Abstract
The emergence of antimicrobial drug resistance is of enormous public concern due to the increased risk of delayed treatment of infections, the increased length of hospital stays, the substantial increase in the cost of care, and the high risk of fatal outcomes. A prerequisite for the development of effective therapy alternatives is a detailed understanding of the diversity of bacterial mechanisms that underlie drug resistance, especially for problematic gram-negative bacteria such as Pseudomonas aeruginosa. This pathogen has impressive chromosomally encoded mechanisms of intrinsic resistance, as well as the potential to mutate, gaining resistance to current antibiotics. In this study we have screened the comprehensive nonredundant Harvard PA14 library for P. aeruginosa mutants that exhibited either increased or decreased resistance against 19 antibiotics commonly used in the clinic. This approach identified several genes whose inactivation sensitized the bacteria to a broad spectrum of different antimicrobials and uncovered novel genetic determinants of resistance to various classes of antibiotics. Knowledge of the enhancement of bacterial susceptibility to existing antibiotics and of novel resistance markers or modifiers of resistance expression may lay the foundation for effective therapy alternatives and will be the basis for the development of new strategies in the control of problematic multiresistant gram-negative bacteria.
There is accumulating evidence that appropriate antibacterial therapy administered early in the course of an infection has major implications for the outcomes of severe bacterial diseases (20, 21, 43). Patients have greatly benefited from the introduction of effective antimicrobials in the past decades; however, the frequency and spectrum of antibiotic-resistant infections have increased worldwide, and substantially higher mortality rates have been reported for patients given ineffective empirical therapy, mainly due to resistance to the agents used (39). Today, in many intensive-care units, multidrug-resistant gram-negative bacteria pose the greatest therapeutic challenge. Therefore, understanding the mechanisms of resistance and developing therapy alternatives for problematic gram-negative bacteria are of profound importance.
The increase in the incidence of multidrug resistance has been attributed to a combination of microbial characteristics, the selective pressure of antimicrobial use, and enhanced transmission of resistant organisms. This growing problem requires a comprehensive strategy that includes compliance with infection control principles, rational use of current antimicrobial agents, and the development of new active agents. The diversity of bacterial mechanisms that underlie multidrug resistance makes the development of effective new antimicrobial agents, especially against problematic species such as Pseudomonas aeruginosa, very difficult. This opportunistic gram-negative rod plays a dominant role as an infectious agent in the lungs of cystic fibrosis patients and has emerged as one of the most important human pathogens causing serious nosocomial infections. P. aeruginosa is able to thrive in various environments and utilizes a broad spectrum of virulence factors to infect different hosts, from plants and insects to humans. Due to intrinsic antibiotic resistance, P. aeruginosa infections are difficult to treat and are associated with high mortality rates (10). The intrinsic resistance to antibiotics seems to result mainly from the reduced permeability of the bacterial cell envelope and the activity of multidrug efflux pumps; however, other, yet to be discovered mechanisms might contribute.
Recent publications have aimed at identifying the intrinsic “resistome” of P. aeruginosa by screening transposon mutant libraries for the profiles of resistance to several antibiotics (7, 16, 40, 45). From these studies it has become clear that many previously unidentified genes play a role in antimicrobial resistance and that, depending on the antibiotic, there are many modifiers of the expression of resistance in P. aeruginosa.
In this study we proceeded in the identification of the P. aeruginosa resistome. We made use of a semiautomated antibiotic susceptibility test method and systematically screened the Harvard Medical School PA14 transposon mutant library (27) for mutants with either increased or decreased susceptibility to 19 different antimicrobial agents commonly used in the clinic. This approach not only uncovered novel genetic determinants of resistance in P. aeruginosa to specific antimicrobial compounds but also identified genes whose inactivation sensitized the bacteria to a broad spectrum of different antimicrobials. Our results provide valuable information for further studies aiming at predicting antibiotic resistance based on genotype and have also led to the identification of promising novel drug targets. Targets that enhance bacterial susceptibility could be the basis for the development of drugs that potentate existing antimicrobials and thus may function as chemosensitizers. Examples of such novel antimicrobial combinations are the use of inhibitors of efflux pumps in combination with tetracycline (TET) against E. coli (30) or in combination with ciprofloxacin (CIP) against P. aeruginosa (28).
MATERIALS AND METHODS
Bacterial strains.
The PA14 transposon mutant library described by Liberati et al. (27) was used to screen a comprehensive set of mutants for their resistance profiles. This library was constructed using a mariner-based transposon containing the resistance cassette aacC1, which confers resistance to gentamicin but not to tobramycin (TOB). The PAO1 transposon mutant library of the University of Washington (22) was used for verification and cross-referencing purposes. This library was constructed using two transposons derived from the IS50 element of transposon Tn5, containing a tetracycline resistance cassette.
Antimicrobial resistance screening. (i) Semiautomated susceptibility testing with Vitek 2.
PA14 mutants were streaked onto Columbia agar plates and incubated at 37°C overnight. A sufficient number of colonies was suspended in sterile saline (0.45%) and adjusted to a 0.5 McFarland turbidity standard using a DensiChek densitometer (bioMérieux). The inoculated tube was placed in a cassette on the Vitek 2 Smart Carrier Station. The sample number was entered and associated with an antimicrobial susceptibility test (AST) card. The Smart Carrier Station cassette with the cards and the test tubes was placed on the Vitek 2 instrument, where the inoculation of the AST cards was automatically performed by the instrument.
(ii) Susceptibility testing by agar dilution.
For standard agar dilution testing, LB agar plates containing serial twofold dilutions of the antimicrobial agents to be tested were prepared, stored at 4°C, and used within 5 days. Overnight-grown P. aeruginosa mutants were diluted 1:100 in 0.9% NaCl solution and replica plated onto the LB agar containing the respective antibiotics in twofold dilutions, with duplicate plates for each single dilution. For testing of both hypersusceptible and hyperresistant mutants, susceptibility to single antibiotics belonging to six different classes was tested: piperacillin (PIP) (penicillins), meropenem (MEM) (carbapenems), ceftazidime (CAZ) (cephalosporins), CIP (fluoroquinolones), TOB (aminoglyocosides), and TET (tetracyclines). The agar plates were incubated at 37°C and read at 18 to 24 h. The MIC was considered to be the lowest concentration of an antimicrobial agent that completely inhibited growth as detected visually.
Purification of membrane proteins.
Wild-type PA14 and the PA14 oprF transposon mutants were grown overnight in 20 ml LB medium at 37°C. Cells were harvested (at 6,000 × g for 10 min) and resuspended in 100 mM CH3COOK, 5 mM (CH3COO)2Mg·4H2O, 0.2% (vol/vol) β-mercaptoethanol, 50 mM HEPES (pH 7.5) containing Complete EDTA-free protease inhibitor cocktail (Roche). Cells were lysed by a French press (four times at 1,000 lb/in2), and unbroken cells were removed by centrifugation. The supernatant was adjusted to 20% sucrose, loaded onto an isopycnic sucrose gradient (3 ml of 80% sucrose, 3 ml of 60% sucrose, 13 ml of 20% sucrose/membrane fraction, 3 ml water cushion), and ultracentrifuged (at 100,000 × g for 1 h). Membranes enriched at the 20/60% and 60/80% sucrose interphases were collected from the gradient using a Pasteur pipette and washed with water in the presence of Complete protease inhibitor cocktail (Roche). Associated proteins were rejected by carbonate extraction (17), and the remaining membrane proteins were precipitated according to the method of reference 44. Five-microgram portions of both wild-type and mutant membrane proteins were suspended in loading buffer, separated on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel, and stained with colloidal Coomassie blue.
RESULTS AND DISCUSSION
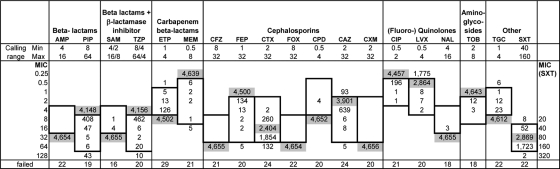
In this study we have screened an ordered, comprehensive PA14 transposon mutant library (27) for mutants exhibiting either increased or decreased susceptibility to various antimicrobial agents. AST was performed with the Vitek 2 system (bioMérieux, Marcy l'Étoile, France). This is an automated, short-incubation broth dilution system capable of performing susceptibility testing based on bacterial growth curves. Twenty different antimicrobial agents were tested using the AST-N063 card at given concentration ranges. These included the two carbapenems ertapenem (ETP) and MEM; the two penicillins ampicillin (AMP) and PIP, both also used in combination with beta-lactam inhibitors in ampicillin-sulbactam (SAM) and piperacillin-tazobactam (TZP); the seven cephalosporins (narrow to extended spectrum), cefazolin (CFZ), cefoxitin (FOX), cefpodoxime (CPD), cefuroxime (CXM), cefotaxime (CTX), CAZ, and cefepime (FEP); the aminoglycoside TOB; the three gyrase inhibitors CIP, levofloxacin (LVX), and nalidixic acid (NAL); one tetracycline, tigecycline (TGC); and trimethoprim-sulfamethoxazole (SXT). Figure 1 summarizes the Vitek 2 MIC results for 4,676 single-gene-knockout PA14 transposon mutants. Since the MICs for wild-type PA14 were lower than the MIC calling ranges of PIP, TZP, MEM, FEP, CIP, and TOB, only mutants that exhibited an increase in the resistance profile could be detected for these antibiotics (Fig. 1). Vice versa, the MICs for wild-type PA14 were greater than the MIC calling ranges of AMP, SAM, ETP, CFZ, FOX, CPD, CXM, NAL, and TGC. Thus, for these antibiotics, only mutants that exhibited enhanced susceptibility could be detected. The MICs of CTX, CAZ, LVX, and SXT were within the given MIC calling range, and therefore both mutants that exhibited increased susceptibility and those with decreased susceptibility were identified. Additionally, the AST-N063 card includes the aminoglycoside gentamicin, which could not be used for the analysis because the transposon applied to create the PA14 mutant library carries a gentamicin resistance cassette (27).
FIG. 1.
AST of the Harvard P. aeruginosa PA14 transposon mutant library using the Vitek 2 system. Min and Max, minimum and maximum MICs (in micrograms per milliliter) that can be determined with the Vitek 2 system. Calling ranges are also indicated by a box in each column. The MIC of each antibiotic for wild-type PA14 is given by aligning the number of wild-type PA14 isolates (shaded) horizontally with the applicable MIC on the left. Similarly, the distribution of the MICs of a particular antibiotic across all PA14 mutants is given by aligning the numbers of mutant isolates horizontally with the applicable MICs on the left. A MIC below the calling range should be read as “less than or equal to” the value given, and a MIC above the calling range should be read as “greater than or equal to” the value given. Note that the MICs for SXT are shown in the rightmost column. Failed, number of failed measurements.
Interestingly, while there was significant overlap of mutants identified in this and previous screens for the “intrinsic” resistome of the PA14 strain (7, 40, 44), only four mutants were identified both in our PA14 screen and a published Tb and 59.20 strain screen for antibiotic resistance (16). It is known that, while all P. aeruginosa strains share a large common set of “core genes,” their genetic repertoire exhibits great variation in the “accessory genome,” which has substantial influence on the phenotype of the strain. Although most of the mutations identified in the two studies affect common (core) genes of P. aeruginosa, the (accessory) genetic background might strongly affect the actual resistance phenotype.
Identification of hypersusceptible P. aeruginosa mutants.
Using this global approach, we identified 40 PA14 mutants that exhibited enhanced susceptibility to at least 2 out of 13 antibiotics or antibiotic combinations (Table 1). Since the Vitek 2 system records MICs on the basis of bacterial growth, we included the growth rates of these mutants in Table 1 (determined from the maximum slope of the growth curve of the AST control; the growth rate of the wild-type control was defined as 1.0). Some mutants exhibited slight growth advantages, whereas others grew more slowly than the wild type and did not reach the maximal optical density (not shown). However, no mutant exhibited serious growth alterations.
TABLE 1.
List of mutants hypersusceptible to at least two different antibiotics
| PA14 gene locus | PAO1 ortholog | Gene name | Growthb | Log2 change in the MICa of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SAMc,d | ETPc,d | CTXc | CAZc | LVXc | NALc,d | TGCc,d | SXTc | ||||
| PA14_05530 | PA0425 | mexA | 1.07 | −1f,g | −3f,g | −1f,g | ≤−1f,g | ≤−1g | −1g | −2f | ≤−2 |
| PA14_05540 | PA0426 | mexB | 1.03 | −2f | −4f | −2f | ≤−1f | ≤−1g | −1g | ≤−2 | |
| PA14_07430 | PA0572 | 1.05 | −1f,g | −1f | ≤−1f | ≤−1g | −1 | ||||
| PA14_07770 | PA0595 | ostA | 0.94 | ≤−1f | −1 | ||||||
| PA14_54410 | PA0764 | mucB | 1.35 | −1f,g | ≤−1f,g | ≤−1g | −1 | ||||
| PA14_54330 | PA0770e | rnc | 0.78 | ≤−1f | ≤−1 | −1f | |||||
| PA14_53970 | PA0794 | 1.19 | −1f | ≤−1f | ≤−1g | −1 | |||||
| PA14_53000 | PA0871 | phhB | 0.66 | −1g | ≤−1f | ≤−2 | |||||
| PA14_52260 | PA0928 | 1.39 | −1f | ≤−1f | ≤−1g | −1 | |||||
| PA14_51780 | PA0967 | ruvB | 1.15 | −1f | ≤−1f | ≤−1 | −1 | ||||
| PA14_51320 | PA1005 | 1.01 | ≤−1f | ≤−1g | −1 | ||||||
| PA14_49320 | PA1167 | 0.95 | −1g | −1f | ≤−1f | ≤−1g | −1 | ||||
| PA14_47230 | PA1316 | 1.01 | −2g | −1f | ≤−1f | −1 | |||||
| PA14_41570 | PA1777 | oprF | 1.17 | −1f,g | −2f | −1f,g | ≤−1f,g | ≤−1f,g | ≤−4f | ≤−2 | |
| PA14_40620 | PA1848 | 1.39 | −1f,g | ≤−1f,g | ≤−1g | −1 | |||||
| PA14_38530 | PA2008 | fahA | 0.77 | −1 | −1g | −1f | ≤−1f | ||||
| PA14_30290 | PA2615e | ftsK | 1.31 | −3f | −2f | ≤−1f | ≤−1f | ||||
| PA14_27920 | PA2800 | 1.18 | ≤−1g | −2f | −1 | ||||||
| PA14_25110 | PA3011 | topA | 0.88 | −2f,g | −1f,g | ≤−1f,g | ≤−4f | ||||
| PA14_23890 | PA3110 | 0.89 | −2f,g | −1f | ≤−1f | ||||||
| PA14_23360 | PA3160 | wzz | 1.12 | ≤−1f | ≤−1g | −1f | |||||
| PA14_22370 | PA3233 | 1.07 | ≤−1g | −2f | −1 | ||||||
| PA14_20730 | PA3351 | flgM | 1.00 | −1 | −1f,g | ≤−1f,g | −1 | ||||
| PA14_17170 | PA3647e | ompH | 1.07 | −1f | ≤−1f | ≤−1f | −2 | −1 | |||
| PA14_16890 | PA3670 | 0.92 | −2 | −1 | ≤−1 | ≤−1g | −1 | ||||
| PA14_14910 | PA3800 | 1.07 | −1f | ≤−1f | ≤−1f,g | −1 | |||||
| PA14_12400 | PA3976 | thiE | 1.12 | −1f,g | ≤−1f,g | ≤−1g | −1 | ||||
| PA14_12030 | PA4005 | 0.85 | −2g | ≤−1f | |||||||
| PA14_08780 | PA4269 | rpoC | 0.96 | −1f,g | ≤−1f,g | ≤−1g | −1f | ||||
| PA14_57690 | PA4441 | 1.00 | −2g | −1f | ≤−1f | ≤−1g | −1 | ||||
| PA14_57880 | PA4456 | 1.05 | ≤−1g | −1 | −1 | ||||||
| PA14_57910 | PA4459e | 1.31 | −1 | ≤−1f | ≤−1 | ≤−4 | −1 | ||||
| PA14_62560 | PA4727 | pcnB | 1.09 | −1f | ≤−1f | ≤−1g | −2 | −1 | |||
| PA14_62770 | PA4745e | nusA | 0.86 | −3g | −1f | ≤−1f | ≤−1f | −1f | −1 | ||
| PA14_62880 | PA4753 | 0.89 | −2g | ≤−1f | |||||||
| PA14_64190 | PA4853e | fis | 1.06 | ≤−1 | −1 | −1 | |||||
| PA14_68670 | PA5198 | 1.22 | −2 | −1 | ≤−1 | ≤−1 | |||||
| PA14_69810 | PA5288 | glnK | 0.71 | −2g | −1 | ||||||
| PA14_70980 | PA5375 | betT1 | 1.23 | −1f,g | ≤−1f,g | ≤−1f,g | −1g | −1 | |||
| PA14_15750 | —e | 0.81 | −1 | −1f | ≤−1f | ≤−1 | −2f | ||||
Data are given only for those antibiotics for which a change in the MIC could be detected; only negative MIC changes are displayed.
Maximum growth rate of the mutant growth control relative to the growth of the wild type.
Retesting by agar dilution was performed using antibiotics of the same class: PIP for β-lactams (SAM), MEM for carbapenems (ETP), CAZ for cephalosporins (CAZ), CIP for quinolones (LVX, NAL), and TET for tetracyclines (TGC). SXT was not retested by agar dilution.
The MIC for the wild-type PA14 strain was above the calling range (Fig. 1); the MIC change was calculated based on an assumed MIC for the wild type of twice the upper limit of the calling range.
No PAO1 transposon mutant available. —, no PAO1 ortholog.
MIC change consistent with agar dilution testing of the PA14 mutant using an antibiotic of the same class.
MIC change consistent with agar dilution testing of the PAO1 mutant using an antibiotic of the same class.
We retested the susceptibility profiles of these 40 mutants for six antimicrobials (PIP, MEM, CIP, TET, CAZ, and TOB) by using the agar dilution method and also confirmed the increased-susceptibility phenotype with the respective independent mutants from the PAO1 transposon mutant library (22). Table 1 lists the MIC changes for these mutants, most of which could be confirmed to be hypersusceptible to several different antibiotics. Five of the 40 mutants could not be confirmed to be hypersusceptible by the agar dilution method (PA3670, PA4456, PA4459, the fis mutant, and PA5198).
Mutations within the MexAB-oprM multidrug efflux pump, several outer membrane proteins, and proteins involved in DNA replication, recombination, and repair confer hypersensitivity.
As expected, the two mutants harboring a transposon insertion within the mexA and mexB genes, encoding the major intrinsic multidrug MexAB-oprM efflux pump in P. aeruginosa (26, 35), exhibited the most pronounced overall susceptibility. They exhibited enhanced sensitivity to all classes of antibiotics tested in this screen. However, we also identified six mutants with insertions in genes involved in cell envelope biogenesis as being hypersusceptible. These included mutants with insertions in wzz, encoding a protein shown to be important for determining the length of the O-antigen side chain attached to lipopolysaccharide (LPS) and to influence serum resistance (24); PA2800, encoding a conserved hypothetical protein (putative lipoprotein); and genes encoding the outer membrane proteins OstA, OprF, OmpH, and BetT1. Outer membrane proteins are key molecules with regard to the interface between the cell and its environment, and these proteins have been suggested to influence the intrinsic resistance to antibiotics. Using a proteomic approach, P. aeruginosa OmpH has been demonstrated to be an antibiotic resistance-related protein (32), and a Vibrio vulnificus ompH mutant showed enhanced sensitivity to SDS and polymyxin B (2).
Ethanol tolerance in Escherichia coli appears to involve increased production of the betT-encoded choline transporter (18), which also confers general protection against osmotic stress by mediating the uptake of choline, the precursor of the compatible solute glycine betaine (46). OstA has been suggested to contribute to n-hexane resistance by reducing the influx of n-hexane in E. coli (1). Furthermore, disruption of the OstA protein in Helicobacter pylori resulted in altered membrane permeability, sensitivity to organic solvents, and susceptibility to antibiotics (11).
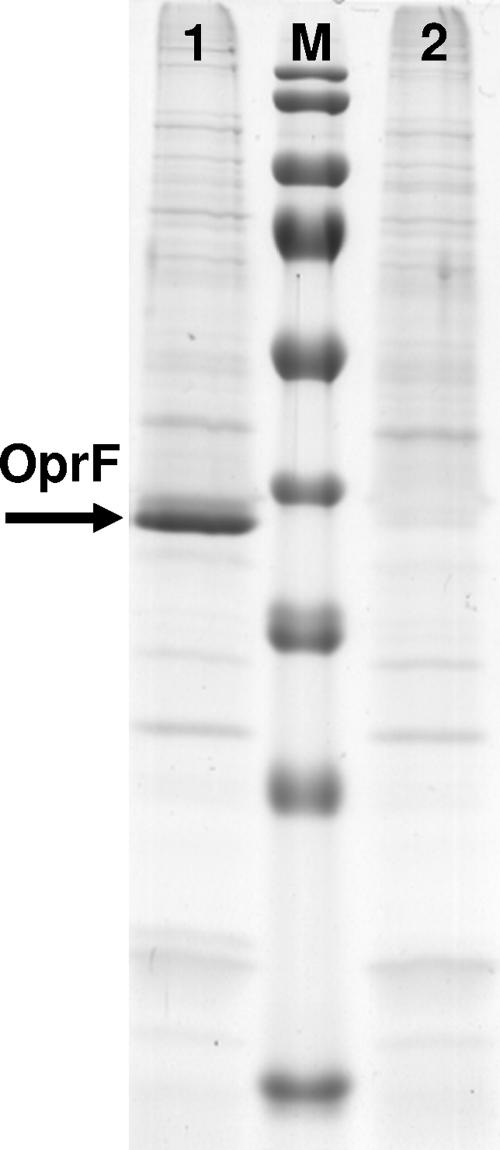
OprF is the most common outer membrane protein in P. aeruginosa and has been studied extensively. It is a nonspecific porin, permitting passive diffusion of small polar nutrients. Since the pore changes its channel size according to the growth conditions, this could affect outer-membrane permeability. OprF is also a promising candidate for vaccine development and is involved in maintaining cell shape and growth in a low-osmolarity environment (38, 48). Furthermore, it has recently been reported that the binding of human gamma interferon to OprF activates quorum-sensing-dependent expression of virulence determinants (49). The role of OprF in antibiotic resistance remains controversial (6). It has been suggested that loss of this protein may be involved in the multiple-antibiotic-resistance phenotype (33, 37), and it has been proposed that OprF has a role in antibiotic uptake in P. aeruginosa (9, 19, 47). As shown in this study, loss of OprF induced susceptibility to a very broad spectrum of antimicrobials, such as SAM, ETP, CTX, CAZ, LVX, SXT, and TGC, whereas growth was not affected (Table 1). As shown in Fig. 2 the OprF protein is completely absent from the outer membrane proteins, confirming that the oprF transposon mutant in the PA14 strain background has lost expression of a functional OprF protein.
FIG. 2.
SDS-polyacrylamide gel electrophoresis of membrane proteins. In contrast to wild-type PA14 (lane 1), the PA14 oprF mutant (lane 2) is missing the OprF band at 37.6 kDa (arrow). Lane M, prestained PageRuler protein ladder (Fermentas).
Within this global screen for hypersensitive P. aeruginosa mutants, we also identified a mucB mutant exhibiting enhanced susceptibility to CTX, CAZ, LVX, and TGC. Inactivation of mucB causes conversion to mucoidy (29), and mucoid clinical isolates have been shown to tend toward more susceptible phenotypes (4).
In accordance with our findings, and despite the use of a different screening method, a recent study of the complex CIP resistome has identified the PA14 outer membrane protein betT1 and oprF transposon mutants as CIP hypersusceptible (7). Furthermore, the study revealed that several strains with mutations of genes involved in DNA replication and repair (ruvA, recG, xerD, sss) display increased CIP sensitivity, as do strains with mutations of ftsK, involved in cell division and chromosome partitioning. Our screen also revealed three genes encoding proteins involved in DNA replication, recombination, and repair (topA, fis, ruvB), mutations of which confer broad-spectrum hypersensitivity, and we also identified the ftsK mutant, exhibiting hypersusceptibility to ETP, CTX, CAZ, and LVX. It has recently been suggested that bactericidal antibiotics induce cellular death by a common mechanism involving the generation of deleterious hydroxyl radicals (25). It thus seems reasonable that the intrinsic “resistome” involves not only multidrug efflux pumps and proteins that maintain the integrity of the cellular outer membrane but also proteins that shield DNA from damaging agents and are involved in DNA repair processes. Proteins of similar functional categories have recently been determined to be of major importance for the E. coli and P. aeruginosa intrinsic resistomes (16, 42).
Identification of antibiotic-resistant P. aeruginosa mutants.
We also screened the library for mutants that exhibited increases in the MICs of the 10 antibiotics where resistance could be determined, because the MIC for the wild-type PA14 strain was within or below the MIC calling range. A total of 193 mutants showing increased resistance to at least one antibiotic are listed in Table 2.
TABLE 2.
List of mutants showing increased resistance
| PA14 gene locus | PAO1 ortholog | Gene name | Growthb | Log2 change in the MICa of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIPc | TZPc | MEMc | FEPc | CTX | CAZ | CIPc | LVX | TOBc | SXT | ||||
| PA14_01080 | PA0089 | 0.71 | ≥2 | 1 | |||||||||
| PA14_01100 | PA0090 | 0.90 | ≥5 | ≥5 | 2 | 1 | 1 | ||||||
| PA14_01720 | PA0140 | ahpF | 1.14 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_02770 | PA0227 | 1.00 | 2 | 1 | 1 | 1 | |||||||
| PA14_03050 | PA0247 | pobA | 0.97 | 2 | * | ||||||||
| PA14_03760 | PA0287 | 1.04 | 1 | 1 | 1 | ≥2 | 1 | 1 | 1 | ||||
| PA14_04110 | PA0316 | serA | 1.18 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_04430 | PA0339 | 0.86 | ≥5 | 4 | 1 | 1 | |||||||
| PA14_04980 | PA0381 | thiG | 0.74 | ≥5 | 4 | 1 | |||||||
| PA14_05050 | PA0387 | 1.10 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_05310 | PA0407 | gshB | 0.63 | 1 | 1 | 2 | 1 | ||||||
| PA14_05410 | PA0415 | 0.84 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_05620 | PA0432 | sahH | 0.45 | 1 | 1 | 1 | 1 | 3 | 1 | ||||
| PA14_06010 | PA0460 | 0.99 | ≥5 | 4 | 1 | 1 | |||||||
| PA14_06260 | PA0479 | 1.40 | 1 | ≥2 | 1 | ||||||||
| PA14_06950 | PA0533 | 0.98 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_07070 | PA0545 | 0.90 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_07780 | PA0596 | 1.18 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_07790 | PA0597 | 1.15 | 2 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_07950 | PA0610 | prtN | 0.85 | 2 | 1 | 1 | |||||||
| PA14_08520 | PA0666 | 1.15 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_08540 | PA0667 | 0.92 | 2 | 1 | 1 | ≥2 | 1 | 1 | 1 | ||||
| PA14_53820 | PA0807 | 0.94 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_53380 | PA0842 | 1.03 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_52510 | PA0908 | 1.02 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_52050 | PA0944 | purN | 0.31 | 2 | * | 1 | 1 | 1 | |||||
| PA14_51880 | PA0958 | oprD | 0.99 | 3 | 1 | ||||||||
| PA14_50980 | PA1032 | pac | 0.82 | ≥5 | 1 | 1 | |||||||
| PA14_50250 | PA1095 | 1.07 | ≥5 | 4 | 2 | ≥2 | 1 | ||||||
| PA14_50140 | PA1101 | fliF | 0.98 | 1 | 1 | 2 | ≥2 | 1 | |||||
| PA14_49320 | PA1167 | 0.95 | ≥5 | ||||||||||
| PA14_49280 | PA1171 | 0.75 | ≥5 | 2 | 1 | 1 | 1 | ||||||
| PA14_48840 | PA1195 | 0.85 | ≥5 | 1 | 1 | 1 | 1 | ||||||
| PA14_47930 | PA1259 | 0.83 | ≥5 | 4 | 1 | 1 | 1 | ||||||
| PA14_47300 | PA1310 | phnW | 1.15 | 1 | 1 | 1 | ≥2 | 1 | 1 | 1 | |||
| PA14_47230 | PA1316 | 1.01 | ≥5 | ||||||||||
| PA14_46960 | PA1338 | ggt | 1.07 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_46850 | PA1347 | 0.99 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_46840 | PA1348 | 0.89 | 1 | 1 | 3 | 1 | ≥2 | 1 | 1 | ||||
| PA14_45980 | PA1428 | 1.07 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_45580 | PA1459 | 0.65 | 1 | 1 | ≥2 | 1 | * | ||||||
| PA14_44440 | PA1549 | 0.85 | 2 | 2 | 2 | ≥2 | 2 | 1 | |||||
| PA14_43950 | PA1588 | sucC | 0.74 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_43920 | PA1590 | braB | 1.03 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_43900 | PA1592 | 0.88 | 4 | 1 | |||||||||
| PA14_43670 | PA1611 | 1.05 | ≥5 | 2 | 1 | ||||||||
| PA14_43310 | PA1639 | 0.85 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_43270 | PA1643 | 1.11 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_41710 | PA1767 | 1.12 | 2 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_41530 | PA1781 | nirB | 1.21 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_40980 | PA1821 | 1.12 | ≥5 | 4 | 1 | ||||||||
| PA14_39420 | PA1942 | 0.85 | 2 | 1 | 1 | 1 | 1 | ||||||
| PA14_38350 | PA2023 | galU | 1.16 | ≥5 | ≥5 | 1 | 3 | ≥2 | 2 | ||||
| PA14_37550 | PA2085 | 0.82 | ≥5 | 4 | 1 | 1 | 1 | ||||||
| PA14_37530 | PA2086 | 1.23 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_37310 | PA2110 | 0.97 | 2 | 1 | 1 | ||||||||
| PA14_37030 | PA2130 | cupA3 | 1.26 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_36280 | PA2198 | 0.81 | 2 | 1 | |||||||||
| PA14_36170 | PA2207 | 0.94 | 4 | 1 | |||||||||
| PA14_34540 | PA2326 | 0.93 | ≥5 | 1 | 2 | 1 | 1 | ||||||
| PA14_33720 | PA2394 | pvdN | 1.05 | ≥5 | 1 | 1 | |||||||
| PA14_32860 | PA2455 | 0.90 | ≥5 | 4 | 1 | 1 | |||||||
| PA14_32590 | PA2478 | 1.11 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_32470 | PA2487 | 1.14 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_31850 | PA2529 | 1.08 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_31820 | PA2531 | 1.24 | ≥5 | 1 | |||||||||
| PA14_31810 | PA2532 | tpx | 1.11 | 2 | 1 | 2 | ≥2 | 1 | |||||
| PA14_31400 | PA2561 | 1.01 | ≥5 | 1 | 1 | ||||||||
| PA14_30840 | PA2571 | 1.01 | ≥5 | 4 | 1 | 1 | 1 | ||||||
| PA14_29970 | PA2641 | nuoF | 0.91 | 2 | 1 | 1 | 1 | ||||||
| PA14_29930 | PA2643 | nuoH | 0.96 | 2 | 1 | ≥2 | 1 | ||||||
| PA14_29920 | PA2644 | nuoI | 1.03 | 2 | 1 | 1 | ≥2 | 1 | |||||
| PA14_29900 | PA2645 | nuoJ | 0.91 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_29880 | PA2647d | nuoL | 0.90 | 2 | 1 | ≥2 | 1 | ||||||
| PA14_29880 | PA2647d | nuoL | 1.02 | 2 | 1 | ≥2 | 1 | 1 | |||||
| PA14_29320 | PA2691 | 1.14 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_29290 | PA2693 | 0.84 | ≥5 | ≥5 | 2 | 4 | 1 | 2 | |||||
| PA14_29220 | PA2700 | 1.10 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_27950 | PA2797 | 0.77 | 1 | 1 | 1 | ≥2 | 1 | 1 | 1 | ||||
| PA14_26960 | PA2871 | 1.11 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_25880 | PA2951 | etfA | 0.74 | 4 | 1 | ≥2 | 1 | ||||||
| PA14_25840 | PA2953 | 1.05 | 2 | ||||||||||
| PA14_25820 | PA2955 | 0.90 | 2 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_25630 | PA2970 | rpmF | 0.97 | 2 | |||||||||
| PA14_25420 | PA2989 | 0.92 | ≥5 | 4 | 1 | 1 | 1 | ||||||
| PA14_25080 | PA3014 | foaA | 0.95 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_25060 | PA3015 | 0.87 | 2 | 1 | |||||||||
| PA14_25050 | PA3016 | 1.05 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_24690 | PA3047 | 0.92 | 2 | 1 | 1 | ≥2 | 2 | 1 | |||||
| PA14_24490 | PA3063 | pelB | 0.93 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_24150 | PA3093 | 1.23 | 2 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_23670 | PA3127 | 1.01 | 2 | 1 | 1 | 1 | 1 | ||||||
| PA14_23470 | PA3141 | wbpM | 0.88 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_23240 | PA3170 | 0.93 | ≥5 | 1 | 1 | ||||||||
| PA14_23210 | PA3172 | 0.90 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_22910 | PA3194d | edd | 0.46 | 1 | 2 | ||||||||
| PA14_22910 | PA3194d | edd | 0.42 | 1 | 2 | ||||||||
| PA14_22480 | PA3224 | 1.01 | 2 | ||||||||||
| PA14_22330 | PA3236 | 1.03 | 1 | ≥2 | 1 | ||||||||
| PA14_21990 | PA3247 | 0.93 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_21860 | PA3259 | 1.26 | 1 | 1 | ≥2 | 1 | |||||||
| PA14_21410 | PA3296 | phoA | 1.01 | ≥2 | |||||||||
| PA14_21050 | PA3324 | 0.83 | ≥5 | 4 | 1 | 1 | |||||||
| PA14_20730 | PA3351 | flgM | 1.00 | 3 | |||||||||
| PA14_19910 | PA3416 | 1.24 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_19340 | PA3462 | 0.97 | 2 | 1 | |||||||||
| PA14_19170 | PA3472 | 0.87 | ≥5 | 1 | |||||||||
| PA14_18080 | PA3574 | 0.88 | 2 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_18060 | PA3575 | 1.07 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_18050 | PA3576 | 0.93 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_17500 | PA3620 | mutS | 1.26 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_16890 | PA3670 | 0.92 | ≥5 | ||||||||||
| PA14_16500 | PA3702 | wspR | 0.81 | ≥5 | ≥5 | 1 | ≥2 | 1 | |||||
| PA14_16470 | PA3704 | 1.05 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_16280 | PA3721 | 1.05 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_16130 | PA3733 | 1.13 | 1 | 1 | 1 | ≥2 | 1 | 1 | 1 | ||||
| PA14_14520 | PA3826 | 0.90 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_14470 | PA3831 | pepA | 0.67 | 2 | 1 | 1 | ≥2 | 1 | 2 | 1 | |||
| PA14_14380 | PA3837 | 0.93 | ≥5 | 3 | 1 | 1 | |||||||
| PA14_11900 | PA4016 | 1.16 | 2 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_11760 | PA4025 | 0.92 | 2 | 1 | 1 | ||||||||
| PA14_10800 | PA4109 | ampR | 1.08 | 3 | 3 | 1 | ≥2 | 3 | 1 | 1 | 1 | ||
| PA14_09550 | PA4204 | 1.09 | ≥2 | ||||||||||
| PA14_09480 | PA4210 | phzA1 | 1.21 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_09300 | PA4223 | 0.99 | 1 | ≥2 | |||||||||
| PA14_55770 | PA4292 | 1.11 | 1 | ≥2 | 1 | ||||||||
| PA14_56620 | PA4354 | 1.09 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_56640 | PA4355 | 1.13 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_57080 | PA4392 | 1.03 | ≥2 | ||||||||||
| PA14_57210 | PA4402 | argJ | 1.00 | 1 | 1 | 1 | ≥2 | 3 | 1 | ||||
| PA14_57260 | PA4406 | lpxC | 0.98 | ≥2 | |||||||||
| PA14_57540 | PA4429 | 0.87 | 2 | 2 | ≥2 | 2 | 1 | ||||||
| PA14_57560 | PA4430 | 0.77 | 1 | 1 | ≥2 | 1 | |||||||
| PA14_57570 | PA4431 | 0.84 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_57850 | PA4454 | 1.06 | ≥5 | 4 | 1 | 1 | 1 | ||||||
| PA14_57950 | PA4463 | 1.09 | ≥5 | 1 | 1 | 1 | |||||||
| PA14_58260 | PA4490 | 0.85 | 2 | 1 | 1 | 1 | |||||||
| PA14_58850 | PA4536 | 0.83 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_60860 | PA4600 | nfxB | 0.79 | 1 | 3 | 3 | 1 | ||||||
| PA14_60990 | PA4609 | radA | 0.89 | ≥ | |||||||||
| PA14_62630 | PA4733 | acsB | 1.02 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_63040 | PA4766 | 0.91 | 2 | 1 | 1 | 1 | |||||||
| PA14_63210 | PA4781 | 0.88 | ≥5 | 4 | 1 | ||||||||
| PA14_63580 | PA4811 | fdnH | 0.97 | 2 | 1 | 1 | |||||||
| PA14_63970 | PA4838 | 1.07 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_65250 | PA4939 | 1.11 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_65280 | PA4942 | hflK | 1.05 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_65320 | PA4945 | miaA | 0.65 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | |||
| PA14_65350 | PA4946d | mutL | 1.30 | 2 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_65350 | PA4946d | mutL | 1.12 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_65750 | PA4974 | 1.01 | 2 | ||||||||||
| PA14_66120 | PA5001 | 0.89 | 2 | 1 | 1 | ≥2 | 3 | 1 | |||||
| PA14_66150 | PA5003 | 1.11 | 2 | 1 | 1 | ≥2 | 2 | 1 | |||||
| PA14_66480 | PA5028 | 1.16 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_66580 | PA5037 | 0.96 | 2 | 1 | 1 | ||||||||
| PA14_66600 | PA5038 | aroB | 0.55 | 4 | 4 | 3 | ≥2 | 3 | 1 | 2 | |||
| PA14_67270 | PA5094 | 0.99 | ≥2 | ||||||||||
| PA14_67530 | PA5114 | 1.09 | ≥5 | 4 | 1 | 1 | |||||||
| PA14_67670 | PA5124 | ntrB | 0.93 | ≥5 | 4 | 1 | |||||||
| PA14_68580 | PA5192 | pckA | 1.05 | 2 | 1 | 1 | 1 | 1 | |||||
| PA14_68610 | PA5193 | hslO | 0.78 | ≥5 | ≥5 | 1 | 1 | ||||||
| PA14_68680 | PA5199 | envZ | 1.06 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||
| PA14_68730 | PA5203 | gshA | 0.59 | 2 | |||||||||
| PA14_68800 | PA5208 | 0.91 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_69000 | PA5224 | pepP | 0.72 | 2 | 1 | 2 | 1 | 1 | 1 | ||||
| PA14_69170 | PA5238 | 0.89 | 1 | 1 | ≥2 | ||||||||
| PA14_69270 | PA5246 | 0.87 | ≥5 | 1 | |||||||||
| PA14_70860 | PA5369 | 0.84 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_71630 | PA5427 | adhA | 1.10 | 1 | 1 | 1 | ≥2 | 1 | |||||
| PA14_71700 | PA5433 | 1.09 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_72140 | PA5466 | 1.11 | 1 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_73200 | PA5551 | 1.19 | 1 | 1 | 1 | ≥2 | 1 | ||||||
| PA14_73370 | PA5565d | gidA | 0.62 | 2 | |||||||||
| PA14_73370 | PA5565d | gidA | 0.78 | 1 | 1 | 2 | 1 | 1 | |||||
| PA14_73410 | PA5568 | 0.81 | 2 | 1 | 1 | 1 | 1 | ||||||
| PA14_14420 | 1.03 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_15540 | 1.13 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_15600 | 0.93 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_18070 | 0.92 | 2 | 2 | 1 | 2 | ≥2 | 1 | 1 | |||||
| PA14_23420 | 0.83 | 2 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_23430 | 1.21 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_23460 | orfN | 0.92 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_28520 | 0.91 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_43090 | 0.91 | 1 | 1 | 1 | 2 | 1 | 1 | ||||||
| PA14_44230 | 1.19 | 1 | 1 | 1 | ≥2 | 1 | |||||||
| PA14_51950 | 1.04 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_54750 | 1.17 | 1 | 1 | 1 | ≥2 | 1 | |||||||
| PA14_58760 | pilC | 0.73 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_66100 | 1.08 | 1 | 1 | 1 | ≥2 | 1 | 1 | ||||||
| PA14_30210d | clpS | 1.20 | 3 | 4 | 1 | ≥2 | 3 | 1 | |||||
| PA14_30210d | clpS | 1.18 | 3 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_60280d | fimU | 0.82 | 2 | 1 | 1 | ≥2 | 1 | 1 | |||||
| PA14_60280d | fimU | 0.80 | 1 | 1 | 1 | ≥2 | 1 | ||||||
Only those mutants for which at least a fourfold increase in the MIC was obtained are listed; only positive MIC changes are displayed. *, failed measurement.
Maximum growth rate of the mutant growth control relative to the growth of the wild type.
The MIC for the wild-type PA14 strain was below the calling range (Fig. 1); the change in the MIC was calculated based on an assumed MIC for the wild type of 0.5 times the lower limit of the calling range.
Independent strains with the transposon in different positions within the gene.
Nineteen of those mutants were retested for antibiotic resistance using the agar dilution method with PIP, MEM, CIP, TET, CAZ, and TOB, and we also confirmed the increased resistance profile with the respective independent isolates from the PAO1 transposon mutant library (Table 3).
TABLE 3.
Retesting of mutants exhibiting increased resistance
| PA14 gene locus | PAO1 ortholog | Gene name | Log2 change in the MICa of:
|
||||
|---|---|---|---|---|---|---|---|
| PIPb | MEMb | CAZ | CIPb | TOBb | |||
| PA14_01100 | PA0090 | ≥5d,e | 1 | ||||
| PA14_51880 | PA0958 | oprD | 3d | ||||
| PA14_50250 | PA1095 | ≥5d,e | 1d | ||||
| PA14_46840 | PA1348 | 1d,e | 3d | 1 | |||
| PA14_44440 | PA1549 | 2d,e | 2d | 1d,e | |||
| PA14_38350 | PA2023 | galU | ≥5d,e | 1d | 2d | ||
| PA14_29290 | PA2693 | ≥5d,e | 2 | 2 | |||
| PA14_22480 | PA3224 | 2d,e | |||||
| PA14_16500 | PA3702 | wspR | ≥5d,e | 1d | |||
| PA14_14470 | PA3831 | pepA | 2d,e | 1d,e | |||
| PA14_10800 | PA4109 | ampR | 3d,e | 3d | 1d | ||
| PA14_57540 | PA4429c | 2 | 2 | 1d | |||
| PA14_57570 | PA4431 | 1d,e | 1d | 1d | |||
| PA14_58260 | PA4490 | 2d,e | 1e | ||||
| PA14_60860 | PA4600 | nfxB | 3d,e | ||||
| PA14_66600 | PA5038 | aroB | 4d,e | 3d | 1d | ||
| PA14_68610 | PA5193 | hslO | ≥5d,e | ||||
| PA14_18070 | —c | 2 | 1d | 1d | |||
| PA14_30210 | —c | clpS | 3d,e | 3d | |||
Only positive MIC changes are displayed.
The MIC for the wild-type PA14 strain was below the calling range (Fig. 1); the MIC change was calculated based on an assumed MIC for the wild type of 0.5 times the lower limit of the calling range.
No PAO1 transposon mutant available. —, no PAO1 ortholog.
MIC change consistent with agar dilution testing of the PA14 mutant with the same antibiotic.
MIC change consistent with agar dilution testing of the PAO1 mutant with the same antibiotic.
As expected, in our Vitek 2 screen we were able to identify various genes that have previously been reported to be involved in the alteration of the bacterial susceptibility profile. Among these genes were nfxB, whose inactivation has been shown to lead to MexCD-OprJ-overproducing efflux mutants exhibiting resistance to gyrase inhibitors and FEP (36); oprD, involved in carbapenem resistance (34); ampR, encoding the transcriptional regulator of chromosomal AmpC beta-lactamase in P. aeruginosa (3); and lpxC. lpxC mutants exhibit increased resistance to CTX, and LpxC inhibitors have been demonstrated to show antibiotic properties. A recently discovered LpxC inhibitor was proven to control the growth of E. coli and P. aeruginosa with an efficacy rivaling that of CIP (5). We furthermore identified a galU mutant exhibiting increased cephalosporin and PIP resistance. A galU mutant was previously identified in a screen for aminoglycoside resistance (15), and it has been speculated that loss of the A- and B-band polysaccharides due to the impaired conversion of glucose-1-phosphate to UDP glucose (14) results in reduced antibacterial activity due to a lack of aminoglycoside binding to the bacterial cell (23). Interestingly, we also identified a wbpM mutant that exhibits increased resistance toward the cephalosporins and PIP. Insertional inactivation of wbpM has been shown to result in mutants exhibiting three distinct LPS phenotypes (14), suggesting that an altered LPS phenotype affects susceptibility not only to aminoglycosides but also to beta-lactam antibiotics.
We furthermore identified mutS and mutL mutants that exhibited increased resistance profiles. Mutation within mutS or mutL has been reported to lead to hypermutable phenotypes exhibiting a 100- to 1,000-fold-increased rate of spontaneous mutation (31). These mutants show enhanced acquisition of antibiotic resistance through the accumulation of secondary mutations and are frequently found in chronic lung infections in cystic fibrosis (12, 31). The radA, PA1767, PA5001, and ahpF mutants also exhibited antibiotic resistance in our screen; all of them were picked in a previous screen of the PA14 mutant library for decreased susceptibility to different antibiotics (CIP, TOB, and CAZ). Among those genes, only radA has been shown to be involved in the expression of an enhanced mutation frequency (45).
Aminoglycoside antibiotics must traverse the bacterial cytoplasmic membrane prior to initiating lethal effects, a process that seems to be dependent on the proton motive force (8, 13, 41). Thus, resistance may be the result of a decreased aminoglycoside uptake. We found that a strain with a mutation in a gene encoding a protein involved in the production and maturation of cytochrome c oxidase (PA1549) exhibited enhanced resistance to tobramycin. The respective PA14 transposon mutant has already been identified in a global screen for novel genetic determinants of aminoglycoside resistance (40). The reason that the screen by Schurek et al. identified a significantly larger number of mutants (40) might be that the tobramycin MIC for the wild-type PA14 strain was below the calling range in the present study, and therefore small MIC changes might have been missed.
Inactivation of the nuoABDEFGHIJKLMN operon, coding for the proton translocating type I NADH oxidoreductase, did not result in enhanced resistance to TOB, as observed by Schurek et al. (40); however, we found enhanced resistance to the cephalosporins (especially CTX) and PIP.
Concluding remarks.
One approach to producing a new generation of useful antimicrobial compounds involves sensitization of the bacteria to existing antibiotics by identifying targets for increasing susceptibility. Our finding that not only the loss of efflux pumps but also the loss of, e.g., OprF entails pronounced hypersusceptibility to various antibiotics belonging to different classes identifies OprF as a possible target candidate for the development of an attractive chemosensitizer.
We acknowledge that the approach of screening a transposon mutant library for mutants exhibiting increased resistance profiles is far from being comprehensive. Only nonessential genes can be identified, and this screen will miss small mutations or a combination of several mutations that lead to the evolution of antibiotic-resistant strains in the clinical setting. Nevertheless, screening of the Harvard PA14 transposon mutant library proved to be sufficient to detect many genes already predicted to play a role in P. aeruginosa resistance, and we identified a large number of previously unknown genes that modified the expression of resistance to many different antibiotics and that might turn out to be targets of clinical relevance. As the technology of high-throughput sequencing and comparative genome hybridization evolves very rapidly, it will be a very challenging task for the near future to identify novel genetic markers of antibiotic resistance and modifiers of resistance expression via genomewide association studies and to combine these data with the “resistome” data from our study and other studies. The prediction of the antibiotic resistance profiles of clinical strains based on their genotypes will be a major advance for the rapid and reliable detection of antimicrobial resistance in modern microbiological diagnostics and will contribute significantly to the efficient control of multiresistance.
Acknowledgments
Perfect technical assistance by Katharina Smaluch is gratefully acknowledged. We thank Jörg Overhage for helpful discussions.
The financial support of the Helmholtz-Gemeinschaft is gratefully acknowledged. A.D. is a recipient of a predoctoral stipend provided by the DFG-sponsored International Research Training Group “Pseudomonas: Pathogenicity and Biotechnology”.
Footnotes
Published ahead of print on 30 March 2009.
REFERENCES
- 1.Abe, S., T. Okutsu, H. Nakajima, N. Kakuda, I. Ohtsu, and R. Aono. 2003. n-Hexane sensitivity of Escherichia coli due to low expression of imp/ostA encoding an 87 kDa minor protein associated with the outer membrane. Microbiology 149:1265-1273. [DOI] [PubMed] [Google Scholar]
- 2.Alice, A., H. Naka, and J. Crosa. 2008. Global gene expression as a function of the iron status of the bacterial cell: influence of differentially expressed genes in the virulence of the human pathogen Vibrio vulnificus. Infect. Immun. 76:4019-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagge, N., O. Ciofu, M. Hentzer, J. Campbell, M. Givskov, and N. Hoiby. 2002. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob. Agents Chemother. 46:3406-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balke, B., L. Hoy, H. Weissbrodt, and S. Haussler. 2004. Comparison of the Micronaut Merlin automated broth microtiter system with the standard agar dilution method for antimicrobial susceptibility testing of mucoid and nonmucoid Pseudomonas aeruginosa isolates from cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 23:765-771. [DOI] [PubMed] [Google Scholar]
- 5.Barb, A., and P. Zhou. 2008. Mechanism and inhibition of LpxC: an essential zinc-dependent deacetylase of bacterial lipid A synthesis. Curr. Pharm. Biotechnol. 9:9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bratu, S., D. Landman, J. Gupta, and J. Quale. 2007. Role of AmpD, OprF and penicillin-binding proteins in beta-lactam resistance in clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 56:809-814. [DOI] [PubMed] [Google Scholar]
- 7.Breidenstein, E., B. Khaira, I. Wiegand, J. Overhage, and R. Hancock. 2008. A complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob. Agents Chemother. 52:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan, L., and S. Kwan. 1983. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob. Agents Chemother. 23:835-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberland, S., F. Malouin, H. Rabin, T. Schollaardt, T. R. Parr, Jr., and L. Bryan. 1990. Persistence of Pseudomonas aeruginosa during ciprofloxacin therapy of a cystic fibrosis patient: transient resistance to quinolones and protein F-deficiency. J. Antimicrob. Chemother. 25:995-1010. [DOI] [PubMed] [Google Scholar]
- 10.Cheong, H., C. Kang, Y. Wi, E. Kim, J. Lee, K. Ko, D. Chung, N. Lee, J. Song, and K. Peck. 2008. Clinical significance and predictors of community-onset Pseudomonas aeruginosa bacteremia. Am. J. Med. 121:709-714. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, H., T. Lin, and J. Wang. 2007. Identification and characterization of an organic solvent tolerance gene in Helicobacter pylori. Helicobacter 12:74-81. [DOI] [PubMed] [Google Scholar]
- 12.Ciofu, O., B. Riis, T. Pressler, H. Poulsen, and N. Hoiby. 2005. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob. Agents Chemother. 49:2276-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damper, P., and W. Epstein. 1981. Role of the membrane potential in bacterial resistance to aminoglycoside antibiotics. Antimicrob. Agents Chemother. 20:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, C., and J. Goldberg. 2002. Pseudomonas aeruginosa galU is required for a complete lipopolysaccharide core and repairs a secondary mutation in a PA103 (serogroup O11) wbpM mutant. FEMS Microbiol. Lett. 210:277-283. [DOI] [PubMed] [Google Scholar]
- 15.El'Garch, F., K. Jeannot, D. Hocquet, C. Llanes-Barakat, and P. Plesiat. 2007. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 51:1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fajardo, A., N. Martinez-Martin, M. Mercadillo, J. Galan, B. Ghysels, S. Matthijs, P. Cornelis, L. Wiehlmann, B. Tummler, F. Baquero, and J. Martinez. 2008. The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 3:e1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiki, Y., S. Fowler, H. Shio, A. Hubbard, and P. Lazarow. 1982. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J. Cell Biol. 93:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez, R., H. Tao, J. Purvis, S. York, K. Shanmugam, and L. Ingram. 2003. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant). Biotechnol. Prog. 19:612-623. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh, N., H. Wakebe, E. Yoshihara, T. Nakae, and T. Nishino. 1989. Role of protein F in maintaining structural integrity of the Pseudomonas aeruginosa outer membrane. J. Bacteriol. 171:983-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harbarth, S., J. Garbino, J. Pugin, J. Romand, D. Lew, and D. Pittet. 2003. Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am. J. Med. 115:529-535. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahim, E., G. Sherman, S. Ward, V. Fraser, and M. Kollef. 2000. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118:146-155. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs, M., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadurugamuwa, J., J. Lam, and T. Beveridge. 1993. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effect. Antimicrob. Agents Chemother. 37:715-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kintz, E., and J. Goldberg. 2008. Regulation of lipopolysaccharide O antigen expression in Pseudomonas aeruginosa. Future Microbiol. 3:191-203. [DOI] [PubMed] [Google Scholar]
- 25.Kohanski, M., D. Dwyer, B. Hayete, C. Lawrence, and J. Collins. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797-810. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberati, N., J. Urbach, S. Miyata, D. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomovskaya, O., M. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, D., M. Schurr, M. Mudd, and V. Deretic. 1993. Differentiation of Pseudomonas aeruginosa into the alginate-producing form: inactivation of mucB causes conversion to mucoidy. Mol. Microbiol. 9:497-506. [DOI] [PubMed] [Google Scholar]
- 30.Nelson, M., and S. Levy. 1999. Reversal of tetracycline resistance mediated by different bacterial tetracycline resistance determinants by an inhibitor of the Tet(B) antiport protein. Antimicrob. Agents Chemother. 43:1719-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 32.Peng, X., C. Xu, H. Ren, X. Lin, L. Wu, and S. Wang. 2005. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Pseudomonas aeruginosa responding to ampicillin, kanamycin, and tetracycline resistance. J. Proteome Res. 4:2257-2265. [DOI] [PubMed] [Google Scholar]
- 33.Piddock, L., M. Hall, F. Bellido, M. Bains, and R. Hancock. 1992. A pleiotropic, posttherapy, enoxacin-resistant mutant of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirnay, J., D. De Vos, D. Mossialos, A. Vanderkelen, P. Cornelis, and M. Zizi. 2002. Analysis of the Pseudomonas aeruginosa oprD gene from clinical and environmental isolates. Environ. Microbiol. 4:872-882. [DOI] [PubMed] [Google Scholar]
- 35.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poole, K., N. Gotoh, H. Tsujimoto, Q. Zhao, A. Wada, T. Yamasaki, S. Neshat, J. Yamagishi, X. Li, and T. Nishino. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21:713-724. [DOI] [PubMed] [Google Scholar]
- 37.Pumbwe, L., M. Everett, R. Hancock, and L. Piddock. 1996. Role of gyrA mutation and loss of OprF in the multiple antibiotic resistance phenotype of Pseudomonas aeruginosa G49. FEMS Microbiol. Lett. 143:25-28. [DOI] [PubMed] [Google Scholar]
- 38.Rawling, E., F. Brinkman, and R. Hancock. 1998. Roles of the carboxy-terminal half of Pseudomonas aeruginosa major outer membrane protein OprF in cell shape, growth in low-osmolarity medium, and peptidoglycan association. J. Bacteriol. 180:3556-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, L. B. 2007. Emerging issues in the management of infections caused by multidrug-resistant gram-negative bacteria. Cleve. Clin. J. Med. 74(Suppl. 4):S12-S20. [DOI] [PubMed] [Google Scholar]
- 40.Schurek, K., A. Marr, P. Taylor, I. Wiegand, L. Semenec, B. Khaira, and R. Hancock. 2008. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:4213-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taber, H., J. Mueller, P. Miller, and A. Arrow. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamae, C., A. Liu, K. Kim, D. Sitz, J. Hong, E. Becket, A. Bui, P. Solaimani, K. Tran, H. Yang, and J. Miller. 2008. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190:5981-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallés, J., J. Rello, A. Ochagavia, J. Garnacho, and M. Alcala. 2003. Community-acquired bloodstream infection in critically ill adult patients: impact of shock and inappropriate antibiotic therapy on survival. Chest 123:1615-1624. [DOI] [PubMed] [Google Scholar]
- 44.Wessel, D., and U. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 45.Wiegand, I., A. Marr, E. Breidenstein, K. Schurek, P. Taylor, and R. Hancock. 2008. Mutator genes giving rise to decreased antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:3810-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood, J. M., E. Bremer, L. N. Csonka, R. Kraemer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A 130:437-460. [DOI] [PubMed] [Google Scholar]
- 47.Woodruff, W., and R. Hancock. 1988. Construction and characterization of Pseudomonas aeruginosa protein F-deficient mutants after in vitro and in vivo insertion mutagenesis of the cloned gene. J. Bacteriol. 170:2592-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodruff, W., and R. Hancock. 1989. Pseudomonas aeruginosa outer membrane protein F: structural role and relationship to the Escherichia coli OmpA protein. J. Bacteriol. 171:3304-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, L., O. Estrada, O. Zaborina, M. Bains, L. Shen, J. Kohler, N. Patel, M. Musch, E. Chang, Y. Fu, M. Jacobs, M. Nishimura, R. Hancock, J. Turner, and J. Alverdy. 2005. Recognition of host immune activation by Pseudomonas aeruginosa. Science 309:774-777. [DOI] [PubMed] [Google Scholar]