Abstract
Escherichia coli clonal group A (CGA) commonly exhibits a distinctive multidrug antimicrobial resistance phenotype—i.e., resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, tetracycline, and trimethoprim (ACSSuTTp)—and has accounted for up to 50% of trimethoprim-sulfamethoxazole-resistant E. coli urinary tract infections in some locales. Annotation of the whole-genome sequencing of UMN026, a reference CGA strain, clarified the genetic basis for this strain's ACSSuTTp antimicrobial resistance phenotype. Most of the responsible genes were clustered in a unique 23-kbp chromosomal region, designated the genomic resistance module (GRM), which occurred within a 105-kbp genomic island situated at the leuX tRNA. The GRM is characterized by numerous remnants of mobilization and rearrangement events suggesting multiple horizontal transfers. Additionally, comparative genomic analysis of the leuX tRNA genomic island in 14 sequenced E. coli genomes showed that this region is a hot spot of integration, with the presence/absence of specific subregions being uncorrelated with either the phylogenetic group or the pathotype. Our data illustrate the importance of whole-genome sequencing in the detection of genetic elements involved in antimicrobial resistance. Additionally, this is the first documentation of the blaTEM and dhfrVII genes in a chromosomal location in E. coli strains.
Emerging antimicrobial resistance among uropathogens increases the complexity of management for acute uncomplicated cystitis. The recently described Escherichia coli clonal group A (CGA) causes infections of the urinary tract and other extraintestinal sites in diverse host populations. CGA is associated with trimethoprim-sulfamethoxazole resistance and, although most highly prevalent in the United States, exhibits a worldwide distribution (6, 7, 12, 13). This clonal group has accounted for up to 50% of trimethoprim-sulfamethoxazole-resistant urinary tract infections (UTIs) due to E. coli in some U.S. locales (13), illustrating the importance of the dissemination of this clone.
Although CGA belongs to phylogenetic group D, which overall is less closely associated with extraintestinal virulence than group B2, CGA exhibits typical extraintestinal virulence traits (the F16 papA allele, papG allele II, iutA, kpsMII, traT, and ompT) and can be as pathogenic as classic group B2 strains in a mouse UTI model (7). Many CGA strains exhibit distinctive O antigens (O11, O17, O73, and O77) and a distinctive multidrug antimicrobial resistance phenotype: resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, tetracycline, and trimethoprim (i.e., ACSSuTTp) (6, 13). The finding of highly similar or indistinguishable pulsed-field gel electrophoresis profiles among isolates from multiple locations in the United States suggests a recent dissemination of CGA (13). This expansion may have been due to the combination of CGA's virulence potential and its antimicrobial resistance capabilities, in contrast to the historically noted negative association between resistance and virulence in E. coli (1, 5, 11).
Using an approach of whole-genome sequencing and comparative genomic analysis, the goals of this study were (i) to identify the genetic basis for the antimicrobial resistance of a representative CGA strain and (ii) to understand the evolutionary history of these genetic determinants.
MATERIALS AND METHODS
Bacterial strain.
Strain UMN026 is a CGA urine isolate from a woman with acute cystitis at the University of Minnesota Student Health Service in the late 1990s (8, 15). Consistent with its CGA background, UMN026 exhibits the O17 O antigen, a typical CGA-associated virulence gene profile that includes pap (P fimbriae, with the F16 papA variant and papG allele II), iutA (aerobactin receptor), and kpsMII (group 2 capsule), and the ACSSuTTp antimicrobial resistance profile.
Sequencing of the E. coli UMN026 genome.
The UMN026 genome was sequenced as part of a whole-genome sequencing project (ColiScope; www.genoscope.cns.fr) at the Genoscope Sequencing Center (Evry, France) (16). Sequencing and assembly of the UMN026 genome were performed as follows. The complete genome sequence of E. coli UMN026 was determined using the whole-genome shotgun method (10× coverage). Three libraries were made. Two plasmid libraries of 3 kbp and 10 kbp, obtained by mechanical shearing, were constructed in plasmid pcDNA2.1 (Invitrogen) and in home vector pCNS (pSU18 modified), respectively. One bacterial artificial chromosome library with an average insert size of 30 kbp was constructed by enzymatic digestion (HindIII) and ligation into pBeloBacII (CalTech). The Phred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment. To resolve contigs, sequence finishing was performed by PCR amplification, primer walking, and/or transposition.
Annotation of the E. coli UMN026 genome.
The MaGe (Magnifying Genome) software program (17) was used for gene annotation and comparative analysis of the UMN026 genome. Using the AMIGene (annotation of microbial genes) (17) program, open reading frames (ORFs) were predicted (assigned a unique identifier prefixed with ECUMN_) and submitted to automatic functional annotation, including gene order conservation analysis (i.e, assessment of conservation of the chromosomal colocalization between pairs of orthologous genes from different genomes). Manual validation of the automatic annotation was performed only on the UMN026 genes/regions without orthologues in the E. coli K-12 genome, with the help of the MaGe interface, which allows graphic visualization of the annotations, enhanced by a synchronized representation of synteny groups in other genomes chosen for comparisons (17). Sequence data for comparative analyses were obtained from the NCBI databases (ftp://ftp.ncbi.nlm.nih.gov). Annotations of the chromosomal genomic resistance module (GRM [see below]) gene cluster described in this paper range from ECUMN_4803 (intB) to ECUMN_4832 (a transposase gene fragment) and are available in public databanks (accession no. CU928163).
Identification of subregions (modules) within the leuX tRNA genomic islands.
The genomic islands located at the leuX tRNA were defined using the synteny breaks between 14 E. coli strains. Of these, 10 were pathogenic strains, including UMN026 and IAI39 (two extraintestinal pathogenic group D strains), E2348/69 (an enteropathogenic group B2 strain), S88, APECO1, CFT073, UTI89, and 536 (five extraintestinal pathogenic group B2 strains), 042 (an enteroaggregative group D strain), O157:H7 Sakaï (an enterohemorrhagic group E strain), and 55989 (an enteroaggregative group B1 strain), whereas 4 were commensal strains, including ED1a (a group B2 strain), IAI1 (a group B1 strain), and K-12 and HS (two group A strains). It appeared that these genomic islands have a composite structure: e.g., they are made up of regions that are partially conserved but are found in different synteny groups (i.e., in different genomic locations) in the different E. coli strains. Thus, the predicted genomic islands were manually divided into subregions (or modules) which are found in only a subset of the compared E. coli strains.
RESULTS AND DISCUSSION
The UMN026 genome includes a chromosome of 5,202,090 bp with 5,086 ORFs, plus two plasmids, p1ECUMN, comprising 122,301 bp for 157 ORFs, and p2ECUMN comprising 33,809 bp for 49 ORFs (16). All but one of the resistance genes corresponding to the strain's characteristic ACSSuTTp phenotype were identified within a single chromosomal region. The sole exception, resistance to tetracycline, was attributable to both the mar operon, located on the chromosome at bp 1,827,757 (origin upstream of thrA), which enhances impermeability to tetracycline, and the tet operon, located on plasmid p1ECUMN, which encodes an efflux pump (see Table S1 in the supplemental material). In contrast, all of the remaining resistance genes, i.e., those coding for resistance to chloramphenicol, trimethoprim, streptomycin, sulfonamides, and ampicillin, are clustered in a unique 105,152-bp chromosomal region located at bp 4977946 (Fig. 1; and see Table S2 in the supplemental material). Within this chromosomal region, the resistance genes are located in the upstream 20%, within a 22,530-bp region that we named the “genomic resistance module” (GRM).
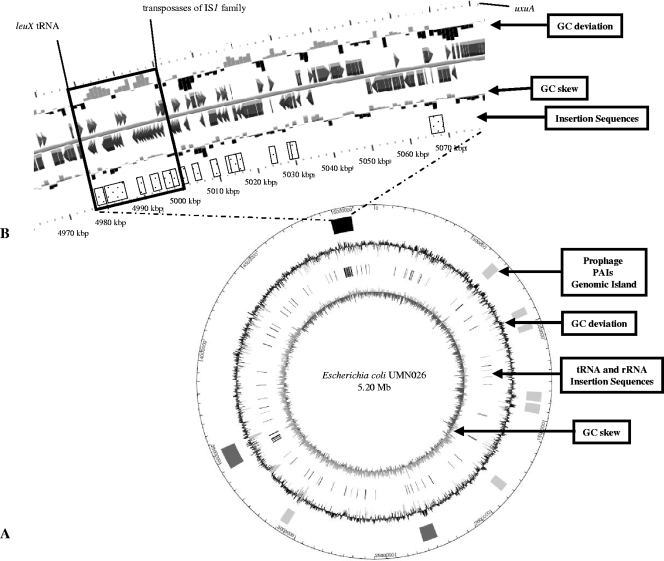
FIG. 1.
Representation of the 105-kbp genomic island of Escherichia coli strain UMN026, bearing the GRM. (A) Circular representation of the E. coli UMN026 chromosome. From the center outward, the concentric circles display the following: (i) GC skew (G + C/G − C using a 1-kbp sliding window); (ii) locations of tRNAs and rRNAs (gray) and insertion sequences (dark gray); (iii) GC deviation (mean GC content in a 1-kbp window − overall mean GC), with gray areas indicating that the deviation is higher than 2 standard deviations; (iv) locations of the genomic islands corresponding to prophage regions (light gray), PAIs (dark gray), and the 105-kbp genomic island containing the 23-kbp GRM described in this study (black); and (v) scale, in which 1 graduation corresponds to 50 kbp. (B) Enlargement of the 105-kbp genomic island. The first and the sixth lines represent the 2-kbp window scale. The second and fifth lines indicated the GC deviation and GC skew in a 1-kbp window. On the third and the fourth lines, genes annotated manually are indicated by arrows. Genes annotated with another E. coli genome serving as the reference genome are represented by rectangles. Insertion sequences are represented in dotted boxes. The leuX tRNA is indicated, as well as the transposases of the IS1 family and the uxuA gene (which flank the downstream part of the GRM and the genomic island, respectively). The plain box indicates the GRM.
The UMN026 GRM encompasses 29 ORFs (25 complete and 4 partial), of which 8 are predicted to be associated with resistance to antimicrobial agents and 2 with resistance to antiseptics or heavy metals. Genes responsible for five of the six constituent resistance markers of the ACSSuTTp phenotype are present, including the chloramphenicol acetyltransferase gene, cat; the dhfrVII trimethoprim resistance gene, encoding a dihydrofolate reductase; the adenyl transferase gene, aadA, conferring resistance to streptomycin; the sulfonamide resistance gene, sul1, encoding a dihydropteroate synthetase, and the blaTEM gene, encoding a penicillinase. Additionally, three genes belong to the mph cluster and encode a phosphotransferase that confers high-level resistance to erythromycin (MIC, >256 mg/liter [data not shown]). Likewise, two genes associated with resistance to antiseptics or heavy metals are present: the ebr gene, which confers ethidium bromide resistance; and a putative chromate transporter gene, which has been defined based on protein domains and could induce resistance to chromate (Fig. 2). This finding illustrates the importance of whole-genome sequencing in the elucidation of the genetic underpinnings of antimicrobial resistance, as previously reported for the plasmidic region in E. coli SMS-3-5, a multidrug-resistant environmental group D strain (4), and for the genomic island reported in an Acinetobacter baumanii strain (3).
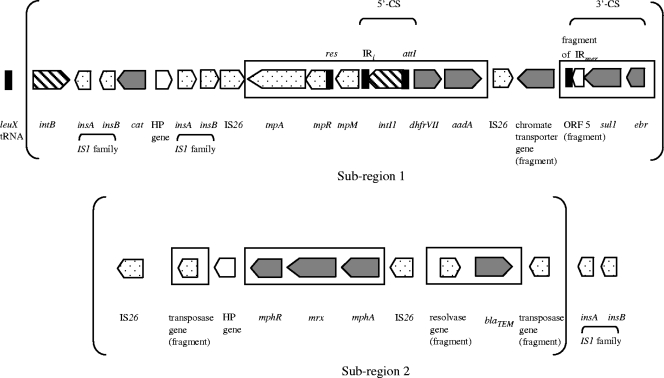
FIG. 2.
Representation of the genes annotated in the Escherichia coli UMN026 GRM. ORF categories are indicated as follows. Genes coding for resistance to antibiotics, antiseptics, or heavy metals are in gray, transposition elements are dotted, integrases have diagonal lines, and a hypothetical protein (HP) is in white. Boxes indicate the rearranged Tn21-like element. The partly inverted repeats (IRs), as well as the res and attI sites of the Tn21-like element and of the class 1 integron, respectively, are indicated in black. The ORFs and noncoding sequences are not presented to scale. Subregions 1 and 2 are indicated within brackets (Fig. 4).
The larger 105-kbp chromosomal region itself can be classified as a genomic island (2) based on the following four characteristics. First, the region's size is over 10 kbp. Second, although its overall G+C content does not vary statistically from the rest of the genome (50% versus 50.7%), its GC deviation (i.e., the mean GC content in a 1-kbp window − the overall mean GC) is frequently higher than 2 standard deviations (Fig. 1). Third, it is located next to a tRNA gene (leuX). Fourth, it includes mobility genes such as integrases, transposases, resolvases, and phage-like elements (Fig. 1; and see Table S2 in the supplemental material).
The GRM, located in the upstream part of this genomic island, is characterized by a high G+C content (55% versus 50.7% for the whole genome, P < 0.001, using a chi-square test). The GRM is flanked upstream by the leuX tRNA gene, and downstream by transposases of the IS1 family (Fig. 1B; and see Table S2 in the supplemental material). The GRM includes many of these mobility-associated genes. We observed two integrase genes: i.e., intB, a phage-like element gene, and intI1, which codes for the integrase of class 1 integrons. Moreover, 10 transposases or transposase fragments, including four IS26 and two IS1 copies, are present (Fig. 2; and see Table S2 in the supplemental material). In addition, we discovered the presence of a rearranged Tn21-like element, with the transposition region comprising tnpA, tnpR, and tnpM, the resolution site (res), and, downstream, a class 1 integron. The class 1 integron contained the integrase intI1, the attI site, and resistance gene cassettes dhfrVII and aadA. The last genes of the integron—i.e., ebr, sul1, and a fragment of ORF5 (which corresponded to the 3′- conserved segment)—are inverted. However, flanking inverted repeats (IRs), as usually observed around transposons and integrons, are partly present. That is, IRtnp (which usually is localized in the upstream part of Tn21) is absent; only 30 of the 38 bp of IRmer (which usually is localized in the downstream part of Tn21) are present; and only IRi is found upstream of intI1 in the class 1 integron, whereas IRt, which usually is localized in the downstream part of class 1 integrons, is missing (Fig. 2).
The Tn21-like element of the GRM is homologous to other Tn21-like elements described in strains of Shigella flexneri (GenBank accession no. AF188331 [no corresponding publication]) and Aeromonas hydrophila (14), in which a macrolide inactivation gene cluster and blaTEM are associated with the Tn21-like element. However, the tni module and the mer operon, which occur in the above-described Tn21-like elements, are lacking in the Tn21-like element of the UMN026 GRM. The large number of transposases observed in this GRM could explain these rearrangements, which may have eliminated the mobility of the Tn21-like element by interruption or deletion of IRs. Additional elements involved in DNA mobilization, as well as the intB gene, flank the GRM, suggesting that the region has been intensively rearranged. This abundance of mobility elements may explain the observed chromosomal location of these resistance genes, which usually are found in plasmids. Indeed, to our knowledge, this represents the first report in E. coli/Shigella strains of a chromosomal location for dhfrVII, the mph cluster, a chromate resistance gene, and blaTEM (Fig. 1B and 2).
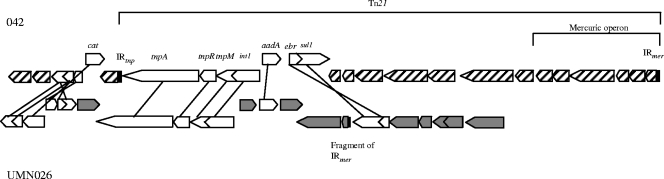
A search for the presence of these GRM genes in other organisms was performed with BLAST, using a criterion of nucleotide sequence similarities greater than 90%. Most of the GRM genes, i.e., 27 (93%), were present in other E. coli/Shigella, genomes, in either a plasmid or chromosomal location (see Table S1 in the supplemental material). Additionally, 11 (38%) of the genes were shared with E. coli 042, a group D diarrheal disease isolate, in which they were localized elsewhere in the chromosome (Fig. 3). Hypotheses regarding the evolutionary history of UMN026 can be derived by noting that another strain bears, in a different chromosomal location, some genes of subregion 1 of the GRM (see Table S2 in the supplemental material). Indeed, E. coli 042, shares a part of the resistance phenotype of CGA, including resistance to chloramphenicol, streptomycin, sulfonamides, and tetracycline (i.e., CSSuT [data not shown]). All of the genes involved in this phenotype are present in strain 042, but clustered elsewhere in the chromosome within the complete Tn21 (Fig. 3). This suggests a possibility of multiple recent events of acquisition of this resistance phenotype in these two virulent, but pathotypically distinct, group D strains, indicating a selective pressure in their natural niche.
FIG. 3.
Representation of the ORFs of the UMN026 genomic resistance module that are shared with Escherichia coli strain 042. In both strains, the corresponding regions are situated on the chromosome. Shared ORFs are indicated in white, whereas 042- and UMN026-specific ORFs are represented by diagonal lines and gray shading, respectively. IRs are represented in black.
Despite the use of a fairly strict similarity criterion (i.e., 90% identity), we found 21 of the GRM genes (73%) in Salmonella enterica, 15 (52%) in other Enterobacteriaceae, 5 (17%) in Haemophilus spp., 2 (7%) in Neisseria spp., 1 (3%) in Pasteurella spp., 3 (10%) in Corynebacterium spp., 3 (10%) in Streptococus spp., and 1 (3%) in Staphylococcus aureus. As for environmental species, 13 (45%) of the GRM genes were present in Acinetobacter baumanii, 7 (24%) in Pseudomonas aeruginosa or other Pseudomonas spp., 7 (23%) in Aeromonas spp. including Aeromonas hydrophila, 2 (6%) in Achromobacter spp., and 1 (3%) in Stenotrophomonas maltophilia (see Table S1 in the supplemental material). Finding of homolog genes in other, phylogenetically distant species suggests multiple horizontal transfers allowed by the numerous mobility-associated genes described above.
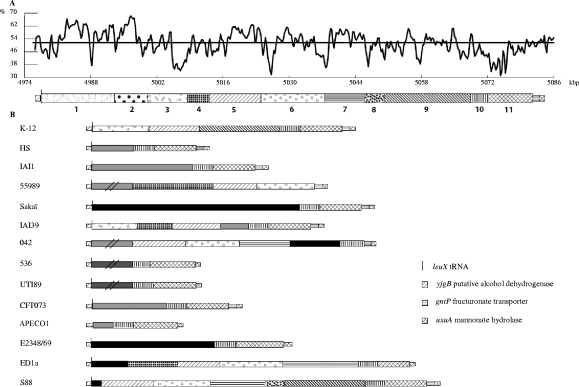
We next compared the entire 105-kbp leuX tRNA genomic island of UMN026 to the sequences occurring at the same chromosomal location in 14 other sequenced E. coli genomes (Fig. 4). As previously reported (16), we find that the leuX tRNA region was a hot spot of insertion. The E. coli genomic regions inserted at the leuX tRNA have a composite structure; indeed, a total of 11 subregions were defined, the first and second of which corresponded to the UMN026 GRM (Fig. 2). The composition of these subregions (i.e., a listing of the constituent genes with their functional descriptions) is available in Table S2 in the supplemental material. The presence/absence in different strains of specific subregions is uncorrelated by eye either to phylogenetic group or pathotype (Fig. 4). For example, parts of the KpLE2 phage-like element (i.e., subregions 3, 5, and 9 [Fig. 4; and see Table S2 in the supplemental material]) are present in the commensal-derived laboratory strain K-12, from phylogenetic group A, but are not found in commensal strains HS and IAI1, from phylogenetic groups A and B1, respectively. Likewise, two other subregions—i.e., regions 10 (which contains hypothetical proteins and N-acetylneuraminic acid outer membrane channel protein) and 11 (which encompasses the fim operon coding for fimbrial proteins)—are found at this insertion point in all analyzed E. coli strains, except for the enteroaggregative E. coli strains 55989 (group B1) and 042 (group D). Moreover, apart from the GRM region, the organization of the genomic island subregions in UMN026 (group D, pathogenic) is similar to that of phylogenetic group B2 strains S88 and ED1a, which are pathogenic (newborn meningitis) and commensal strains, respectively. Interestingly, at the same position downstream of leuX tRNA and intB, in strains UTI89 and 536, which are two group B2 strains associated with UTI, pathogenicity island II (PAI II) is present (Fig. 4). PAI II comprises such virulence-associated genes as hek and the pap operon (which contribute to adherence); hly (which lyses erythrocytes and impairs phagocyte function); and, in strain UTI89, cnf1 (which is cytotoxic for epithelial cells). Furthermore, the leuX tRNA region has been demonstrated to be involved in the virulence attenuation of asymptomatic bacteriuria strains (18). Indeed, in some asymptomatic bacteriuria strains, the end of the leuX tRNA genomic island encompassing the fim operon is lacking or exhibits point mutations that inactivate the fim operon, which is known to be involved in adherence (18). These observations underscore the importance of the leuX tRNA-associated region in the plasticity of the genome by modulating the acquisition and loss of genes of virulence or resistance.
FIG. 4.
Genomic islands at the leuX tRNA insertion hot spot in 15 Escherichia coli strains. This region was defined using the synteny breaks between 15 pathogenic and commensal E. coli strains from various phylogenetic groups, including CGA strain UMN026. In all of the E. coli strains, the gene just before the leuX tRNA gene corresponds to the ECK4262 gene (yjgB, coding for a putative alcohol dehydrogenase), whereas the beginning of the core genome immediately after the leuX tRNA gene corresponds to the ECK4313 gene (uxuA, coding for mannonate hydrolase). In strains APECO1 and 042, the leuX tRNA gene is absent. (A) The genomic island from UMN026, with its GC deviation (mean GC content in a 500-bp window − overall mean GC of complete genome [50.7%; indicated by the horizontal line]) is represented by the GC content (%) along the position in kbp (origin at the thrA gene). The organization of the various modules is also indicated along the genomic island. A total of 11 subregions (for which the complete list of genes and their functional descriptions are available in Table S2 in the supplemental material) have been defined, with the first and second of these corresponding to the GRM (Fig. 2) and the last to the fim operon (crossed lines). (B) Organization of the subregions in the 14 compared E. coli strains. Homologous subregions have the same geometric code. Black subregions are strain specific. Gray subregions are found in other strains but at another genomic location. (The ones containing double black slashes are not drawn to scale since they are much longer.) Dark gray subregions indicate PAI II in strains 536 and UTI89.
UMN026 was selected as the representative CGA strain for genome sequencing (16) because of its typical CGA-associated O17 antigen, virulence profile, and ACSSuTTp resistance profile. To clarify the generalizability of the present findings, it will be useful in the future to assess how broadly distributed the UMN026 GRM is within CGA, as well as within non-CGA lineages that are closely related (e.g., ST394) or more distantly related (e.g., the O15:K52:H1 clonal group) (9) to CGA, including isolates with resistance profiles similar or dissimilar to that of UMN026. Likewise, in view of previous evidence that the ACSSuTTp phenotype of certain CGA and O15:K52:H1 strains is conjugally transferable to laboratory strains (6, 10), it will be relevant to assess possible conjugal transfer of the GRM.
In summary, annotation of the whole-genome sequence of strain UMN026 allowed us to define the genetic underpinnings of this strain's antimicrobial resistance. Most of the responsible genes were clustered within a unique 23-kbp chromosomal region, named the GRM, which occurred within a 105-kbp genomic island situated at the leuX tRNA locus. Comparative genome analysis showed that this genomic island is located at a hot spot of gene acquisition and loss within the chromosome. This illustrates the plasticity of E. coli genomes, which supports multiple adaptive paths (16) that allow, at a single locus, the alternative presence of resistance or virulence genes (1, 5, 11).
Supplementary Material
Acknowledgments
M.L. was funded by the “Fondation pour la Recherche Médicale,” and C.H. was supported by the “Agence Nationale de la Recherche” ANR-05-JC-0136-01. J.R.J. was supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs.
Footnotes
Published ahead of print on 13 April 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Branger, C., O. Zamfir, S. Geoffroy, G. Laurans, G. Arlet, H. V. Thien, S. Gouriou, B. Picard, and E. Denamur. 2005. Genetic background of Escherichia coli and extended-spectrum beta-lactamase type. Emerg. Infect. Dis. 11:54-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 3.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fricke, W. F., M. S. Wright, A. H. Lindell, D. M. Harkins, C. Baker-Austin, J. Ravel, and R. Stepanauskas. 2008. Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J. Bacteriol. 190:6779-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson, J. R., P. Goullet, B. Picard, S. L. Moseley, P. L. Roberts, and W. E. Stamm. 1991. Association of carboxylesterase B electrophoretic pattern with presence and expression of urovirulence factor determinants and antimicrobial resistance among strains of Escherichia coli that cause urosepsis. Infect. Immun. 59:2311-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson, J. R., A. R. Manges, T. T. O'Bryan, and L. W. Riley. 2002. A disseminated multidrug-resistant clonal group of uropathogenic Escherichia coli in pyelonephritis. Lancet 359:2249-2251. [DOI] [PubMed] [Google Scholar]
- 7.Johnson, J. R., A. C. Murray, M. A. Kuskowski, S. Schubert, M. F. Prere, B. Picard, R. Colodner, and R. Raz. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, J. R., K. Owens, A. Gajewski, and M. A. Kuskowski. 2005. Bacterial characteristics in relation to clinical source of Escherichia coli isolates from women with acute cystitis or pyelonephritis and uninfected women. J. Clin. Microbiol. 43:6064-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson, J. R., K. L. Owens, C. R. Clabots, S. J. Weissman, and S. B. Cannon. 2006. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 8:1702-1713. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. R., A. L. Stell, T. T. O'Bryan, M. Kuskowski, B. Nowicki, C. Johnson, J. N. Maslow, A. Kaul, J. Kavle, and G. Prats. 2002. Global molecular epidemiology of the O15:K52:H1 extraintestinal pathogenic Escherichia coli clonal group: evidence of distribution beyond Europe. J. Clin. Microbiol. 40:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, J. R., C. van der Schee, M. A. Kuskowski, W. Goessens, and A. van Belkum. 2002. Phylogenetic background and virulence profiles of fluoroquinolone-resistant clinical Escherichia coli isolates from The Netherlands. J. Infect. Dis. 186:1852-1856. [DOI] [PubMed] [Google Scholar]
- 12.Manges, A. R., P. S. Dietrich, and L. W. Riley. 2004. Multidrug-resistant Escherichia coli clonal groups causing community-acquired pyelonephritis. Clin. Infect. Dis. 38:329-334. [DOI] [PubMed] [Google Scholar]
- 13.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 14.Poole, T. L., T. R. Callaway, K. M. Bischoff, C. E. Warnes, and D. J. Nisbet. 2006. Macrolide inactivation gene cluster mphA-mrx-mphR adjacent to a class 1 integron in Aeromonas hydrophila isolated from a diarrhoeic pig in Oklahoma. J. Antimicrob. Chemother. 57:31-38. [DOI] [PubMed] [Google Scholar]
- 15.Sannes, M. R., M. A. Kuskowski, and J. R. Johnson. 2004. Antimicrobial resistance of Escherichia coli strains isolated from urine of women with cystitis or pyelonephritis and feces of dogs and healthy humans. J. Am. Vet. Med. Assoc. 225:368-373. [DOI] [PubMed] [Google Scholar]
- 16.Touchon, M., C. Hoede, O. Tenaillon, V. Barbe, S. Baeriswyl, P. Bidet, E. Bingen, S. Bonacorsi, C. Bouchier, O. Bouvet, A. Calteau, H. Chiapello, O. Clermont, S. Cruveiller, A. Danchin, M. Diard, C. Dossat, M. El Karoui, E. Frapy, L. Garry, J. M. Ghigo, A. M. Gilles, J. Johnson, C. Le Bouguenec, M. Lescat, S. Mangenot, V. Martinez-Jehanne, I. Matic, X. Nassif, S. Oztas, M. A. Petit, C. Pichon, Z. Rouy, C. Saint Ruf, D. Schneider, J. Tourret, B. Vacherie, D. Vallenet, C. Medigue, E. P. Rocha, and E. Denamur. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallenet, D., L. Labarre, Z. Rouy, V. Barbe, S. Bocs, S. Cruveiller, A. Lajus, G. Pascal, C. Scarpelli, and C. Medigue. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34:53-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zdziarski, J., C. Svanborg, B. Wullt, J. Hacker, and U. Dobrindt. 2008. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect. Immun. 76:695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.