Abstract
Polymyxin, a long-known peptide antibiotic, has recently been reintroduced in clinical practice because it is sometimes the only available antibiotic for the treatment of multidrug-resistant gram-negative pathogenic bacteria. Lack of information on the biosynthetic genes of polymyxin, however, has limited the study of structure-function relationships and the development of improved polymyxins. During whole genome sequencing of Paenibacillus polymyxa E681, a plant growth-promoting rhizobacterium, we identified a gene cluster encoding polymyxin synthetase. Here, we report the complete sequence of the gene cluster and its function in polymyxin biosynthesis. The gene cluster spanning the 40.6-kb region consists of five open reading frames, designated pmxA, pmxB, pmxC, pmxD, and pmxE. The pmxC and pmxD genes are similar to genes that encode transport proteins, while pmxA, pmxB, and pmxE encode polymyxin synthetases. The insertional disruption of pmxE led to a loss of the ability to produce polymyxin. Introduction of the pmx gene cluster into the amyE locus of the Bacillus subtilis chromosome resulted in the production of polymyxin in the presence of extracellularly added l-2,4-diaminobutyric acid. Taken together, our findings demonstrate that the pmx gene cluster is responsible for polymyxin biosynthesis.
Since polymyxin was first isolated from Bacillus polymyxa in 1947 (1, 4, 47), at least 15 unique polymyxins have been reported (31, 49). Because of its excellent bactericidal activity against gram-negative bacteria, polymyxin antibiotics (polymyxin B and polymyxin E) were used until early 1970 as therapies against many diseases caused by pathogenic microorganisms. However, because they carried serious side effects, including fever, skin eruption, and pain, and also induced severe nephrotoxicity and neurotoxicity (18, 37), it was rapidly replaced by other, better-tolerated antibiotics. In recent years, its application has been restricted to use as an ointment on local surface wounds.
Due to the increased and often unnecessary use of antibiotics, pathogenic microorganisms with resistance to antibiotics have become more widespread (2, 14, 30, 38). Under the limited therapeutic options available to treat multidrug-resistant gram-negative bacteria such as Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae, polymyxins are sometimes the only available active antibiotics and have now become important therapeutic agents (13, 25, 28, 29, 55). Many recent reports have shown that patients infected with multidrug-resistant gram-negative pathogens improved upon treatment with polymyxins (19, 27, 44, 48). In addition, polymyxins have been applied to prevent septic shock by removing circulating endotoxin to polystyrene fibers in an immobilized form (8). Therefore, the clinical value of polymyxin, an antibiotic discovered 6 decades ago, is currently being reappraised. However, until now, we have had a very limited understanding of various characteristics of this agent, especially its biosynthetic genes.
To analyze structure-function relationships and to develop improved polymyxins with lowered toxicities, novel polymyxin derivatives must be generated. Recently, total or semisynthesis or modifications of polymyxins was performed chemically or enzymatically, and the resulting products were effectively used for structure-function study (6, 20, 36, 45, 50, 52). There is a limitation to obtaining diverse derivatives by using chemical or enzymatic approaches, however, and this limitation is related to the structural complexity of polymyxin. The basic structure of polymyxin is a cyclic heptapeptide with a tripeptide side chain acylated by a fatty acid at the amino terminus (49). Normally, 6-methyloctanoic acid or 6-methylheptanoic acid is attached to the side chain. This structure favors solubility of polymyxin in both water and organic solvent. Unlike other general ribosomally translated peptides, polymyxin is produced by a nonribosomal peptide synthetase (NRPS) (22, 31). NRPSs are multienzyme complexes that have modular structures (35, 46). A module is a distinct section of the multienzyme that is responsible for the incorporation of one or more specific amino acids into the final product. Each module can be divided into different domains, each of which is responsible for a specific biochemical reaction. Three types of domains, the adenylation (A), thiolation (T; also referred to as the peptidyl carrier protein, PCP), and condensation (C) domains, are essential for nonribosomal peptide synthesis. The A-domain plays a role in the selection and activation of an amino acid monomer, the T-domain is responsible for transportation of substrates and elongation intermediates to the catalytic centers, and the C-domain catalyzes peptide bond formation. In addition to these core domains, there are the thioesterase domain (TE-domain), the epimerization domain (E-domain), and some other modification domains. Many NRPS gene clusters have been reported, but no polymyxin biosynthetic gene cluster has been reported to date.
During whole genome sequencing of Paenibacillus polymyxa E681, a plant growth-promoting rhizobacterium, we found a gene cluster encoding polymyxin synthetase. In this study, the complete sequences of the polymyxin synthetase genes and the function of the gene cluster have been identified and analyzed by domain analysis, insertional mutagenesis, and heterologous expression of the genes, as well as by antibacterial assay and liquid chromatography-mass spectrometry (LC/MS) analysis of the strains and their culture supernatants. The genome information and the heterologous expression of the polymyxin synthetase gene cluster will be useful for further studies of the regulation of pmx genes, their structure-function relationships, and the improvement of polymyxins.
MATERIALS AND METHODS
Strains and culture conditions.
P. polymyxa E681 was isolated from the roots of winter barley in the Republic of Korea (41). Escherichia coli DH5α and BW25113 carrying the Red recombinase of pKD46 (9) were used for cloning and λ Red recombination, respectively. Bacillus subtilis 168 was used as a host for heterologous expression of the pmx genes. P. polymyxa E681 was grown in Tryptic soy broth (Difco) for general purposes, brain heart infusion (Difco) containing 10% sucrose for transformation, and glucose-starch-CaCO3 (GSC) medium (10) for analysis of polymyxin. B. subtilis strains were grown in LB medium for general purposes, and in GSC medium with or without 200 μg/ml of l-2,4-diaminobutyric acid (l-Dab; Sigma-Aldrich) for analysis of polymyxin.
LC/MS analysis.
P. polymyxa E681 was grown in GSC medium under aerobic conditions at 30°C for 3 days and then centrifuged at 5,000 × g for 10 min to obtain the supernatant. LC/MS was performed with the supernatant using a high-pressure liquid chromatography system provided by Thermo Electron Co. and an ion spectrometer. The sample was injected into a reverse-phase column, YMC Pack Pro C18 (10 by 250 mm, 5 μm) or Terra MS C18 (2.1 by 50 mm, 3.5 μm), and was analyzed in a mixed solvent of acetonitrile and water containing 0.1% formic acid (0.2 ml/min). Analysis of metabolites from recombinant B. subtilis was conducted after solid-phase extraction using the general protocol. Bacillus cells were grown in 200 ml GSC medium with or without l-Dab (200 μg/ml) for 2 days. After the culture supernatant was extracted using the same volume of butanol, the butanol phase was evaporated and reextracted with methanol. The final methanol extract was evaporated and dissolved in 2 ml water. After the concentrated sample was passed through a C18 column (SiliCycle Inc., Quebec, Canada), it was eluted using 3 ml of water-methanol gradient (10, 20, 40, 60, 80, and 100%). A 50-μl aliquot of each fraction was used to assay antimicrobial activity against E. coli, and the active fraction was subsequently used for LC/MS analysis.
PCR-targeted mutagenesis.
The PCR primers used in this study are listed in Table 1. A deletion mutant of the pmxE gene was constructed using an E. coli fosmid clone. In brief, the fosmid DNA (PP12G04) harboring truncated pmxA and complete pmxB, pmxC, pmxD, and pmxE in a 38.1-kbp chromosomal DNA fragment cloned into pCC1fos (Epicentre Biotechnologies) was introduced into E. coli BW25113 carrying the Red recombinase expression plasmid, pKD46 (9). The chloramphenicol acetyltransferase (cat) gene of fosmid PP12G04 was replaced with a tetracycline resistance gene (Tc) using a λ Red recombination system to construct fosmid pPmx-Tc. The Tc gene was amplified from pBC16 (5) with the Foscm-TCF and Foscm-TCR primers bearing 70-bp side arms that bind to the flanking regions of the cat gene of pCC1fos. For inactivation of the pmxE gene, a chloramphenicol resistance gene-kanamycin resistance gene (cat-kan) cassette was introduced into the pmxE structural gene of pPmx-Tc using a λ Red recombination system. The cat-kan cassette was constructed as follows. The cat gene was amplified by PCR with primers CatF and CatR from pDG1661 (15) and was then introduced into pGem7zf(+) (Invitrogen Inc.) with EcoRI and BamHI cleavage sites. The resulting plasmid was digested with the NarI restriction enzyme and was then ligated with the PCR product containing the kanamycin resistance gene that was amplified from pKD4 (9) by using the Kd4kanF and Kd4kanR primer set. The constructed cat-kan cassette was amplified with primers PmxEckF and PmxEckR, yielding 60-bp homologous arms of the target site to each of the ends. The amplified cat-kan cassette was inserted into pPmx-Tc to construct the pDpmxE fosmid. To remove the pKD46 plasmid completely, kanamycin-resistant transformants were transferred onto fresh agar medium containing kanamycin and were subsequently incubated at 37°C. The disruption of pmxE with the cat-kan cassette was confirmed by PCR with primers pmxEdelF and pmxEdelR, which bind to the outer regions of the homologous arm. The pDpmxE fosmid was introduced into P. polymyxa E681 to generate a polymyxin-defective mutant. The mutant was also confirmed by PCR using the pmxEdelF and pmxEdelR primers. Transformation of P. polymyxa was performed according to a previously reported method (7).
TABLE 1.
Primers used in this study
| Primer | Oligonucleotide sequencea |
|---|---|
| Foscm-TCF | 5′-TATCGAGATTTTCAGGAGCTAAGGAAGCTAAAATGGAGAAAAAAATCACTGGATATACCACCGTTGATAGATACAAGAGAGGTCTCTCG-3′ |
| Foscm-TCR | 5′-GGCACCAATAACTGCCTTAAAAAAATTACGCCCCGCCCTGCCACTCATCGCAGTACTGTTGTAATTCATAACAAACGGGCCATATTGTTG-3′ |
| CatF | 5′-AAAGGATCCTCATGTTTGACAGCTTATCATCG-3′ |
| CatR | 5′-AAAGAATTCCCACGCCGAAACAAGCGCTC-3′ |
| Kd4kanF | 5′-CCATCGATGTGTAGGCTGGAGCTGCTTC-3′ |
| Kd4kanR | 5′-CCATCGATATGGGAATTAGCCATGGTCC-3′ |
| PmxEckF | 5′-GCATTCAATAACAAAGATTATGCCGTTTGGCAGCATTCCCGAAGCTTACGGGCAGATGCTCCAGCCGCAGAATCATGTTTGACAGCTTATCATCG-3′ |
| PmxEckR | 5′-GCAGCACGTCCATGGAAAGGCCGTCGGAACCAATATGATGAATATCCAGCGCGAGCAGAAATTTCTCTTTCCACGCCGAAACAAGCGCTC-3′ |
| PmxEdelF | 5′-GTCTCGGATGGCATTTCGACAG-3′ |
| PmxEdelR | 5′-AGAAGTCGAGAGGCAGCTCAAG-3′ |
| ydiO-up-F | 5′-TAATGAGTTAGATGAAATACC-3′ |
| ydiO-up-R | 5′-TTTGGATCCTTATCATTCCTAGTATTACAC-3′ |
| ydjA-down-F | 5′-TTTGGATCCTGTTATTAGTCGGAATGAATG-3′ |
| ydjA-down-R | 5′-TATCTGCAGGAATAAACAGAAAGGAAAGACTG-3′ |
| 1730-12DF | 5′-ACTGCATGTCCCCAGTGCATCGGTCCCCATACGGATTTATACGGGTAATGTTGATAGAACAAGTGATATTGGTATGTTTCTCTTTGATGTC-3′ |
| 1730-12DR | 5′-AGATTATCGGCTGAACTACCATTTAATGGCTGAATTGGGCTGGATGAATGATCCGAACGGCTTCATTCAAGAATGGCGATTTTCGTTCGTG-3′ |
| pmxAF | TAACGTTTTCACCCCATTGG |
| pmxAR | GGGAGCTTGGAGCTTTGCTG |
| pmxBF | TCCACAACTCGAGCTAAGCC |
| pmxBR | ACTTACCGCTCCAGTACTGTTC |
| pmxCF | GAACAAGTCAAGCGGCAGATC |
| pmxCR | CTTTCACTTGCGAGAGCCATC |
| pmxDF | CAGGAATTTACCGAGTCTGCC |
| pmxDR | GTCGCATTCGCAAGCAGGAAG |
| pmxEF | GAGCGGCTGAAACGTCAGGAAGCC |
| pmxER | CTGCTTCGCCTGTATGATTGTC |
The underlined sequences indicate the targeted regions for Red recombinase, and the italicized residues indicate the synthetic restriction sites.
Heterologous expression of the pmx gene cluster in B. subtilis.
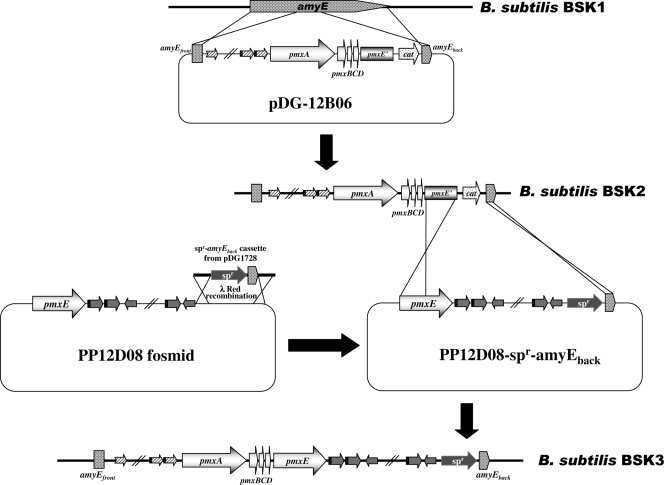
For the efficient transformation of the Bacillus host strain with large DNA fragments, the genes responsible for BsuM restriction and modification (RM) (16) were removed from B. subtilis 168 as follows. DNA fragments upstream of ydiO and downstream of ydjA were amplified by PCR with primers ydiO-up-F and ydiO-up-R and primers ydjA-down-F and ydjA-down-R, respectively. The DNA fragments were inserted into the EcoRI and PstI sites of plasmid pBGSC6 (12) in tandem to construct plasmid pDBSUM. Transformation of B. subtilis was conducted using a previously reported method (17). After single-crossover integration of the pDBSUM plasmid into the chromosome of B. subtilis 168, cells were grown in LB medium without antibiotics and then screened for chloramphenicol-sensitive colonies. BSK1, a resultant recombinant strain with a disrupted RM system, was constructed without any marker gene. Integration of the pmx gene cluster into the chromosome of BSK1 was conducted in two steps, using fosmid clones, as shown in Fig. 3. Fosmid PP12B06 containing pmxABCD, a truncated pmxE, and a 5′-flanking region was digested with BamHI, and the DNA fragment containing pmx genes was ligated into the BamHI site of integration plasmid pDG1662 (15) to construct pDG-12B06. The pmx genes of pDG-12B06 were introduced into the amyE locus of strain BSK1 by homologous recombination to construct strain BSK2. To restore truncated pmxE, the PP12D08 fosmid containing an entire pmxE gene and a 3′-flanking region was used. A recombinant fosmid, PP12D08-Spr-amyEback, was constructed by integration of the Spr-amyEback cassette amplified from plasmid pDG1730 (15) by PCR with primers 1730-12DF and 1730-12DR using a λ Red recombination system. Strain BSK3 containing the entire pmxABCDE sequence was constructed by homologous recombination between PP12D08-Spr-amyEback and the chromosome of BSK2. Functional sfp was introduced into BSK1 and BSK3 by transferring the chromosomal DNA of B. subtilis CB114 (26) to construct BSK1S and BSK3S, respectively. Construction of BSK2, BSK3, and BSK3S was confirmed by PCRs with the primer sets of pmxAF/pmxAR, pmxBF/pmxBR, pmxCF/pmxCR, pmxDF/pmxDR, and pmxEF/pmxER. Introduction of the functional sfp was confirmed by observing reduced surface tension of the culture broth as described in a previous study (26).
FIG. 3.
Scheme for the transfer of pmx genes into B. subtilis. The pmx gene cluster was integrated into the amyE locus of B. subtilis BSK1 containing a deleted BsuM RM system from B. subtilis 168. The detailed protocol is described in Materials and Methods.
Antibacterial activity assay.
The antibacterial activity was analyzed using freshly prepared E. coli plates. E. coli cells grown overnight in 3 ml of LB medium at 37°C were mixed with 300 ml of LB agar, autoclaved, and cooled below 50°C to prepare the plates. When necessary, l-Dab was added at a final concentration of 200 μg/ml. To analyze the antibacterial activity of culture supernatants of P. polymyxa strains and their extracts, 50 μl of each sample was loaded onto a paper disk and transferred to the E. coli plates. Recombinant B. subtilis cells grown overnight in 3 ml of LB medium at 37°C were inoculated directly onto the E. coli plates by dropping 5 μl of the culture onto plates. Each plate was then incubated at 37°C for 24 h to observe the growth inhibition effect.
Nucleotide sequence accession number.
The GenBank accession number for the polymyxin synthetase gene cluster is EU371992.
RESULTS
Domain analysis of the polymyxin synthetase.
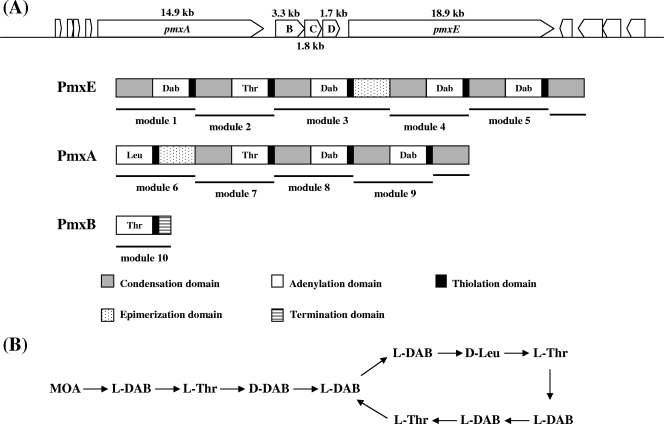
During the whole genome sequencing of P. polymyxa E681 that was recently completed in our laboratory (J. F. Kim et al., unpublished results), an NRPS gene cluster was identified as a potential polymyxin synthetase gene cluster. The gene cluster consisted of five open reading frames, pmxA, pmxB, pmxC, pmxD, and pmxE (Fig. 1A). The results of a BLAST search suggested that pmxC and pmxD may encode membrane transporters, while pmxA, pmxB, and pmxE encode polymyxin synthetase.
FIG. 1.
The pmx gene cluster. (A) Genetic structure of pmx genes and domain organization of the Pmx enzymes. (B) Primary structure of polymyxin A. MOA, 6-methyloctanoic acid.
The domains of the polymyxin synthetase were analyzed based on the method of Ansari et al. (3). PmxA, containing 4,953 amino acids, comprises four modules and a C-domain (Fig. 1A). The substrate specificities of the four PmxA A-domains were predicted to activate the amino acid substrates, Leu, Thr, Dab, and Dab, respectively (Table 2). PmxB, a 1,102-amino-acid polypeptide, comprises one module containing A-T-TE domains. The predicted amino acid specificity of the A-domain of PmxB was Thr (Table 2). Due to the presence of the TE-domain, PmxB may contribute to the termination of polymyxin synthesis. PmxE, a 6,312-amino-acid polypeptide, has five modules and a C-domain. The substrate specificities of the five PmxE A-domains were predicted to activate the amino acid substrates Dab, Thr, Dab, Dab, and Dab, respectively (Table 2). Based on the polymyxin structure, the order of modules for amino acid assembly during polymyxin synthesis should be PmxE-PmxA-PmxB, and the last C-domains of PmxE and PmxA should become one module with the A-T-E domains of PmxA and the A-T-TE domains of PmxB, respectively. The third module of PmxE contains an E-domain, which suggests that the third amino acid, Dab, may be a d form in polymyxin produced by the E681 strain. Taken together, these findings suggest that the polymyxin synthetase of P. polymyxa E681 may synthesize polymyxin A (Fig. 1B), the structure of which was reported by Wilkinson and Lowe in 1966 (54).
TABLE 2.
Specificity-conferring amino acids of adenylation domains in the polymyxin synthetase
| A-domain | Active site residue at position:
|
Amino acid specificity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | ||
| PmxA A1 | D | A | W | I | V | G | A | I | V | K | Leu |
| PmxA A2 | D | F | W | N | I | G | M | V | H | K | Thr |
| PmxA A3 | D | V | G | E | I | S | A | I | D | K | Dab |
| PmxA A4 | D | V | G | E | I | S | A | I | D | K | Dab |
| PmxB A1 | D | F | W | N | I | G | M | V | H | K | Thr |
| PmxE A1 | D | V | G | E | I | S | S | I | D | K | Dab |
| PmxE A2 | D | F | W | N | I | G | M | V | H | K | Thr |
| PmxE A3 | D | V | G | E | I | S | S | I | D | K | Dab |
| PmxE A4 | D | V | G | E | I | S | A | I | D | K | Dab |
| PmxE A5 | D | V | G | E | I | S | A | I | D | K | Dab |
Analysis of polymyxin in P. polymyxa E681.
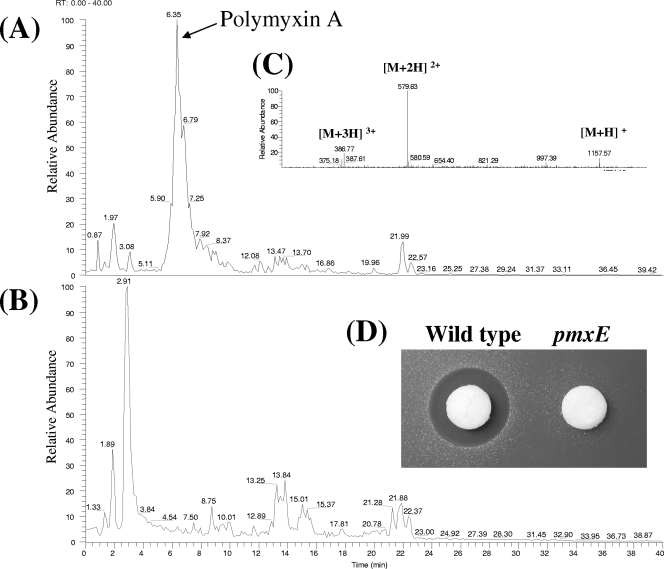
The composition of the supernatant of P. polymyxa E681 grown in GSC medium was analyzed using an LC/MS system (Fig. 2A and C). The (M+H)+, (M+2H)2+, and (M+3H)3+ ion peaks were observed at 1,157, 579, and 386, respectively (Fig. 2C). The molecular weight of the polymyxin was the same as those of polymyxins A and M (31, 54). The only difference between polymyxin A and M is the d/l configuration of the third amino acid, Dab; a d-Dab is present in polymyxin A, and an l-Dab is found in polymyxin M. From the results of domain analysis, we concluded that the polymyxin produced by P. polymyxa E681 is polymyxin A.
FIG. 2.
Analysis of polymyxin synthesis in P. polymyxa E681. LC analysis of culture supernatants of E681 (A) and the pmxE mutant (B), respectively, using a YMC Pack Pro C18 column. (C) MS data for polymyxin A produced by P. polymyxa E681. The arrow indicates the peak for polymyxin A. (D) Antibacterial activities of the culture supernatants of wild-type E681 and the pmxE mutant strains against E. coli DH5α.
Insertional disruption of the polymyxin synthetase gene cluster.
To confirm that the pmx gene cluster is involved in polymyxin biosynthesis, we constructed and characterized a pmxE mutant strain. The antibacterial activity of the pmxE mutant of P. polymyxa E681 was completely abolished in a bioassay against E. coli (Fig. 2D). LC/MS data supported our earlier results by showing that the peak corresponding to polymyxin could not be detected in the pmxE mutant (Fig. 2B). Taken together, these results demonstrated that the pmx gene cluster is essential for polymyxin biosynthesis.
Heterologous expression of the pmx gene cluster in B. subtilis.
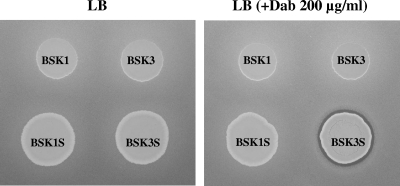
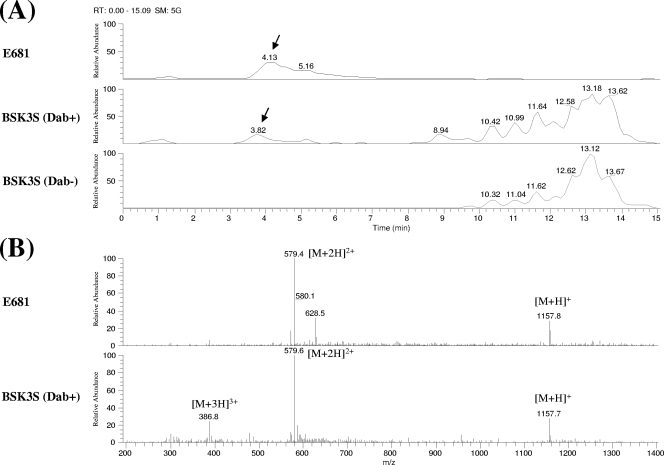
As described in Materials and Methods, a recombinant B. subtilis strain BSK1 having a disrupted RM system was constructed and showed at least 100-times-higher transformation efficiency than did the parent 168 strain with large DNA fragments 30 to 70 kb in length (data not shown). For heterologous expression, the entire pmx gene cluster was integrated into the amyE locus of B. subtilis BSK1, using fosmid clones containing pmx genes. The integration was carried out in two steps because no fosmid clone containing the entire pmx gene cluster was present in our fosmid library. The scheme of the integration is shown in Fig. 3. In the first step, a DNA fragment (36.8 kb) containing pmxABCD, truncated pmxE, and an 8.2-kb upstream region was introduced into the amyE locus of B. subtilis BSK1 by homologous recombination using a recombinant plasmid, pDG-12B06, containing the pmx genes and a flanking region. The resulting strain, BSK2, was then transformed with a recombinant fosmid, PP12D08-Spr-amyEback, which contained an intact pmxE and its 22.7-kb downstream region. Through this second step of homologous recombination, strain BSK3 containing the entire pmxABCDE and its flanking regions was constructed. Strain BSK3, however, did not show antibacterial activity against E. coli (Fig. 4). For the synthesis of nonribosomal peptide antibiotics, functional Sfp, a phosphopantetheinyl transferase, is required (24). Because Sfp in B. subtilis 168 is nonfunctional due to a mutation of the sfp gene (51), a functional sfp gene from B. subtilis CB114 (26) was introduced into BSK3 to construct strain BSK3S. However, the introduction of intact sfp still did not induce antibacterial activity (Fig. 4). We found that the synthetic mechanism of an amino acid, Dab, which is a major amino acid in polymyxin, was absent in B. subtilis 168. When Dab was added extracellularly in growth medium, the antimicrobial activity of strain BSK3S against E. coli was successfully detected (Fig. 4). LC/MS analysis of the supernatant of BSK3S grown in GSC medium containing Dab showed that the polymyxin peak of BSK3S had the same mass profile as that of P. polymyxa E681, thus demonstrating that B. subtilis BSK3S produced polymyxin (Fig. 5).
FIG. 4.
Antibacterial activities of recombinant B. subtilis strains against E. coli under conditions with or without l-Dab. B. subtilis BSK1 derived from B. subtilis 168 contains a deleted BsuM RM system. Strain BSK1S was constructed by introducing a functional sfp from B. subtilis CB114 into BSK1. BSK3 contains complete pmx genes (pmxABCDE) in the amyE locus of BSK1. BSK3S was constructed by introducing a functional sfp into BSK3.
FIG. 5.
Biosynthesis of polymyxin in B. subtilis. (A) LC analysis of culture supernatants of P. polymyxa E681 and B. subtilis BSK3S grown in GSC medium with or without l-Dab, using a Terra MS C18 column. Arrows indicate the peaks to be analyzed by MS. (B) MS data for polymyxins produced by P. polymyxa E681 and B. subtilis BSK3S.
DISCUSSION
The excellent antibacterial activities of polymyxins against multidrug-resistant, pathogenic, gram-negative bacteria have led to its reemergence among the antibiotics currently used in clinical practice in order to cope with such bacteria. However, widespread use of these antibiotics has been limited by their severe side effects, which include nephrotoxicity and neurotoxicity (18, 37). The development of polymyxin analogues with reduced toxicity has been limited because of the structural complexity of polymyxin and the lack of information on relevant biosynthetic genes. This report represents the complete sequence of the polymyxin synthetase gene cluster. Information on the sequence of the gene cluster may facilitate the development of a polymyxin analogue with reduced toxicity, as well as novel polymyxin-based antibiotics.
An interesting feature of the polymyxin gene cluster is the presence of pmxC and pmxD genes encoding transporter-like proteins within the gene cluster. The deduced gene products, PmxC (608 amino acids) and PmxD (577 amino acids), are 32.4% identical. PmxC and PmxD share 40.5% and 43.5% identities, respectively, with TycD and TycE, members of the ABC transporter family, of Brevibacillus brevis (34). Analysis of PmxC and PmxD with the Transporter Classification database (42) showed the presence of five and seven transmembrane helices, respectively. The locations of the two tandem transporters within the polymyxin gene cluster suggest a role in conferring resistance against polymyxin via secretion by the producer cell. Work is in progress to clarify the potential roles of the ABC transporters PmxC and PmxD in the secretion of polymyxin.
One of the greatest concerns in polymyxin biosynthesis is the mechanism of incorporation of the fatty acid moiety to the peptide. The N-terminal C-domains (named starter C-domains) in first subunits of NRPSs clearly distinguishable from the other downstream C-domains were proposed to have a role in coupling a fatty acid to an amino acid (32, 33). Recent phylogenetic studies of C-domains showed that many other NRPSs have these starter C-domains (39, 40). PmxE also contains a starter C-domain, which suggests that the C-domain may mediate a fatty acyl tailing of polymyxin. In contrast, Komura and Kurahashi suggested that a separate acyltransferase is necessary for the fatty acyl tailing of polymyxin (21, 23). In this study, the pmx gene cluster was introduced into the amyE locus of the B. subtilis chromosome, with 8.2 kb of upstream flanking region and 22.7 kb of downstream flanking region. The upstream and downstream flanking regions contain 7 and 24 putative open reading frames, respectively (see Table S1 in the supplemental material). Among them, we could not find any gene that was potentially involved in the incorporation of a fatty acyl group into the polymyxin. If the suggestion of Komura and Kurahashi is correct, P. polymyxa E681 and B. subtilis may contain acyltransferases with the same specificity, because polymyxins produced by the two species showed the same mass profiles (Fig. 5).
Synthesis of polymyxin in B. subtilis 168 harboring entire pmx genes was induced only in Dab-containing medium (Fig. 4), which suggests that there is no synthetic mechanism of Dab in the strain. Synthesis of Dab is mediated by 2,4-diaminobutyrate aminotransferase encoded by ectB (43). There is no homologue of ectB in B. subtilis 168. The ectB is composed of an operon structure with ectA and ectC encoding 2,4-diaminobutyrate acetyltransferase and ectoine synthase, respectively, in Halobacillus halophilus (43). The ectABC genes responsible for ectoine biosynthesis have usually been found in halophilic bacteria. The order of these genes was found to be highly conserved, even in a gram-negative bacterium, Halomonas elongate (43). In P. polymyxa E681, the amino acid sequence of the ectB homologue shares 51% identity with that of H. halophilus. Interestingly, the ectB gene of P. polymyxa E681 is not part of an operon. There is no homologue of ectA or ectC in the genome, which suggests that P. polymyxa E681 does not produce ectoine. Therefore, in P. polymyxa E681, Dab synthesized by the EctB may not be used as an intermediate for the synthesis of ectoine, resulting in an increase in its concentration in the cell. This condition may be favorable to the cell in terms of the synthesis of polymyxin.
Many bacterial isolates producing natural products such as peptide antibiotics are usually difficult to handle because of our lack of knowledge of their physiological and genetic traits and the low transformation efficiencies of these isolates. Therefore, studies of the production of natural products and development of novel analogues through biosynthetic engineering often encounter difficulties from their initiation. Many reports have dealt with the heterologous expressions of natural product pathways from the original microbial organisms to well-developed surrogate hosts (11, 51, 53). Although we succeeded in constructing a pmxE knockout mutant in this study, the low level of transformation efficiency of P. polymyxa E681 remains a bottleneck in genetic studies. Therefore, heterologous expression of the polymyxin biosynthetic gene cluster in B. subtilis may accelerate structure-function study and engineering of pmx genes for the generation of novel analogues.
Supplementary Material
Acknowledgments
This research was supported by the 21C Frontier Microbial Genomics and Applications Center Program of the Ministry of Education, Science and Technology, and the KRIBB Research Initiative Program, Republic of Korea.
Footnotes
Published ahead of print on 20 March 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ainsworth, G. C., A. M. Brown, and G. Brownlee. 1947. Aerosporin, an antibiotic produced by Bacillus aerosporus Greer. Nature 160263. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 2007. Molecular mechanisms of antibacterial multidrug resistance. Cell 1281037-1050. [DOI] [PubMed] [Google Scholar]
- 3.Ansari, M. Z., G. Yadav, R. S. Gokhale, and D. Mohanty. 2004. NRPS-PKS: a knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32W405-W413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedict, R. G., and A. F. Langlykke. 1947. Antibiotic activity of Bacillus polymyxa. J. Bacteriol. 5424-25. [PubMed] [Google Scholar]
- 5.Bernhard, K., H. Schrempf, and W. Goebel. 1978. Bacteriocin and antibiotic resistance plasmids in Bacillus cereus and Bacillus subtilis. J. Bacteriol. 133897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chihara, S., T. Tobita, M. Yahata, A. Ito, and Y. Koyamn. 1973. Enzymatic degradation of colistin. Isolation and identification of α-N-acyl-α,Y-diaminobutyric acid and colistin nonapeptide. Agric. Biol. Chem. 372455-2463. [Google Scholar]
- 7.Choi, S.-K., S.-Y. Park, R. Kim, C.-H. Lee, J. F. Kim, and S.-H. Park. 2008. Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 36589-95. [DOI] [PubMed] [Google Scholar]
- 8.Cruz, D. N., M. A. Perazella, R. Bellomo, M. de Cal, N. Polanco, V. Corradi, P. Lentini, F. Nalesso, T. Ueno, V. M. Ranieri, and C. Ronco. 2007. Effectiveness of polymyxin B-immobilized fiber column in sepsis: a systematic review. Crit. Care 11R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eliasson Lantz, A., P. Jorgensen, E. Poulsen, C. Lindemann, and L. Olsson. 2006. Determination of cell mass and polymyxin using multi-wavelength fluorescence. J. Biotechnol. 121544-554. [DOI] [PubMed] [Google Scholar]
- 11.Eppelmann, K., S. Doekel, and M. A. Marahiel. 2001. Engineered biosynthesis of the peptide antibiotic bacitracin in the surrogate host Bacillus subtilis. J. Biol. Chem. 27634824-34831. [DOI] [PubMed] [Google Scholar]
- 12.Fajardo-Cavazos, P., C. Salazar, and W. L. Nicholson. 1993. Molecular cloning and characterization of the Bacillus subtilis spore photoproduct lyase (spl) gene, which is involved in repair of UV radiation-induced DNA damage during spore germination. J. Bacteriol. 1751735-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falagas, M. E., and S. K. Kasiakou. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 401333-1341. [DOI] [PubMed] [Google Scholar]
- 14.Giske, C. G., D. L. Monnet, O. Cars, and Y. Carmeli. 2008. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 52813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guérout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 18057-61. [DOI] [PubMed] [Google Scholar]
- 16.Haima, P., S. Bron, and G. Venema. 1987. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet. 209335-342. [DOI] [PubMed] [Google Scholar]
- 17.Harwood, C. R., and S. M. Cutting (ed.). 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Inc., New York, NY.
- 18.Hermsen, E. D., C. J. Sullivan, and J. C. Rotschafer. 2003. Polymyxins: pharmacology, pharmacokinetics, pharmacodynamics, and clinical applications. Infect. Dis. Clin. North Am. 17545-562. [DOI] [PubMed] [Google Scholar]
- 19.Jiménez-Mejías, M. E., C. Pichardo-Guerrero, F. J. Marquez-Rivas, D. Martin-Lozano, T. Prados, and J. Pachon. 2002. Cerebrospinal fluid penetration and pharmacokinetic/pharmacodynamic parameters of intravenously administered colistin in a case of multidrug-resistant Acinetobacter baumannii meningitis. Eur. J. Clin. Microbiol. Infect. Dis. 21212-214. [DOI] [PubMed] [Google Scholar]
- 20.Kline, T., D. Holub, J. Therrien, T. Leung, and D. Ryckman. 2001. Synthesis and characterization of the colistin peptide polymyxin E1 and related antimicrobial peptides. J. Pept. Res. 57175-187. [DOI] [PubMed] [Google Scholar]
- 21.Komura, S., and K. Kurahashi. 1980. Biosynthesis of polymyxin E by a cell-free enzyme system. II. Synthesis of enzyme-bound octanoyldiaminobutyric acid. J. Biochem. 88285-288. [PubMed] [Google Scholar]
- 22.Komura, S., and K. Kurahashi. 1980. Biosynthesis of polymyxin E. III. Total synthesis of polymyxin E by a cell-free enzyme system. Biochem. Biophys. Res. Commun. 951145-1151. [DOI] [PubMed] [Google Scholar]
- 23.Komura, S., and K. Kurahashi. 1985. Biosynthesis of polymyxin E by a cell-free enzyme system. IV. Acylation of enzyme-bound l-2,4-diaminobutyric acid. J. Biochem. 971409-1417. [DOI] [PubMed] [Google Scholar]
- 24.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3923-936. [DOI] [PubMed] [Google Scholar]
- 25.Landman, D., C. Georgescu, D. A. Martin, and J. Quale. 2008. Polymyxins revisited. Clin. Microbiol. Rev. 21449-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, Y. K., S. B. Kim, C. S. Park, J. G. Kim, H. M. Oh, B. D. Yoon, and H. S. Kim. 2005. Chromosomal integration of sfp gene in Bacillus subtilis to enhance bioavailability of hydrophobic liquids. Appl. Microbiol. Biotechnol. 67789-794. [DOI] [PubMed] [Google Scholar]
- 27.Levin, A. S., A. A. Barone, J. Penco, M. V. Santos, I. S. Marinho, E. A. Arruda, E. I. Manrique, and S. F. Costa. 1999. Intravenous colistin as therapy for nosocomial infections caused by multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Clin. Infect. Dis. 281008-1011. [DOI] [PubMed] [Google Scholar]
- 28.Li, J., R. L. Nation, J. D. Turnidge, R. W. Milne, K. Coulthard, C. R. Rayner, and D. L. Paterson. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect. Dis. 6589-601. [DOI] [PubMed] [Google Scholar]
- 29.Li, J., R. L. Nation, R. W. Milne, J. D. Turnidge, and K. Coulthard. 2005. Evaluation of colistin as an agent against multi-resistant gram-negative bacteria. Int. J. Antimicrob. Agents 2511-25. [DOI] [PubMed] [Google Scholar]
- 30.Livermore, D. M. 2007. Introduction: the challenge of multiresistance. Int. J. Antimicrob. Agents 29(Suppl. 3)S1-S7. [DOI] [PubMed] [Google Scholar]
- 31.Martin, N. I., H. Hu, M. M. Moake, J. J. Churey, R. Whittal, R. W. Worobo, and J. C. Vederas. 2003. Isolation, structural characterization, and properties of mattacin (polymyxin M), a cyclic peptide antibiotic produced by Paenibacillus kobensis M. J. Biol. Chem. 27813124-13132. [DOI] [PubMed] [Google Scholar]
- 32.Miao, V., R. Brost, J. Chapple, K. She, M. F. Gal, and R. H. Baltz. 2006. The lipopeptide antibiotic A54145 biosynthetic gene cluster from Streptomyces fradiae. J. Ind. Microbiol. Biotechnol. 33129-140. [DOI] [PubMed] [Google Scholar]
- 33.Miao, V., M. F. Coëffet-Legal, P. Brian, R. Brost, J. Penn, A. Whiting, S. Martin, R. Ford, I. Parr, M. Bouchard, C. J. Silva, S. K. Wrigley, and R. H. Baltz. 2005. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 1511507-1523. [DOI] [PubMed] [Google Scholar]
- 34.Mootz, H. D., and M. A. Marahiel. 1997. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 1796843-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 3490-504. [DOI] [PubMed] [Google Scholar]
- 36.Okimura, K., K. Ohki, Y. Sato, K. Ohnishi, and N. Sakura. 2007. Semi-synthesis of polymyxin B (2-10) and colistin (2-10) analogs employing the trichloroethoxycarbonyl (Troc) group for side chain protection of alpha, gamma-diaminobutyric acid residues. Chem. Pharm. Bull. (Tokyo) 551724-1730. [DOI] [PubMed] [Google Scholar]
- 37.Pedersen, M. F., J. F. Pedersen, and P. O. Adsen. 1971. A clinical and experimental comparative study of sodium colistimethate and polymyxin B sulfate. Investig. Urol. 9234-237. [PubMed] [Google Scholar]
- 38.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 513471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rausch, C., I. Hoof, T. Weber, W. Wohlleben, and D. H. Huson. 2007. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol. Biol. 778-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roongsawang, N., S. P. Lim, K. Washio, K. Takano, S. Kanaya, and M. Morikawa. 2005. Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol. Lett. 252143-151. [DOI] [PubMed] [Google Scholar]
- 41.Ryu, C.-M., J. Kim, O. Choi, S.-Y. Park, S.-H. Park, and C.-S. Park. 2005. Nature of a root-associated Paenibacillus polymyxa from field-grown winter barley in Korea. J. Microbiol. Biotechnol. 15984-991. [Google Scholar]
- 42.Saier, M. H., Jr., C. V. Tran, and R. D. Barabote. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saum, S. H., and V. Muller. 2008. Growth phase-dependent switch in osmolyte strategy in a moderate halophile: ectoine is a minor osmolyte but major stationary phase solute in Halobacillus halophilus. Environ. Microbiol. 10716-726. [DOI] [PubMed] [Google Scholar]
- 44.Segal-Maurer, S., N. Mariano, A. Qavi, C. Urban, and J. J. Rahal, Jr. 1999. Successful treatment of ceftazidime-resistant Klebsiella pneumoniae ventriculitis with intravenous meropenem and intraventricular polymyxin B: case report and review. Clin. Infect. Dis. 281134-1138. [DOI] [PubMed] [Google Scholar]
- 45.Sharma, S. K., A. D. Wu, N. Chandramouli, C. Fotsch, G. Kardash, and K. W. Bair. 1999. Solid-phase total synthesis of polymyxin B1. J. Pept. Res. 53501-506. [DOI] [PubMed] [Google Scholar]
- 46.Sieber, S. A., and M. A. Marahiel. 2005. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105715-738. [DOI] [PubMed] [Google Scholar]
- 47.Stansly, P. G., R. G. Shepard, and H. J. White. 1947. Polymyxin: a new chemotherapeutic agent. Bull. Johns Hopkins Hosp. 8143-54. [PubMed] [Google Scholar]
- 48.Stein, A., and D. Raoult. 2002. Colistin: an antimicrobial for the 21st century? Clin. Infect. Dis. 35901-902. [DOI] [PubMed] [Google Scholar]
- 49.Storm, D. R., K. S. Rosenthal, and P. E. Swanson. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46723-763. [DOI] [PubMed] [Google Scholar]
- 50.Tsubery, H., I. Ofek, S. Cohen, M. Eisenstein, and M. Fridkin. 2002. Modulation of the hydrophobic domain of polymyxin B nonapeptide: effect on outer-membrane permeabilization and lipopolysaccharide neutralization. Mol. Pharmacol. 621036-1042. [DOI] [PubMed] [Google Scholar]
- 51.Tsuge, K., S. Inoue, T. Ano, M. Itaya, and M. Shoda. 2005. Horizontal transfer of iturin A operon, itu, to Bacillus subtilis 168 and conversion into an iturin A producer. Antimicrob. Agents Chemother. 494641-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaara, M., J. Fox, G. Loidl, O. Siikanen, J. Apajalahti, F. Hansen, N. Frimodt-Mφller, J. Nagai, M. Takano, and T. Vaara. 2008. Novel polymyxin derivatives carrying only three positive charges are effective antibacterial agents. Antimicrob. Agents Chemother. 523229-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenzel, S. C., and R. Muller. 2005. Recent developments towards the heterologous expression of complex bacterial natural product biosynthetic pathways. Curr. Opin. Biotechnol. 16594-606. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson, S., and L. A. Lowe. 1966. Structures of the polymyxins A and the question of identity with the polymyxins M. Nature 212311. [DOI] [PubMed] [Google Scholar]
- 55.Zavascki, A. P., L. Z. Goldani, J. Li, and R. L. Nation. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 601206-1215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.