit growth (1 μg/ml [2 μM] for fusidic acid, 2 μg/ml [3.1 μM] for novobiocin, 10 μg/ml [17 μM] for streptomycin, and 0.2 μg/ml [0.16 μM] for actinomycin). Total RNA was extracted from cells just before addition of the drug or after 30 min of incubation and subjected to RT-qPCR analysis. In control cells (without antibiotic), the level of yusP transcript was decreased to 25% during the 30-min incubation. This is not surprising, because the yusOP expression was decreased abruptly upon entry of the cells into stationary phase (Fig. 2B and C). As expected, yusP expression was induced by adding 1 μg/ml fusidic acid (Fig. 6), with lower concentrations being less effective (data not shown). Importantly, the concentration of fusidic acid required for yusOP induction (1 μg/ml = 2 μM) was much lower than the Ki value (1.8 to 3.7 mM) found in gel mobility shift assay. In contrast, no significant induction of yusP was detected in cells treated with novobiocin, streptomycin, or actinomycin, indicating that these antibiotics do not induce yusOP expression.
FIG. 5.
Effect of antibiotics on the binding of YusO to the yusOP promoter region. DIG-labeled yusOP probe A (2.3 nM) was incubated with YusO (146 nM as a dimer) in the presence of fusidic acid (A) or novobiocin (B). Each antibiotic was diluted stepwise by twofold and added to the mixture.
FIG. 6.

Inducibility of yusOP transcription by various antibiotics. Strain 168 was grown in L medium until the OD650 reached 0.4, and antibiotics were added to final concentrations of 1 μg/ml for fusidic acid, 2 μg/ml for novobiocin, 10 μg/ml for streptomycin, and 0.2 μg/ml for actinomycin D. “None” represents no addition of antibiotics. Total RNAs were extracted from cells just before and after 30 min of treatment and used for RT-qPCR analysis. The transcription level was normalized relative to the amount of 16S rRNA in each RNA sample. The relative expression of yusP was calculated by dividing the relative amount of yusP in cells after treatment by that in cells before treatment. The average values (with standard deviations) from three independent experiments are shown. FA, fusidic acid; NB, novobiocin; SM, streptomycin; ACT, actinomycin D.
DISCUSSION
We have described here the identification and characterization of a novel multidrug resistance operon, yusOP, of B. subtilis. We demonstrated that the YusO protein binds directly to the yusOP promoter region, leading to the repression of its expression. In addition, YusP was found to contribute to resistance to several antibiotics, including fusidic acid, novobiocin, streptomycin, and actinomycin, possibly by pumping these structurally unrelated antibiotics out of cells. Consequently, the loss of YusO function caused increased expression of yusOP, resulting in the multidrug resistance phenotype by extruding antibiotics through YusP. Thus, the YusO protein acts as a repressor of the yusOP operon, and the YusP protein functions as a multidrug efflux transporter. We therefore propose renaming yusO as a multidrug transporter regulator, mdtR, and yusP as a multidrug transporter protein, mdtP.
The MdtR protein belongs to the MarR family of transcriptional regulatory proteins, each of which contains a “winged-helix” DNA binding motif in its central domain. The crystal structure of MdtR has been deposited in the Protein Data Bank (ID 1s3j) by other investigators. By isolating mutants with low-level resistance to fusidic acid, we obtained several MdtR mutants that exhibit a multidrug resistance phenotype. Our results indicated that the mutations, A67T and R83K, markedly reduced the binding affinity of MdtR to the mdtRP promoter region, leading to derepression of mdtRP transcription. Similar to the case for wild-type MdtR, both MdtR variants (A67T and R83K) formed a dimer in solution, indicating that these mutations had no effect on protein dimerization. Based on the structures of B. subtilis MdtR and E. coli MarR, residues Ala67 and Arg83 in the MdtR protein correspond to E. coli MarR Ala70 (in the α4 region) and Arg86 (in the β2 region), respectively. In E. coli, these regions are known to contribute to the DNA binding activity of MarR (3). Consistent with our observation, an amino acid change of Ala70 to Thr in E. coli MarR has also been reported to abolish its ability to bind to DNA (2). Although there is no experimental evidence that residue Arg86 of E. coli MarR plays a critical role in its DNA binding activity, this residue is highly conserved in other MarR family repressors. Therefore, MdtR residues Ala67 and Arg83 likely play essential roles in the DNA binding activity of the MdtR protein. Similarly, an amino acid change of Lys78 to Glu also led to a multidrug resistance phenotype, indicating that this residue also participates, directly or indirectly, in MdtR activity. As predicted from DNA sequencing, the LF02 mutant contains a frameshift mutation followed by a stop codon, thus lacking the C-terminal helix (α6). This C-terminal domain may be involved in dimer formation.
We successfully identified the cis-acting elements for MdtR. We found that the MdtR protein recognized and bound specifically to DNA containing a 31-bp imperfect inverted repeat sequence, IR1 (AAaTgCGAATAAaTataAaTTATTCGtAaTT), which overlaps with the −10 region of the mdtRP promoter. Gel mobility shift assays showed that the binding of MdtR to the mdtRP promoter region was severely inhibited by adding excess concentrations of fusidic acid or novobiocin but not by actinomycin or streptomycin. Since the MICs of fusidic acid and novobiocin were 0.4 μg/ml (0.77 μM) and 0.8 μg/ml (1.3 μM), respectively, the Ki values observed in gel mobility shift assays are too high to permit these antibiotics to release MdtR from the mdtRP promoter at physiological concentrations. In fact, novobiocin failed to induce mdtRP expression at a concentration close to its MIC. Although fusidic acid did induce mdtRP expression at low concentrations, the level of mdtRP expression was much lower than that in the mdtR disruptant. Therefore, it is likely that another compound, which could be a fusidic acid analogue, plays a role in inducing multidrug resistance, perhaps by interacting efficiently with MdtR. In this regard, the mdtP protein is similar to EmrB/QacA family transporters. The E. coli EmrB confers resistance to hydrophobic uncouplers such as carbonyl cyanide m-chlorophenylhydrazone and tetrachlorosalicylanilide, to organomercurials, and to some hydrophobic antibiotics such as thiolactomycin (8, 23). In addition, Staphylococcus aureus QacA confers resistance to organic cations such as ethidium, benzalkonium, cetrimide, chlorhexidine, and pentamidine (22). These compounds, together with the fusidic acid analogue, could be inducers for the mdtRP system.
Interestingly, we found that mdtRP expression was decreased during stationary phase, even in mdtR mutants, indicating that an additional regulatory mechanism may act in stationary-phase repression of mdtRP. Thus, further analyses are required to understand the overall mechanism regulating mdtRP transcription.
Acknowledgments
This work was supported by grants from the Effective Promotion of Joint Research of Special Coordination Funds (to K.O.) and a Grant-in Aid for Scientific Research on Priority Areas (to Y.F.) from the Ministry of Education, Culture, Sports, and Technology of the Japanese Government.
We thank Roche NimbleGen, Inc., Madison, WI, for supporting the mutation search using the comparative genome sequencing technique.
Footnotes
Published ahead of print on 13 March 2009.
REFERENCES
- 1.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2951-953. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., Y. S. Kim, and S. B. Levy. 2000. Mutational analysis of MarR, the negative regulator of marRAB expression in Escherichia coli, suggests the presence of two regions required for DNA binding. Mol. Microbiol. 351394-1404. [DOI] [PubMed] [Google Scholar]
- 3.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8710-714. [DOI] [PubMed] [Google Scholar]
- 4.Besier, S., A. Ludwig, V. Brade, and T. A. Wichelhaus. 2003. Molecular analysis of fusidic acid resistance in Staphylococcus aureus. Mol. Microbiol. 47463-469. [DOI] [PubMed] [Google Scholar]
- 5.Bodley, J. W., F. J. Zieve, L. Lin, and S. T. Zieve. 1969. Formation of the ribosome-G factor-GDP complex in the presence of fusidic acid. Biochem. Biophys. Res. Commun. 37437-443. [DOI] [PubMed] [Google Scholar]
- 6.Dzidic, S., J. Suskovic, and B. Kos. 2008. Antibiotic resistance mechanisms in bacteria: biochemical and genetic aspects. Food Technol. Biotechnol. 4611-21. [Google Scholar]
- 7.Fujita, Y., and Y. Miwa. 1989. Identification of an operator sequence for the Bacillus subtilis gnt operon. J. Biol. Chem. 2644201-4206. [PubMed] [Google Scholar]
- 8.Furukawa, H., J. T. Tsay, S. Jackowski, Y. Takamura, and C. O. Rock. 1993. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J. Bacteriol. 1753723-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 591-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirooka, K., S. Kunikane, H. Matsuoka, K. Yoshida, K. Kumamoto, S. Tojo, and Y. Fujita. 2007. Dual regulation of the Bacillus subtilis regulon comprising the lmrAB and yxaGH operons and yxaF gene by two transcriptional repressors, LarA and YxaF, in response to flavonoids. J. Bacteriol. 1895170-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosaka, T., N. Tamehiro, N. Chumpolkulwong, C. Hori-Takemoto, M. Shirouzu, S. Yokoyama, and K. Ochi. 2004. The novel mutation K87E in ribosomal protein S12 enhances protein synthesis activity during the late growth phase in Escherichia coli. Mol. Genet. Genomics 271317-324. [DOI] [PubMed] [Google Scholar]
- 12.Hosaka, T., J. Xu, and K. Ochi. 2006. Increased expression of ribosome recycling factor is responsible for the enhanced protein synthesis during the late growth phase in an antibiotic-overproducing Streptomyces coelicolor ribosomal rpsL mutant. Mol. Microbiol. 61883-897. [DOI] [PubMed] [Google Scholar]
- 13.Inaoka, T., K. Kasai, and K. Ochi. 2001. Construction of an in vivo nonsense readthrough assay system and functional analysis of ribosomal proteins S12, S4, and S5 in Bacillus subtilis. J. Bacteriol. 1834958-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaoka, T., and K. Ochi. 2007. Regulation of secondary metabolism in Bacillus subtilis, p. 143-154. In Y. Fujita (ed.), Global regulatory networks in Bacillus subtilis. Transworld Research Network, Kerala, India.
- 15.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 2782169-2176. [DOI] [PubMed] [Google Scholar]
- 16.Inaoka, T., K. Takahashi, H. Yada, M. Yoshida, and K. Ochi. 2004. RNA polymerase mutation activates the production of a dormant antibiotic 3,3′-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 2793885-3892. [DOI] [PubMed] [Google Scholar]
- 17.Inaoka, T., T. Satomura, Y. Fujita, and K. Ochi. 2009. Novel gene regulation mediated by overproduction of secondary metabolite neotrehalosadiamine in Bacillus subtilis. FEMS Microbiol. Lett. 291151-156. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, H., K. Kobayashi, and Y. Kobayashi. 1977. Isolation and characterization of fusidic acid-resistant, sporulation-defective mutants of Bacillus subtilis. J. Bacteriol. 132262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 1004678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurosawa, K., T. Hosaka, N. Tamehiro, T. Inaoka, and K. Ochi. 2006. Improvement of α-amylase production by modulation of ribosomal component protein S12 in Bacillus subtilis 168. Appl. Environ. Microbiol. 7271-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurberg, M., O. Kristensen, K. Martemyanov, A. T. Gudkov, I. Nagaev, D. Hughes, and A. Liljas. 2000. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J. Mol. Biol. 303593-603. [DOI] [PubMed] [Google Scholar]
- 22.Littlejohn, T. G., I. T. Paulsen, M. T. Gillespie, J. M. Tennent, M. Midgley, I. G. Jones, A. S. Purewal, and R. A. Skurray. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 95259-266. [DOI] [PubMed] [Google Scholar]
- 23.Lomovskaya, O., and K. Lewis. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 898938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriya, S., E. Tsujikawa, A. K. Hassan, K. Asai, T. Kodama, and N. Ogasawara. 1998. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29179-187. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura, K., T. Hosaka, S. Tokuyama, S. Okamoto, and K. Ochi. 2007. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J. Bacteriol. 1893876-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimura, K., S. K. Johansen, T. Inaoka, T. Hosaka, S. Tokuyama, Y. Tahara, S. Okamoto, F. Kawamura, S. Douthwaite, and K. Ochi. 2007. Identification of the RsmG methyltransferase target as 16S rRNA nucleotide G527 and characterization of Bacillus subtilis rsmG mutants. J. Bacteriol. 1896068-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochi, K. 2007. From microbial differentiation to ribosome engineering. Biosci. Biotechnol. Biochem. 711373-1386. [DOI] [PubMed] [Google Scholar]
- 28.Ochi, K., S. Okamoto, Y. Tozawa, T. Inaoka, T. Hosaka, J. Xu, and K. Kurosawa. 2004. Ribosome engineering and secondary metabolite production. Adv. Appl. Microbiol. 56155-184. [DOI] [PubMed] [Google Scholar]
- 29.Ochi, K., J. Y. Kim, Y. Tanaka, G. Wang, K. Masuda, H. Nanamiya, S. Okamoto, S. Tokuyama, Y. Adachi, and F. Kawamura. 2009. Inactivation of KsgA, a 16S rRNA methyltransferase, causes vigorous emergence of mutants with high-level kasugamycin resistance. Antimicrob. Agents Chemother. 53193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto, S., A. Tamaru, C. Nakajima, K. Nishimura, Y. Tanaka, S. Tokuyama, Y. Suzuki, and K. Ochi. 2007. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 631096-1106. [DOI] [PubMed] [Google Scholar]
- 31.Okamoto-Hosoya, Y., T. Hosaka, and K. Ochi. 2003. An aberrant protein synthesis activity is linked with antibiotic overproduction in rpsL mutants of Streptomyces coelicolor A3(2). Microbiology 1493299-3309. [DOI] [PubMed] [Google Scholar]
- 32.Satomura, T., D. Shimura, K. Asai, Y. Sadaie, K. Hirooka, and Y. Fujita. 2005. Enhancement of glutamine utilization in Bacillus subtilis through the GlnK-GlnL two-component regulatory system. J. Bacteriol. 1874813-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamehiro, N., T. Hosaka, J. Xu, H. Hu, N. Otake, and K. Ochi. 2003. Innovative approach for improvement of an antibiotic-overproducing industrial strain of Streptomyces albus. Appl. Environ. Microbiol. 696412-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 1443097-3104. [DOI] [PubMed] [Google Scholar]
- 35.Walsh, C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406775-781. [DOI] [PubMed] [Google Scholar]
- 36.Wang, G., T. Hosaka, and K. Ochi. 2008. Dramatic activation of antibiotic production in Streptomyces coelicolor by cumulative drug resistance mutations. Appl. Environ. Microbiol. 742834-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]