Abstract
In view of the threat of the potential use of variola virus in a terrorist attack, considerable efforts have been performed to develop new antiviral strategies against orthopoxviruses. Here we report on the use of RNA interference, either alone or in combination with cidofovir, as an approach to inhibit orthopoxvirus replication. Two selected small interfering RNAs (siRNAs), named siB1R-2 and siG7L-1, and a previously reported siRNA, i.e., siD5R-2 (which targets the viral D5R mRNA), were evaluated for antiviral activity against vaccinia virus (VACV) by plaque reduction and virus yield assays. siB1R-2 and siG7L-1, administered before or after viral infection, reduced VACV replication by more than 90%. Also, these two siRNAs decreased monkeypox virus replication by 95% at a concentration of 1 nM. siB1R-2 and siG7L-1 were demonstrated to specifically silence their corresponding transcripts, i.e., B1R and G7L mRNAs, without induction of a beta interferon response. Strong synergistic effects were observed when siB1R-2, siG7L-1, or siD5R-2 was combined with cidofovir. In addition, the antiviral activities of these three siRNAs were evaluated against VACV resistant to cidofovir and other acyclic nucleoside phosphonates. siG7L-1 and siD5R-2 remained active against four of five VACV mutants, while siB1R-2 showed activity against only one of the mutants. Our results showed that siRNAs are potent inhibitory agents in vitro, not only against wild-type VACV but also against several cidofovir-resistant VACV. Furthermore, we showed that a combined therapy using siRNA and cidofovir may be useful in the treatment of poxvirus infections.
Following the eradication of smallpox in the late 1970s, vaccination against smallpox was discontinued, resulting in an increasing number of nonimmunized individuals in today's population, who are susceptible to poxvirus infections. Currently, the potential release of the etiological agent of smallpox, variola virus (VARV), by bioterrorists (5) has prompted renewed interest in the development of safer smallpox vaccines, as well as new therapeutic molecules that inhibit poxvirus replication. Today, there are no U.S. Food and Drug Administration-approved drugs for the treatment of smallpox, and only vaccinia virus immunoglobulins are available for complications related to smallpox vaccination. Cidofovir [(S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine (HPMPC)], approved for treatment of cytomegalovirus retinitis in AIDS patients, is permitted for use as an emergency treatment in the case of a smallpox outbreak (13). Several in vitro and in vivo studies have indeed demonstrated the inhibitory potency of cidofovir against poxviruses by interfering with their viral DNA polymerase (16). However, its use in humans would be limited since the compound needs to be administered intravenously, with a risk of nephrotoxicity (37, 38). Recently, a compound with a novel mechanism of action, ST-246 [4-trifluoromethyl-N-(3,3a,4,4a,5,5a,6,6a-octahydro-1,3-dioxo-4,6-ethenocycloprop[f] isoindol-2(1H)-yl-benzamide], proved to be a potent and specific inhibitor of orthopoxvirus egress in vitro (72). In addition, orally given ST-246 protected mice from lethal orthopoxvirus challenge (54) and nonhuman primates from lethal monkeypox virus (MPXV) challenge. This orally bioavailable molecule also demonstrated its safety and tolerability in a phase I clinical trial (33) and is currently being developed further.
However, there is still a need to enlarge the armamentarium of antiviral molecules active against poxviruses with a different and original mechanism of action. The selection of new antipoxviral compounds with different targets could also be useful in the potential case of emergence of drug resistance to the reference antipoxvirus agents (4, 35, 62). In this context, RNA interference (RNAi) represents a promising strategy (60). Numerous studies have demonstrated the antiviral efficacy of small interfering RNAs (siRNAs), not only in cell cultures but also in animal models, against several viruses associated with human diseases, including human immunodeficiency virus, hepatitis C and B viruses (28, 30, 31, 36, 42, 43, 45, 46, 73), severe acute respiratory syndrome (SARS) coronavirus (39), herpes simplex virus (53), influenza virus (26, 66), parainfluenza virus, respiratory syncytial virus (10, 17), and Ebola virus (27). Previous studies also reported the inhibitory effect of siRNAs against orthopoxvirus replication in vitro by targeting the E3L, F11L, and D5R genes (15, 67, 68). The Poxviridae family comprises large double-stranded DNA viruses (47). The Orthopoxvirus genus includes VARV, MPXV, vaccinia virus (VACV; used as a smallpox vaccine), and cowpox virus (CPXV). Orthopoxviruses carry approximately 200 open reading frames, which can be divided into early, intermediate, and late gene classes, with expression of each gene class dependent upon prior expression of proteins of the preceding class. We focused our present work on the inhibition of expression of an early gene (B1R) and a late gene (G7L) that are essential for VACV replication. The B1 protein is involved in viral DNA replication, and the G7 protein is required for virus morphogenesis.
B1 is an essential serine/threonine protein kinase that is present in infecting virions (7, 56) and is required for viral DNA synthesis, as shown by the phenotype of two temperature-sensitive mutants expressing a very labile B1 protein without kinase activity (11, 55, 56). The mechanism through which B1 contributes to DNA replication remains elusive, as the protein does not appear to phosphorylate any of the known components of the viral replication machinery (11). So far, the B1 substrates identified are viral and cellular proteins. B1 phosphorylates the viral H5 protein, which is a late transcription factor (11), and cellular ribosomal proteins, as well as the cellular DNA-binding protein named BAF (50) and p53 proteins (57). Recent data suggested that the poxviral B1 protein has evolved to usurp defense signaling pathways of the host cell. Indeed, it was shown that phosphorylation of BAF or p53 mediated by B1 could contribute to the survival of infected cells, thus allowing the course of the infection to progress (57, 71).
The late, 42-kDa G7 protein is a component of the core of the intracellular mature virus required for the stability and maintenance of virosomes as well as for the attachment and uptake of virosomal material by viral crescents to form immature virions (IV) (44). Phenotypic analyses of G7 mutant viruses have shown that G7 is essential for VACV morphogenesis. Inhibition of G7 protein expression had no effect on the synthesis of viral proteins, but the proteolytic processing of certain structural proteins was severely inhibited. It has been shown that such inhibition of proteolytic processing is commonly associated with a block of VACV morphogenesis at a stage prior to the formation of intracellular mature virions (64).
In the present study, we demonstrated the inhibitory activities of two siRNAs, i.e., siB1R-2 and siG7L-1, against VACV in cell cultures. We further analyzed the antiviral effects of these siRNAs and of a previously described siRNA which targets the D5R transcript of VACV (68), used in combination with the reference compound cidofovir. Moreover, we evaluated the antiviral potencies of these three siRNAs against five VACV strains bearing mutations in the viral DNA polymerase gene (E9L) (4, 25) which are known to confer resistance to cidofovir and other acyclic nucleoside phosphonates (ANPs). Our results give novel insights into the inhibition of orthopoxvirus replication in cell cultures and highlight the antiviral potential of RNAi.
MATERIALS AND METHODS
Cells and viruses.
A549 cells (human lung carcinoma; ATCC CCL-185) were grown in F12K medium (Gibco, Invitrogen Corporation, United Kingdom) containing 10% heat-inactivated fetal calf serum (FCS). Human embryonic lung (HEL) fibroblasts (HEL-299; ATCC CCL-137) were cultured in Earle's minimal essential medium (Gibco, Invitrogen Corporation, Paisley, United Kingdom) supplemented with 10% FCS, 1% l-glutamine, 1% nonessential amino acids, 1% sodium pyruvate, and 1% HEPES at 37°C in a 5% CO2 atmosphere. The following viral strains were used: VACV strain Western Reserve (VACV-WR), MPXV strain Copenhagen (MPXV-Cop; kindly provided by Hermann Meyer, Bundeswehr Institute of Microbiology, Munich, Germany), and CPXV strain Brighton (CPXV-BR; ATCC VR 302). The drug-resistant VACV-WR strains bearing single mutations in the viral DNA polymerase gene (E9L) were A314T, A684V, and S851Y mutants, and those harboring double mutations were A314T+A684V and A684V+S851Y mutants. These amino acid changes have been shown to occur following selection with cidofovir and/or HPMPDAP {(S)-9-[3-hydroxy-2-(phosphonomethoxy)propyl]-2,6-diaminopurine} (4, 25).
Compounds.
The sources of compounds were as follows: cidofovir (HPMPC), Gilead Sciences, Foster City, CA; and HPMPDAP, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Prague, Czech Republic.
Design and synthesis of siRNAs.
The genome sequence of VACV-WR (based on GenBank accession no. AY243312) was used as a template for the design of siRNAs targeting B1R and G7L mRNAs. siRNAs were double-stranded RNAs of 21 nucleotides containing dTdT overhangs at both 3′ ends, according to the rules suggested by Elbashir and collaborators (20). siRNA duplexes were chemically synthesized by Qiagen (Courtaboeuf, France) or Applied Biosystems (Courtaboeuf, France). The antiviral effects of four individual B1R- and G7L-specific siRNAs were tested, and one siRNA for each targeted gene, i.e., siB1R-2 for the B1R gene and siG7L-1 for the G7L gene, was selected based on antiviral potency against VACV-WR replication. The siRNA sequences were as follows: siB1R-2, 5′-AAAGGTGGATTCGGTAGTATT-3′ (sense strand; corresponds to positions 163948 to 163969 of the GenBank VACV-WR complete genome); and siG7L-1, 5′-GCGTCGTTCTACAATTTTT-3′ (sense strand; corresponds to positions 51652 to 51671 of the GenBank VACV-WR complete genome). A previously described siRNA, siD5R-2, which targets the viral D5 protein and inhibits VACV replication, was also included in this study (68). The siControl nontargeting siRNA (siNT) (Perbio Science, Brebieres, France), which does not have any significant homology to the orthopoxvirus genomes or to known gene sequences of mice, rats, or humans, was used in this study as a negative control. The siControl TOX (siTOX) siRNA (Perbio Science, Brebieres, France) was used to optimize the relative siRNA uptake and to control transfection efficiency.
Antiviral assays. (i) A549 cells.
The siRNAs were added to approximately 60% confluent A549 cell monolayers grown in 24-well microtiter plates. siRNAs (siB1R-2, siG7L-1, and siNT) were used at various concentrations, ranging from 1 to 100 nM, and mixed with the transfection reagent Lipofectamine 2000 (Invitrogen, Cergy Pontoise, France). The siRNA-Lipofectamine complex was then incubated for 20 min at room temperature, and 100 μl of this mixture was added per well. At 24 hours posttransfection, cells were washed twice and infected with VACV-WR, CPXV-BR, or MPXV-Cop at a multiplicity of infection (MOI) of 0.1. Following an incubation period of 2 h at 37°C, the inoculum was removed and the cells were washed twice and further incubated in F12K medium containing 0.4% FCS. Cells were harvested at 24 h postinfection, and viral titers were determined. For therapeutic studies, 60% confluent A549 cells were infected with VACV-WR at an MOI of 0.0001 for 1 hour. The inoculum was then removed, and cells were washed twice and incubated with fresh medium. Transfections of siNT, siB1R-2, or siG7L-1 were then performed at 1 h, 8 h, or 24 h postinfection by adding 100 μl of the transfection mixture (containing 100 nM of the siRNA of interest with 1.5 μl of Lipofectamine) to each well. At 24 hours posttransfection, cells were washed, cultured in F12K medium with 0.4% FCS, and harvested at 48 h postinfection. For all experiments performed with A549 cells, virus titers were determined in Vero cells as previously described (23). Transfection efficiency was monitored in each experiment by using siTOX, and approximately 90% of the cells were successfully transfected.
(ii) HEL cells.
The antiviral activities of the siRNAs against VACV-WR and the five drug-resistant viruses listed above were evaluated on HEL cells. Cells were plated at a density of 1 × 104 cells per well in 96-well microtiter plates and then cultured for 1 day. Seventy percent confluent HEL cell monolayers were then transfected in duplicate with the siRNA of interest, i.e., siB1R-2, siG7L-1, siD5R-2, or siNT, at various concentrations ranging from 1 nM to 100 nM. Thirty microliters of siRNA-Lipofectamine mixture was added to each well. One day after transfection, cells were washed twice and infected with each viral strain at an MOI of 0.01. At 1 hour postinfection, residual virus was removed, and cells were washed twice and immediately replenished with fresh medium (transfected wells) or with medium containing serial dilutions of cidofovir or HPMPDAP (nontransfected wells). After 2 days, HEL cells were fixed with ethanol and stained with Giemsa solution. Viral cytopathic effect (CPE) was recorded using a scale of 0 to 5 (where 5 equals 100% CPE and 0 is no CPE), and the 50% effective concentration (EC50) was defined as the concentration of compound required to reduce viral CPE by 50%. The EC50s of the siRNAs and the compounds tested against each strain were calculated as the means for three independent experiments. Of note, the CPE recorded for the siNT-transfected infected cells was similar to that for the nontransfected infected cells. Transfection efficiency was determined in each experiment by using siTOX, and approximately 90% of the cells were successfully transfected.
Combination assays.
HEL cells were seeded 1 day prior to transfection at a density of 1 × 104 cells per well in 96-well microtiter plates. At the time of transfection, the cells were approximately 70% confluent. Lipofectamine 2000 was mixed with various concentrations of the siRNA of interest. At 24 hours posttransfection, cells were washed twice and infected with VACV-WR at an MOI of 0.01. At 1 hour postinfection, the inoculum was removed, and cells were washed twice. Serial dilutions of cidofovir were then added to each concentration of the siRNA of interest. After 2 days at 37°C, the CPE was recorded as described for the antiviral assays. The interpretation of the combination assays with HEL cells was done as described below. Of note, the CPE recorded for the siNT-transfected infected cells was similar to that for the nontransfected infected cells. For the combination assays done with A549 cells grown in 24-well microtiter plates, siRNAs (siD5R-2, siB1R-2, and siG7L-1) were used at a concentration of 1 nM. At 24 hours posttransfection, cells were washed twice and infected with VACV-WR at an MOI of 0.0001. At 2 hours postinfection, the inoculum was removed as described above, and transfected or nontransfected cells were cultured in the absence or presence of 31.75 μM cidofovir. Cells were harvested at 48 h postinfection, and the antiviral effect of the combination of cidofovir and the siRNA of interest on VACV-WR growth was determined by virus titers determined with Vero cells due to the fact that we cannot observe CPE in A549 cells. The virus titers recorded for the siNT-transfected infected cells were similar to that for the nontransfected infected cells.
Interpretation of drug interactions in combinations.
The effects of treatment of VACV-WR-infected cells with cidofovir and siRNA, alone or in combination, were analyzed according to the isobologram method (12, 24, 29, 58, 59, 70). For each drug combination, the EC50s were determined. The fractional inhibitory concentration (FIC) was calculated by use of the following formulas: FICsiRNA = EC50 (siRNA combined)/EC50 (siRNA alone) and FICcidofovir = EC50 (cidofovir combined)/EC50 (cidofovir alone). The combined effects of the two drugs were assessed by calculation of the FIC index (FICi), as previously described (2, 21, 32, 65), from the graphic interpretation of isobolograms. Curves were generated by pairing the data points of FICsiRNA with those of FICcidofovir. The diagonal line connecting the FICsiRNA of 1 on the ordinate axis with the FICcidofovir of 1 on the abscissa axis corresponds to the line of additivity (unity line). Also, the FICi was obtained by adding both FIC values (FICcidofovir + FICsiRNA). A synergistic effect, resulting in a stronger antiviral effect than the sum of the individual effects, was interpreted as follows: (i) FIC values falling below the unity line on the isobolograms or (ii) FICi of ≤0.5. An additive effect, computed as the sum of the individual effects of the drugs, was interpreted as follows: (i) FIC values falling on the unity line or (ii) 0.5 ≥ FICi ≤ 2. An antagonistic effect, where the two compounds in combination have an overall effect that is less than that predicted from their individual effects (60, 71), was interpreted as follows: (i) FIC values falling above the unity line or (ii) FICi of >2. The FICi values were calculated from three independent combination experiments.
Cytotoxicity assays.
The toxicities of the siRNAs and the compounds on HEL cells were evaluated based on inhibition of cell growth. Cells were seeded into 96-well microtiter plates at a density of 1 × 104 cells in 150 μl of medium per well. After 1 day, cells were transfected with serial dilutions of the siRNA-Lipofectamine mixture. At 1 day posttransfection, cells were washed twice and replaced with fresh medium or with medium containing serial dilutions of cidofovir or HPMPDAP. Following 3 days of incubation, the cells were trypsinized, and the number of cells was determined as previously described (19). The cytotoxic effects of the molecules were expressed as the 50% cytostatic concentration, defined as the concentration required to reduce cell growth by 50%.
Relative quantification by RT-PCR.
A549 cells (60% confluent) grown in 24-well microtiter plates were transfected with 100 nM of siB1R-2 or siG7L-1 for 24 h and infected with VACV-WR at an MOI of 3 for 3 h (siB1R-2) or 10 h (siG7L-1). Total mRNAs were extracted using Trizol reagent (Invitrogen) following the manufacturer's instructions and then were treated with amplification-grade DNase I (Invitrogen). Real-time one-step reverse transcription-PCR (RT-PCR) was carried out by using a TaqMan Gold RT-PCR kit, including the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control reagent (Applied Biosystems, Courtaboeuf, France). Primer and probe sequences were as follows: B1R forward primer, 5′-AATCAATGGGTCGTTGGACCAT-3′; B1R reverse primer, 5′-AATACATCATTTTTATCTCGGGTTTCGATTGC-3′; B1R MGB probe, 5′-CCTTTTCCACCTAAGCC-6-carboxyfluorescein-3′; G7L forward primer, 5′CTGCAGAACAGCGTCGTT-3′; G7L reverse primer, 5′-TGTATCTCAGGTTTCGATTTGTTAACAC-3′; and G7L MGB probe, 5′-TGTTAAAAACTGTATCAAAGTTT-6-carboxyfluorescein-3′. RT-PCR was performed using an ABI Prism 7000 SDS instrument following the manufacturer's instructions (Applied Biosystems). Results were analyzed with ABI Prism 7000 SDS relative quantification study software. Levels of relative quantities of B1 and G7 mRNAs were normalized with GAPDH.
ELISA for IFN-β analysis.
A549 cells were seeded into 24-well plates 1 day before transfection. Cells were transfected with 100 nM (1.6 μg/ml) of the siRNA of interest (siNT, siG7L-1, or siB1R-2) or with 0.5 μg/ml of poly(I-C) (Sigma-Aldrich, Saint-Quentin Fallavier, France), each of which was mixed with Lipofectamine. At 24 hours posttransfection, cell culture supernatants were collected, and human beta interferon (IFN-β) was measured by a sandwich enzyme-linked immunosorbent assay (ELISA) kit (PBL Biomedical Laboratories Tebu-Bio, Le Perray-en-Yvelines, France) following the manufacturer's instructions.
Statistical analysis.
Differences in the inhibitory effects of the siRNA of interest against virus replication compared to the negative control (siNT-transfected infected cells) were tested for statistical significance by using Student's t test. P values of <0.05 were considered to indicate significant differences between the two groups.
RESULTS
Specific knockdown of B1R and G7L transcripts by siB1R-2 and siG7L-1 in A549 cells.
We investigated the inhibitory potencies of two siRNAs, i.e., siB1R-2 and siG7L-1, against VACV-WR replication. For that purpose, real-time RT-PCR was performed in order to demonstrate the specific knockdown of B1R and G7L transcripts. A549 cells were transfected with 100 nM of siB1R-2, siG7L-1, or siNT, which is a functional nontargeting siRNA used as a negative control for potential off-target effects caused by siRNA transfection. Twenty-four hours later, transfected cells were infected with VACV-WR at an MOI of 3. Total mRNAs were extracted at 3 h and at 10 h postinfection for siB1R-2- and siG7L-1-treated cells, respectively. Notably, these two time points correspond to the peaks of production of B1R and G7L mRNAs following viral infection (22, 64). Relative quantification of the B1R and G7L steady-state mRNA levels was then performed. As shown in Fig. 1, siB1R-2 and siG7L-1 induced significant reductions of B1R and G7L gene transcripts (96% and 92% inhibition, respectively) compared to siNT (P < 0.05). These results confirmed the specific targeting of B1R and G7L mRNAs by their respective siRNAs. Also, no significant differences in levels of B1R and G7L mRNAs were observed between nontransfected infected cells (virus control) and siNT-transfected infected cells.
FIG. 1.
Specific knockdown of B1R and G7L transcripts by siB1R-2 and siG7L-1 in A549 cells. Cells were transfected with siB1R-2, siG7L-1, or siNT (100 nM) and infected 24 h later with VACV-WR (MOI of 3). Real-time RT-PCR was performed as described in Materials and Methods. GAPDH was included as an endogenous reference and used for normalization. Error bars represent the standard deviations (SD) of the means of triplicates for at least two independent experiments. *, siRNAs differ significantly from the siNT control (Student's t test; P < 0.05). There was no significant difference between siNT-transfected infected cells and nontransfected infected cells.
Concentration-dependent inhibition of VACV-WR replication by RNAi in A549 cells.
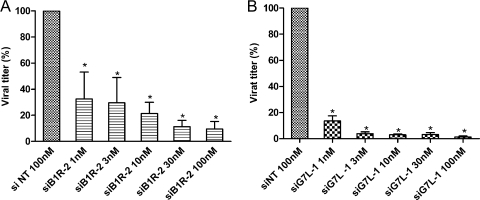
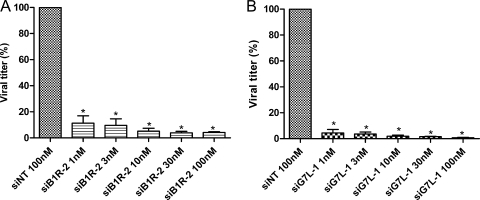
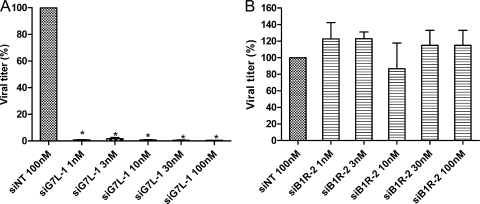
We then evaluated the prophylactic inhibitory potencies of siB1R-2 and siG7L-1 against VACV-WR replication. A549 cells were transfected for 1 day with siB1R-2 or siG7L-1 and then infected with VACV-WR. Following 24 h of incubation, virus production in cultures treated with siRNA concentrations ranging from 1 nM to 100 nM were determined and compared to the virus production obtained in cultures treated with 100 nM of siNT. As illustrated in Fig. 2, siB1R-2 and siG7L-1 showed a concentration-dependent inhibitory effect compared to siNT. siB1R-2 inhibited virus replication by 68% at 1 nM and by 91% at 100 nM (P < 0.05) (Fig. 2A). The virus yields were also significantly decreased, by 86% and 97% at 1 and 100 nM of siG7L-1, respectively, compared to that with siNT (P < 0.05) (Fig. 2B). Thus, both siRNAs exhibited significant antiviral activity at concentrations as low as 1 nM against VACV-WR replication, and they inhibited virus replication in a dose-dependent manner.
FIG. 2.
Dose-dependent inhibition of VACV-WR replication by specific siRNAs in A549 cells. Various concentrations of siB1R-2 (A) and siG7L-1 (B) (1, 3, 10, 30, and 100 nM) or siNT (100 nM) were transfected into A549 cells. Twenty-four hours later, cells were infected with VACV-WR at an MOI of 0.1 and were harvested at 24 h postinfection. Viral titers were determined as described in Materials and Methods. Error bars represent the SD of the means of quadruplicates for at least two independent experiments. *, siB1R-2 or siG7L-1 differs significantly from the siNT control (Student's t test; P < 0.05). There was no significant difference between siNT-transfected infected cells and nontransfected infected cells.
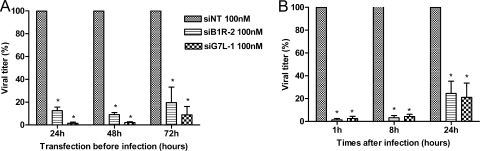
Long-term antiviral potencies of siB1R-2 and siG7L-1 in A549 cells.
In order to examine the duration of siRNA-mediated prophylactic activity, a time course study was conducted. A549 cells were infected with VACV-WR at 24 h, 48 h, or 72 h posttransfection with siB1R-2 or siG7L-1, and virus yields were determined at day 1 postinfection. As shown in Fig. 3A, transfection performed 24 h before infection led to a significant inhibition of virus production (88% and 99% inhibition at a concentration of 100 nM of siB1R-2 and siG7L-1, respectively). A significant reduction of VACV-WR replication was also observed when viral infection was performed 48 h following siRNA transfection (i.e., 91% [siB1R-2] and 98% [siG7L-1] inhibition). Both siB1R-2 and siG7L-1 remained active if infection occurred at 72 h posttransfection, affording decreases in virus production of 81% and 91%, respectively (P < 0.05). Thus, the antiviral activity mediated by these two siRNAs is preserved for at least 72 h following transfection of cells.
FIG. 3.
Prophylactic and therapeutic effects of siRNAs in A549 cells. (A) siRNAs maintain viral inhibition over an extended period. A549 cells were infected with VACV-WR (MOI, 0.1) 24, 48, or 72 h after transfection with 100 nM of siB1R-2 or siG7L-1, and samples were harvested at 24 h postinfection. Error bars represent the SD of the means of quadruplicates for at least two independent experiments. *, P < 0.05 (siRNAs differ significantly from the siNT control) (Student's t test). (B) Therapeutic effects of siB1R-2 and siG7L-1. Cells were infected with VACV-WR (MOI = 0.0001) and transfected with 100 nM of siB1R-2, siG7L-1, or siNT at 1, 8, or 24 h postinfection. Samples were harvested at 48 h postinfection, and viral titers were determined in cell culture. Error bars represent the SD of the means of quadruplicates for at least two independent experiments. *, P < 0.05 (siRNAs differ significantly from the siNT control) (Student's t test). There was no significant difference between siNT-transfected infected cells and nontransfected infected cells.
Therapeutic antiviral effects of siRNAs against VACV-WR-infected A549 cells.
Considering a possible use of siRNAs in vivo, we explored the therapeutic inhibitory potencies of siB1R-2 and siG7L-1. A549 cells were infected with VACV-WR at an MOI of 0.0001 and transfected with 100 nM of the siRNA of interest at 1 h, 8 h, or 24 h postinfection. siB1R-2 decreased virus replication by 98% (P < 0.05) when cells were transfected at 1 h postinfection (Fig. 3B). Antiviral effects were still observed when siB1R-2 was added at 8 h and 24 h postinfection, with 97% and 75.5% reductions, respectively, in virus yield (P < 0.05).
A similar inhibitory profile was observed with siG7L-1 (97%, 96%, and 79% reductions in virus titer when the cells were treated at 1 h, 8 h, and 24 h postinfection, respectively) (P < 0.05). Thus, both siRNAs induced a significant antiviral effect when administered to cell cultures for up to 24 h postinfection.
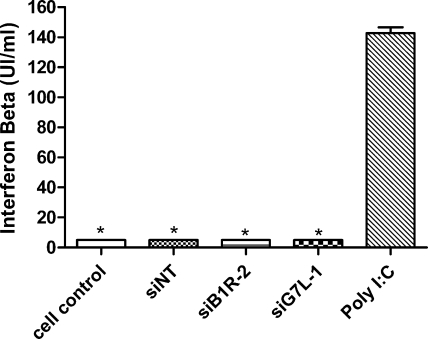
Absence of IFN-β induction in A549 cell cultures following siRNA transfection.
It has been reported that siRNAs can stimulate the IFN pathway under certain circumstances (34, 61). Therefore, we examined whether siB1R-2 and siG7L-1 were able to induce IFN-β production in uninfected A549 cell monolayers. Supernatants were collected at 24 h posttransfection from siRNA-treated A549 cells and analyzed by ELISA for IFN-β induction. As depicted in Fig. 4, 100 nM (1.6 μg/ml) of siNT, siB1R-2, or siG7L-1 failed to trigger an IFN-β response in A549 cells, while 0.5 μg/ml of poly(I-C) induced a strong IFN response. These results demonstrated that siB1R-2 and siG7L-1 act in a highly sequence-specific manner without inducing an IFN-β response.
FIG. 4.
Absence of IFN-β induction in A549 cell cultures following siRNA transfection. IFN-β production levels were determined by ELISA for supernatants of nontransfected cells (cell control) and cells transfected with either siNT, siB1R-2, or siG7L-1 (100 nM = 1.6 μg/ml) or with poly(I-C) (0.5 μg/ml). Error bars represent the SD of the means of duplicates for at least two independent experiments. *, P < 0.05 [samples differ significantly from poly(I-C)] (Student's t test).
Antiviral activities of siB1R-2 and siG7L-1 against other human-pathogenic orthopoxviruses in A549 cells.
We further determined the antiviral potencies of siB1R-2 and siG7L-1 against MPXV-Cop and CPXV-BR growth. The sequences of the targeted genes, i.e., B1R and G7L of MPXV-Cop and CPXV-BR, were subjected to a BLAST search to compare their percentages of homology. The G7L gene sequence showed 98.9% identity between MPXV-Cop and CPXV-BR, and the B1R gene sequence revealed 96% identity. As shown in Fig. 5A, siB1R-2 decreased MPXV-Cop replication by 90% (1 nM) and 96% (100 nM) (P < 0.05). Also, the MPXV-Cop yield was significantly reduced, by 96% and 99% at 1 and 100 nM of siG7L-1, respectively, compared to that with siNT (Fig. 5B). In the case of CPXV-BR, siG7L-1 decreased the virus yield by 99% at a concentration as low as 1 nM (Fig. 6A). In contrast, siB1R-2 was inactive against CPXV-BR replication (Fig. 6B). The sequence of siB1R-2 was designed using the VACV-WR B1R gene as a template. However, alignment of siB1R-2 with the CPXV-BR B1R gene showed one nucleotide difference (C versus T), at position 10 in the 5′ end of the sense strand of siB1R-2. Thus, this change appeared to be responsible for the lack of antiviral activity of siB1R-2 against CPXV-BR (Fig. 6B).
FIG. 5.
Inhibition of MPXV by siB1R-2 and siG7L-1 in A549 cells. Cells were transfected with different concentrations (1, 3, 10, 30, or 100 nM) of siB1R-2 (A), siG7L-1 (B), or siNT (100 nM). At 24 hours posttransfection, cells were infected at an MOI of 0.1 with MPXV-Cop, and samples were harvested at 24 h postinfection. Viral titers were determined in cell culture. Error bars represent the SD of the means of quadruplicates for at least two independent experiments. *, P < 0.05 (siRNAs differ significantly from the siNT control) (Student's t test). There was no significant difference between siNT-transfected infected cells and nontransfected infected cells.
FIG. 6.
Inhibition of CPXV by siG7L-1 but not by siB1R-2 in A549 cells. Cells were transfected with various concentrations (1, 3, 10, 30, or 100 nM) of siG7L-1(A), siB1R-2 (B), or siNT (100 nM). At 24 hours posttransfection, cells were infected at an MOI of 0.1 with CPXV-BR, and samples were harvested at 24 h postinfection. Viral titers were determined in cell culture. Error bars represent the SD of the means of quadruplicates for at least two independent experiments. *, P < 0.05 (siRNAs differ significantly from the siNT control) (Student's t test). There was no significant difference between siNT-transfected infected cells and nontransfected infected cells.
Synergistic inhibitory effects of cidofovir and siRNAs against VACV-WR replication.
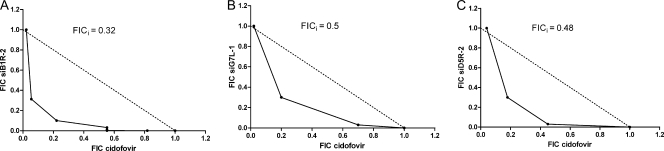
Both cidofovir and siRNAs are potent inhibitors of orthopoxvirus replication. Since these molecules have distinct targets and different mechanisms of action, we hypothesized that combined therapy could result in a synergistic effect. Therefore, the antiviral activity of siB1R-2, siG7L-1, or siD5R-2 (a previously described siRNA) in combination with cidofovir against VACV was determined. The antiviral effects were evaluated using a CPE reduction assay of HEL cells. Combination indexes were calculated using the isobologram method. The FICs obtained for cidofovir and for each siRNA were plotted on the x and y axes, respectively, to generate isobolograms (Fig. 7A to C). The antiviral effects of the different combinations were strongly synergistic against VACV-WR replication (Fig. 7A to C), while no increase in cytotoxicity was noted (data not shown). The mean FICi value calculated from three independent experiments was <0.5 for each combination of cidofovir and siRNA (Student's t test; P < 0.05) (Table 1).
FIG. 7.
siRNAs combined with cidofovir are strongly synergistic against VACV-WR replication in HEL cells. (A to C) Isobolograms showing the effect of a combination of cidofovir with either siB1R-2 (A), siG7L-1 (B), or siD5R-2 (C) against VACV-WR growth in HEL cells. The numbers on the axes represent normalized FIC values (see Materials and Methods). The diagonal line connecting the FICsiRNA of 1 on the ordinate axis with the FICcidofovir of 1 on the abscissa axis corresponds to the line of additivity (unity line). Experimental data points located below, on, and above the unity line indicate synergy, additivity, and antagonism, respectively. Combination measurements were done in three independent experiments, and the most representative data are shown.
TABLE 1.
Determination of FICi values in HEL cells
| Drug-siRNA combination | FICi
|
|||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Mean ± SD | |
| Cidofovir-siB1R-2 | 0.32 | 0.3 | 0.3 | 0.31 ± 0.01 |
| Cidofovir-siG7L-1 | 0.5 | 0.33 | 0.5 | 0.44 ± 0.10 |
| Cidofovir-siD5R-2 | 0.48 | 0.5 | 0.43 | 0.47 ± 0.04 |
The synergistic effect of these combinations was also confirmed in A549 cells. The antiviral effect of cidofovir (32 μM) combined with each siRNA (1 nM) was assessed by virus yield reduction assays with A549 cells. As shown in Table 2, a 1.1-log decrease in virus yield was observed when cidofovir was combined with siB1R-2, whereas only a 0.5-log reduction was seen with each compound alone. The cidofovir-siD5R-2 and cidofovir-siG7L-1 combinations resulted in 2.8- and 3.0-log reductions in VACV yield, respectively, compared to that with siNT (Student's t test; P < 0.05), whereas these molecules alone decreased virus titers by 0.5 log (cidofovir) and 1.2 log (siG7L-1 or siD5R-2) (Student's t test; P < 0.05). These experiments confirmed that the combination of cidofovir and siRNAs resulted in a greater inhibition of VACV-WR growth than that afforded by the sum of effects for each compound alone.
TABLE 2.
Antiviral activities of siRNAs in combination with cidofovir in A549 cellsa
| siRNA | Virus titer reduction (log10 TCID50/ml)b
|
Enhancement of cidofovir activitvc (log10 TCID50/ml) | Synergistic interactiond (log10 TCID50/ml) | |
|---|---|---|---|---|
| siRNA alone | siRNA in combination with cidofovir | |||
| siB1R-2 | 0.5 | 1.1 | +0.6 | +0.1 |
| siG7L-1 | 1.2 | 2.8 | +2.3 | +1.1 |
| siD5R-2 | 1.2 | 3.0 | +2.5 | +1.3 |
| 0.5 | ||||
Cells were transfected with the siRNA of interest (siD5R-2, siB1R-2, or siG7L-1) at a concentration of 1 nM. At 24 hours posttransfection, cells were infected with VACV-WR at an MOI of 0.0001 and then treated or not with cidofovir (32 μM). Samples were harvested at 48 h postinfection, and viral titers (log10 50% tissue culture infective dose [TCID50]/ml) were determined in cell culture.
Means of triplicate experiments.
Enhancement of cidofovir activity was calculated with the following formula: enhancement of cidofovir activity = (virus titer with combination of cidofovir and siRNA) − (virus titer with cidofovir alone).
The synergistic effect of the combination was calculated with the following formula: synergistic interaction = (viral titer with combination of cidofovir and siRNA) − (titer with siRNA alone + titer with cidofovir alone).
Antiviral activities of siRNAs against cidofovir-resistant VACV strains harboring specific mutations in the viral DNA polymerase gene.
We also explored whether siB1R-2, siG7L-1, and siD5R-2 (61) were able to inhibit the growth of five VACV strains bearing single or double mutations in the viral DNA polymerase gene (E9L). These mutations were previously described and have been associated with resistance to the reference antipoxvirus compound cidofovir and other ANPs, including HPMPDAP (4, 25). The efficacy of each siRNA against the different VACV-WR DNA polymerase mutants compared to that against the wild-type virus VACV-WR was evaluated in HEL cell cultures by CPE reduction assay. Cells were transfected 1 day before infection with the siRNA of interest or were treated at 1 hour postinfection with cidofovir or HPMPDAP. Virus-induced CPE was recorded at day 2 postinfection, and EC50s for the different siRNAs, cidofovir, and HPMPDAP were calculated for each viral strain (Table 3). The EC50s for cidofovir and HPMPDAP are in agreement with previously published data for these mutants and confirm their resistance profiles for cidofovir and/or HPMPDAP (4, 25). Mutant viruses bearing single mutations, i.e., A314T, A684V, and S851Y mutants, were less resistant to cidofovir and to HPMPDAP (mean EC50s of 147 μM and 37 μM, respectively) than those carrying double mutations (mean EC50s of >159 μM and 147.5 μM, respectively) and more resistant than the wild-type virus (mean EC50s of 34 μM for cidofovir and 9 μM for HPMPDAP). The EC50s obtained for siD5R-2 and siG7L-1 against the wild-type virus were 0.016 and 0.011 μM, respectively, whereas siB1R-2 proved to be less active, with an EC50 of 0.1 μM (Table 3). siRNAs exhibited three different spectra of antiviral activities (EC50s for mutant viruses were similar to, lower, or higher than that of the wild-type virus), depending on the amino acid substitution(s) in the viral DNA polymerase associated with resistance to the ANPs. As a general conclusion, it appears that siD5R-2 and siG7L-1 affected the replication of these viral mutants in similar ways. Thus, viruses bearing the A314T or S851Y change were more sensitive to the inhibitory action of these two siRNAs, with EC50s that were 3.2-fold (siD5R-2) and 1.6-fold (siG7L-1) lower for the A314T mutant and ∼5-fold lower for the S851Y mutant than for the wild-type virus. In contrast, the virus bearing the A684V change appeared to be less sensitive to siD5R-2 and siG7L-1 (EC50s were 2.5-fold and 6.4-fold higher, respectively, than those for the wild-type virus). Mutant viruses encoding the double substitutions (A314T+A684V and A684V+851Y) exhibited a susceptibility to siD5R-2 or siG7L-1 that was comparable to that of the wild-type strain (P > 0.05). In the case of siB1R-2, no inhibition of virus replication was observed with any of the mutants at the higher concentration of siRNA tested (i.e., 0.1 μM), except for the S851Y mutant, which displayed increased sensitivity (EC50 = 0.02 μM for the S851Y virus, compared to 0.1 μM for the wild-type virus). The presence of the S851Y mutation appeared to significantly enhance the antiviral activities of siD5R-2, siG7L-1, and siB1R-2, while the presence of the A684V mutation resulted in a decreased sensitivity to these siRNAs. In contrast, the A314T mutation affected the activities of these three siRNAs in a different way, i.e., caused increased sensitivity to siD5R-2 and siG7L-1 and decreased sensitivity to siB1R-2. On the other hand, the combination of the A314T+A684V and A684V+S851Y mutations seemed to abrogate the effects of each individual change and resulted in a null effect, since the antiviral activities observed with siD5R-2 and siG7L-1 against these double mutants were similar to that seen against the wild-type virus.
TABLE 3.
Antiviral activities of cidofovir, HPMPDAP, and siRNAs against different VACV-WR DNA polymerase mutants
| Compound | Antiviral activity (EC50)a (μM)
|
|||||
|---|---|---|---|---|---|---|
| VACV-WR | VACV-WR DNA polymerase mutantsb
|
|||||
| Single mutants
|
Double mutants
|
|||||
| A314T | A684V | S851Y | A314T+A684V | A684V+S851Y | ||
| Cidofovir | 34 ± 5 | 139* ± 17 | 151* ± 13 | 151* ± 8 | >159* ± 0 | >159* ± 0 |
| HPMPDAP | 9 ± 2 | 59* ± 26 | 32* ± 5 | 21* ± 5 | 138* ± 13 | >157* ± 0 |
| siD5R-2 | 0.016 ± 0.01 | 0.005* ± 0.001 | 0.04* ± 0.01 | 0.003* ± 0.0003 | 0.018 ± 0.004 | 0.012 ± 0.004 |
| siG7L-1 | 0.011 ± 0.005 | 0.007* ± 0.003 | 0.07* ± 0.03 | 0.002* ± 0.0008 | 0.02 ± 0.01 | 0.03 ± 0.02 |
| siB1R-2 | 0.1 ± 0.0 | >0.1 ± 0.0 | >0.1 ± 0.0 | 0.02* ± 0.008 | >0.1 ± 0.0 | >0.1 ± 0.0 |
The EC50 of each compound represents the mean ± SD of the EC50s from at least three independent experiments. The 50% cytostatic concentrations of cidofovir and HPMPDAP were >159 μM, and that of each siRNA was >0.1 μM.
Mutations were identified in the VACV-WR DNA polymerase gene and were responsible for the resistance phenotypes observed with cidofovir (A314T, A684V, and A314T+A684V) and HPMPDAP (S851Y and A684V+S851Y). *, P < 0.05 (EC50s of siRNAs and compounds against VACV-WR DNA polymerase mutants differ significantly from EC50s against the wild-type strain) (Student's t test).
DISCUSSION
Currently, there is a need to develop novel antipoxvirus drugs due to the threat represented by the potential use of VARV as a bioweapon and the increasing number of poxvirus infections in humans (1, 52). Indeed, no specific medical treatment for smallpox other than supportive care is available, and no approved antiviral treatment exists. However, two antivirals are recommended by the U.S. Centers for Disease Control in case of a smallpox outbreak: they are cidofovir and a drug under development, i.e., ST-246, which is available as an investigational new drug. Novel antiviral approaches are still demanded to enlarge the collection of compounds active against orthopoxviruses. Today, the use of RNAi drugs as a therapeutic strategy to manage viral infections can be considered because multiple siRNA targets can be chosen within conserved viral genes. The in vitro effectiveness of orthopoxvirus siRNA-based treatment by targeting the conserved viral gene was demonstrated in our previous study (68). In this study, we complement the previous study and provide new insights into the antiviral potencies of other siRNAs specifically designed to target essential viral gene transcripts of human-pathogenic orthopoxviruses.
The two novel siRNAs, i.e., siB1R-2 and siG7L-1, assessed in the present study demonstrated their inhibitory activities against VACV-WR in two different cell types, i.e., epithelial and fibroblast cells. Virus yield experiments showed a prophylactic effect of siG7L-1 and siB1R-2, with a dose-dependent inhibition of virus growth, with a concentration of 1 nM of each siRNA still being effective. This prophylactic effect persisted for at least 72 h, showing the relative stability of these siRNAs in the cells, as well as the long-lasting siRNA-mediated silencing effect. A similar degree of antiviral potency has already been reported for siD5R-2, developed in our laboratory (68). Moreover, our results are in line with previous studies demonstrating that the effects of siRNAs have a relatively long half-life (from 5 to 7 days after transfection) (8, 63). The antiviral effects seen with siG7L-1 and siB1R-2 were highly specific because both siRNAs knocked down the transcripts of the genes of interest, as shown by RT-PCR. However, it has to be noticed that poxviruses produce late mRNA transcripts that are unusually long and may overlap several open reading frames (6, 40). These important data suggest that the mRNA levels of several late genes located upstream of G7L, such as L3L and J5L, may also be reduced by treatment with siG7L-1. Such an indirect effect could contribute to an increased antiviral potency of a specific siRNA targeting a late gene. In addition, the inhibitory activities were not related to induction of an IFN response (61). A therapeutic effect of siG7L-1 and siB1R-2 could be observed in cell culture, since viral replication was reduced by 80% when siRNAs were administered to the cells for up to 24 h postinfection. siG7L-1 proved its antiviral activity against the other pathogenic orthopoxviruses tested in this study, i.e., MPXV-Cop and CPXV-BR. In contrast, siB1R-2 reduced MPXV-Cop replication but failed to inhibit CPXV-BR. The inactivity of siB1R-2 against CPXV-BR could be explained by the presence of a central single nucleotide change between the target sequence and the siRNA (at position 10 of the 5′ end of the sense strand). This observation highlights the importance of base paring between the central regions of the siRNA and the target site (3, 14). Several studies demonstrated that many peripheral nucleotide changes can be well tolerated, but a stringent homology for central residues appears to be crucial for conserving the silencing capability (9, 18, 41). It should be noted that both the siB1R-2 and siG7L-1 sequences match perfectly with the target sequences of VARV, suggesting that these two siRNAs should be active in vitro against VARV replication.
Due to the potential nephrotoxic risk associated with cidofovir therapy, combination drug regimens might be a solution for reducing such an adverse effect (37, 38). Previous reports have described the advantages of drug combination for the treatment of viral infections (49, 51) compared with monotherapy. Several factors promote the use of combined therapy, including the reduction of drug dosages, with an equivalent efficacy, lowering the potential risk of toxicity, and delaying or preventing the appearance of drug resistance. The benefit of drug combination has already been demonstrated for orthopoxvirus infections in mice for two compounds, ST-246 and CMX001, an ether-lipid-cidofovir conjugate (hexadecylpropanediol-cidofovir) (54). In the present study, we investigated the antiviral potency of siRNAs combined with cidofovir against VACV-WR growth in cell culture. This investigation was motivated by the fact that these two inhibitors have different mechanisms of action and distinct targets. In our hands, the antiviral effects when siB1R-2, siG7L-1, or siD5R-2 was combined with cidofovir were more potent than those seen with the sum of effects of each drug alone. These results clearly demonstrated the synergism of the siRNA and cidofovir combination. Moreover, combination of these molecules did not result in increased toxicity for the cell cultures. However, further studies are still required to demonstrate the benefit of double therapy with siRNA-cidofovir in vivo.
We finally investigated the activity of siB1R-2, siG7L-1, or siD5R-2 against VACV-WR strains harboring specific mutations in the viral DNA polymerase which are known to confer resistance to cidofovir and HPMPDAP. These two molecules belong to the ANP family and are known to inhibit poxviruses by interfering with the viral DNA polymerase, encoded by the E9L gene in VACV (4, 25). Several mutations conferring a resistance phenotype have been mapped to the E9L gene of VACV (4, 35). Although these viruses were highly resistant in vitro, they also exhibited a decreased pathogenicity in vivo compared to that of the wild-type VACV strain (4). EC50s for each siRNA against mutant and wild-type viruses were expected to be similar since the mechanism of action of siRNAs and their targets differs from those of cidofovir and HPMPDAP. However, interesting differences in the potency of siRNAs against mutant viruses were found. We showed that siG7L-1 and siD5R-2 were active against four of five VACV-WR mutants; only the mutant virus bearing the A684V change proved to be less sensitive to these two siRNAs. On the other hand, siB1R-2 was poorly active against the wild-type virus (EC50 = 0.1 μM) in human fibroblasts, and except for the S851Y mutant, which was hypersensitive to this siRNA, all mutants were not inhibited by siB1R-2 at the higher concentration assessed (0.1 μM). An interesting feature was noticed with the VACV carrying the S851Y mutation, which was hypersensitive to all three siRNAs, i.e., siB1R-2, siG7L-1, and siD5R-2. This phenomenon can be explained by its reduced replicative capacity in vitro, as reported in a previous study (4). In contrast, the A684V mutant virus appeared to be less sensitive to siD5R-2 and siG7L-1. Why the A684V mutation counteracted the antiviral effects of siD5R-2 and siG7L-1 is unclear. In our hands, siD5R-2 silenced the D5 transcript by more than 90% and knocked down the expression of the D5 protein in cells infected with wild-type VACV-WR (68). Looking at the role of the D5 protein in viral replication, we hypothesized that the A684V mutation could alter the E9 protein by rendering it less dependent on the role of D5. Indeed, D5 is included within the replication complex of orthopoxviruses and interacts with the A20 protein, the stoichiometric component of the processivity factor of the catalytic subunit of the viral DNA polymerase (E9). We suggested that the A684V mutation, located in the DNA polymerase catalytic domain, might lead to the production of a mutated E9 protein which is less dependent on the interaction between D5 and A20 proteins. In the case of siG7L-1, we cannot explain the resistance phenotype seen with the virus bearing the A684V mutation: the G7 protein is required in the early stages of VACV morphogenesis and is not recognized to play a role in viral DNA synthesis. Interestingly, no cross-resistance to siRNAs was seen with the double mutant viruses (A314T+A684V and A684V+S851Y). The presence of the A314T and S851Y mutations, located in the 3′→5′ proofreading exonuclease domain and in the C-terminal DNA polymerase domain of the E9 protein, respectively, seemed to abrogate the siRNA resistance phenotypes observed with the single A684V mutation. These results show the importance of testing new drug candidates such as siRNAs against well-characterized drug-resistant viruses.
The most common route of VARV infection is via aerosol (48), with the mouth, trachea, and lungs being the sites of primary replication of the virus. siRNAs have been reported to be effective in vivo following intranasal or intravenous administration in several models of viral respiratory diseases, such as those associated with influenza virus, respiratory syncytial virus, paramyxovirus, and SARS coronavirus (10, 66, 74). Therefore, the use of siRNAs to inhibit orthopoxviruses in vivo appears to be conceivable, although our previous results on the efficacy of naked siRNAs in CPXV-infected mice are not reproducible (69). This lack of reproducibility can be explained by the use of naked siRNAs; thus, it seems mandatory to evaluate the efficiency of chemically modified siRNAs. Experiments are currently ongoing to investigate the antiviral activities mediated by chemically modified siG7L-1, siB1R-1, and siD5R-2 and by lipid-based nanoparticles for nucleic acid delivery in different mouse models of orthopoxvirus infections (60).
In conclusion, we have demonstrated the antiviral potencies of siRNAs against wild-type orthopoxviruses and against four of five cidofovir-resistant VACV. Also, this is the first report showing that combined therapies of siRNAs with cidofovir can be a promising approach in antipoxviral chemotherapy. Further studies are necessary to evaluate the antiviral efficacies of chemically modified siG7L-1, siB1R-1, and siD5R-2, as well as of a combined siRNA-cidofovir therapy, in animal models of orthopoxvirus infections.
Acknowledgments
We thank Danielle Gratier, Steven Carmans, Lies Van den Heurck, and Anita Camps for excellent technical assistance.
This work was supported by research grants from the Service de Santé des Armées, the Délégation Générale pour l'Armement, the ARAMI association, and the NIH (grant GAI062540.01).
Footnotes
Published ahead of print on 23 March 2009.
REFERENCES
- 1.Alibek, K. 1999. The Soviet Union's anti-agricultural biological weapons. Ann. N. Y. Acad. Sci. 894:18-19. [DOI] [PubMed] [Google Scholar]
- 2.Allen, L. B., L. K. Vanderslice, C. M. Fingal, F. H. McCright, E. F. Harris, and P. D. Cook. 1982. Evaluation of the anti-herpesvirus drug combinations: virazole plus arabinofuranosylhypoxanthine and virazole plus arabinofuranosyladenine. Antivir. Res. 2:203-216. [DOI] [PubMed] [Google Scholar]
- 3.Amarzguioui, M., T. Holen, E. Babaie, and H. Prydz. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrei, G., D. B. Gammon, P. Fiten, E. De Clercq, G. Opdenakker, R. Snoeck, and D. H. Evans. 2006. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 80:9391-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashford, D. A., R. M. Kaiser, M. E. Bales, K. Shutt, A. Patrawalla, A. McShan, J. W. Tappero, B. A. Perkins, and A. L. Dannenberg. 2003. Planning against biological terrorism: lessons from outbreak investigations. Emerg. Infect. Dis. 9:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assarsson, E., J. A. Greenbaum, M. Sundstrom, L. Schaffer, J. A. Hammond, V. Pasquetto, C. Oseroff, R. C. Hendrickson, E. J. Lefkowitz, D. C. Tscharke, J. Sidney, H. M. Grey, S. R. Head, B. Peters, and A. Sette. 2008. Kinetic analysis of a complete poxvirus transcriptome reveals an intermediate-early class of genes. Proc. Natl. Acad. Sci. USA 105:2140-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banham, A. H., and G. L. Smith. 1992. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology 191:803-812. [DOI] [PubMed] [Google Scholar]
- 8.Bayry, J., and D. F. Tough. 2005. Is RNA interference feasible for the control of foot-and-mouth disease outbreaks? Trends Immunol. 26:238-239. [DOI] [PubMed] [Google Scholar]
- 9.Bitko, V., and S. Barik. 2001. Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitko, V., A. Musiyenko, O. Shulyayeva, and S. Barik. 2005. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 11:50-55. [DOI] [PubMed] [Google Scholar]
- 11.Boyle, K. A., and P. Traktman. 2004. Members of a novel family of mammalian protein kinases complement the DNA-negative phenotype of a vaccinia virus ts mutant defective in the B1 kinase. J. Virol. 78:1992-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briolant, S., D. Garin, N. Scaramozzino, A. Jouan, and J. M. Crance. 2004. Inhibition of Chikungunya and Semliki Forest viruses replication by antiviral compounds: synergistic effect of interferon-alpha and ribavirin combination. Antivir. Res. 61:111-117. [DOI] [PubMed] [Google Scholar]
- 13.Cono, J., C. G. Casey, and D. M. Bell. 2003. Smallpox vaccination and adverse reactions. Guidance for clinicians. MMWR Recomm. Rep. 52:1-28. [PubMed] [Google Scholar]
- 14.Dahlgren, C., H. Y. Zhang, Q. Du, M. Grahn, G. Norstedt, C. Wahlestedt, and Z. Liang. 2008. Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 36:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dave, R. S., J. P. McGettigan, T. Qureshi, M. J. Schnell, G. Nunnari, and R. J. Pomerantz. 2006. siRNA targeting vaccinia virus double-stranded RNA binding protein [E3L] exerts potent antiviral effects. Virology 348:489-497. [DOI] [PubMed] [Google Scholar]
- 16.De Clercq, E. 2002. Cidofovir in the treatment of poxvirus infections. Antivir. Res. 55:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeVincenzo, J., J. E. Cehelsky, R. Alvarez, S. Elbashir, J. Harborth, I. Toudjarska, L. Nechev, V. Murugaiah, A. Van Vliet, A. K. Vaishnaw, and R. Meyers. 2008. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV). Antivir. Res. 77:225-231. [DOI] [PubMed] [Google Scholar]
- 18.Du, Q., H. Thonberg, J. Wang, C. Wahlestedt, and Z. Liang. 2005. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 33:1671-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duraffour, S., R. Snoeck, M. Krecmerova, J. van Den Oord, R. De Vos, A. Holy, J. M. Crance, D. Garin, E. De Clercq, and G. Andrei. 2007. Activities of several classes of acyclic nucleoside phosphonates against camelpox virus replication in different cell culture models. Antimicrob. Agents Chemother. 51:4410-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elion, G. B., S. Singer, and G. H. Hitchings. 1954. Antagonists of nucleic acid derivatives. VIII. Synergism in combinations of biochemically related antimetabolites. J. Biol. Chem. 208:477-488. [PubMed] [Google Scholar]
- 22.Evans, E., and P. Traktman. 1987. Molecular genetic analysis of a vaccinia virus gene with an essential role in DNA replication. J. Virol. 61:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrier, A., D. Garin, and J. M. Crance. 2004. Rapid inactivation of vaccinia virus in suspension and dried on surfaces. J. Hosp. Infect. 57:73-79. [DOI] [PubMed] [Google Scholar]
- 24.Fivelman, Q. L., I. S. Adagu, and D. C. Warhurst. 2004. Modified fixed-ratio isobologram method for studying in vitro interactions between atovaquone and proguanil or dihydroartemisinin against drug-resistant strains of Plasmodium falciparum. Antimicrob. Agents Chemother. 48:4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gammon, D. B., R. Snoeck, P. Fiten, M. Krecmerova, A. Holy, E. De Clercq, G. Opdenakker, D. H. Evans, and G. Andrei. 2008. Mechanism of antiviral drug resistance of vaccinia virus: identification of residues in the viral DNA polymerase conferring differential resistance to antipoxvirus drugs. J. Virol. 82:12520-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge, Q., L. Filip, A. Bai, T. Nguyen, H. N. Eisen, and J. Chen. 2004. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl. Acad. Sci. USA 101:8676-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisbert, T. W., L. E. Hensley, E. Kagan, E. Z. Yu, J. B. Geisbert, K. Daddario-DiCaprio, E. A. Fritz, P. B. Jahrling, K. McClintock, J. R. Phelps, A. C. Lee, A. Judge, L. B. Jeffs, and I. MacLachlan. 2006. Postexposure protection of guinea pigs against a lethal Ebola virus challenge is conferred by RNA interference. J. Infect. Dis. 193:1650-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giladi, H., M. Ketzinel-Gilad, L. Rivkin, Y. Felig, O. Nussbaum, and E. Galun. 2003. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther. 8:769-776. [DOI] [PubMed] [Google Scholar]
- 29.Hall, M. J., R. F. Middleton, and D. Westmacott. 1983. The fractional inhibitory concentration (FIC) index as a measure of synergy. J. Antimicrob. Chemother. 11:427-433. [DOI] [PubMed] [Google Scholar]
- 30.Hamasaki, K., K. Nakao, K. Matsumoto, T. Ichikawa, H. Ishikawa, and K. Eguchi. 2003. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 543:51-54. [DOI] [PubMed] [Google Scholar]
- 31.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jawetz, E. 1967. Combined antibiotic action: some definitions and correlations between laboratory and clinical results. Antimicrob. Agents Chemother. 7:203-209. [DOI] [PubMed] [Google Scholar]
- 33.Jordan, R., D. Tien, T. C. Bolken, K. F. Jones, S. R. Tyavanagimatt, J. Strasser, A. Frimm, M. L. Corrado, P. G. Strome, and D. E. Hruby. 2008. Single-dose safety and pharmacokinetics of ST-246, a novel orthopoxvirus egress inhibitor. Antimicrob. Agents Chemother. 52:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Judge, A. D., V. Sood, J. R. Shaw, D. Fang, K. McClintock, and I. MacLachlan. 2005. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 23:457-462. [DOI] [PubMed] [Google Scholar]
- 35.Kornbluth, R. S., D. F. Smee, R. W. Sidwell, V. Snarsky, D. H. Evans, and K. Y. Hostetler. 2006. Mutations in the E9L polymerase gene of cidofovir-resistant vaccinia virus strain WR are associated with the drug resistance phenotype. Antimicrob. Agents Chemother. 50:4038-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar, P., H. S. Ban, S. S. Kim, H. Wu, T. Pearson, D. L. Greiner, A. Laouar, J. Yao, V. Haridas, K. Habiro, Y. G. Yang, J. H. Jeong, K. Y. Lee, Y. H. Kim, S. W. Kim, M. Peipp, G. H. Fey, N. Manjunath, L. D. Shultz, S. K. Lee, and P. Shankar. 2008. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell 134:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lacy, S. A., M. J. Hitchcock, W. A. Lee, P. Tellier, and K. C. Cundy. 1998. Effect of oral probenecid coadministration on the chronic toxicity and pharmacokinetics of intravenous cidofovir in cynomolgus monkeys. Toxicol. Sci. 44:97-106. [DOI] [PubMed] [Google Scholar]
- 38.Lalezari, J. P., R. J. Stagg, B. D. Kuppermann, G. N. Holland, F. Kramer, D. V. Ives, M. Youle, M. R. Robinson, W. L. Drew, and H. S. Jaffe. 1997. Intravenous cidofovir for peripheral cytomegalovirus retinitis in patients with AIDS. A randomized, controlled trial. Ann. Intern. Med. 126:257-263. [DOI] [PubMed] [Google Scholar]
- 39.Li, B. J., Q. Tang, D. Cheng, C. Qin, F. Y. Xie, Q. Wei, J. Xu, Y. Liu, B. J. Zheng, M. C. Woodle, N. Zhong, and P. Y. Lu. 2005. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in rhesus macaque. Nat. Med. 11:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahr, A., and B. E. Roberts. 1984. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J. Virol. 49:510-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez, J., and T. Tuschl. 2004. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 18:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCaffrey, A. P., L. Meuse, T. T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay. 2002. RNA interference in adult mice. Nature 418:38-39. [DOI] [PubMed] [Google Scholar]
- 43.McCaffrey, A. P., H. Nakai, K. Pandey, Z. Huang, F. H. Salazar, H. Xu, S. F. Wieland, P. L. Marion, and M. A. Kay. 2003. Inhibition of hepatitis B virus in mice by RNA interference. Nat. Biotechnol. 21:639-644. [DOI] [PubMed] [Google Scholar]
- 44.Mercer, J., and P. Traktman. 2005. Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins. J. Virol. 79:7146-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrissey, D. V., K. Blanchard, L. Shaw, K. Jensen, J. A. Lockridge, B. Dickinson, J. A. McSwiggen, C. Vargeese, K. Bowman, C. S. Shaffer, B. A. Polisky, and S. Zinnen. 2005. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 41:1349-1356. [DOI] [PubMed] [Google Scholar]
- 46.Morrissey, D. V., J. A. Lockridge, L. Shaw, K. Blanchard, K. Jensen, W. Breen, K. Hartsough, L. Machemer, S. Radka, V. Jadhav, N. Vaish, S. Zinnen, C. Vargeese, K. Bowman, C. S. Shaffer, L. B. Jeffs, A. Judge, I. MacLachlan, and B. Polisky. 2005. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23:1002-1007. [DOI] [PubMed] [Google Scholar]
- 47.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 48.Moss, B., and B. M. Ward. 2001. High-speed mass transit for poxviruses on microtubules. Nat. Cell Biol. 3:E245-E246. [DOI] [PubMed] [Google Scholar]
- 49.Nakata, H., S. M. Steinberg, Y. Koh, K. Maeda, Y. Takaoka, H. Tamamura, N. Fujii, and H. Mitsuya. 2008. Potent synergistic anti-human immunodeficiency virus (HIV) effects using combinations of the CCR5 inhibitor aplaviroc with other anti-HIV drugs. Antimicrob. Agents Chemother. 52:2111-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nichols, R. J., M. S. Wiebe, and P. Traktman. 2006. The vaccinia-related kinases phosphorylate the N′ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol. Biol. Cell 17:2451-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikonenko, B. V., K. A. Sacksteder, S. Hundert, L. Einck, and C. A. Nacy. 2008. Preclinical study of new TB drugs and drug combinations in mouse models. Recent Patents Anti-Infect. Drug Discov. 3:102-116. [DOI] [PubMed] [Google Scholar]
- 52.Orent, W. 1998. Escape from Moscow. Sciences 3:26-31. [Google Scholar]
- 53.Palliser, D., D. Chowdhury, Q. Y. Wang, S. J. Lee, R. T. Bronson, D. M. Knipe, and J. Lieberman. 2006. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature 439:89-94. [DOI] [PubMed] [Google Scholar]
- 54.Quenelle, D. C., R. M. Buller, S. Parker, K. A. Keith, D. E. Hruby, R. Jordan, and E. R. Kern. 2007. Efficacy of delayed treatment with ST-246 given orally against systemic orthopoxvirus infections in mice. Antimicrob. Agents Chemother. 51:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rempel, R. E., M. K. Anderson, E. Evans, and P. Traktman. 1990. Temperature-sensitive vaccinia virus mutants identify a gene with an essential role in viral replication. J. Virol. 64:574-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rempel, R. E., and P. Traktman. 1992. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J. Virol. 66:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos, C. R., F. M. Vega, S. Blanco, R. Barcia, and P. A. Lazo. 2004. The vaccinia virus B1R kinase induces p53 downregulation by an Mdm2-dependent mechanism. Virology 328:254-265. [DOI] [PubMed] [Google Scholar]
- 58.Selleseth, D. W., C. L. Talarico, T. Miller, M. W. Lutz, K. K. Biron, and R. J. Harvey. 2003. Interactions of 1263W94 with other antiviral agents in inhibition of human cytomegalovirus replication. Antimicrob. Agents Chemother. 47:1468-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin, S., and S. Lim. 2004. Antifungal effects of herbal essential oils alone and in combination with ketoconazole against Trichophyton spp. J. Appl. Microbiol. 97:1289-1296. [DOI] [PubMed] [Google Scholar]
- 60.Shrivastava, N., and A. Srivastava. 2008. RNA interference: an emerging generation of biologicals. Biotechnol. J. 3:339-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 62.Smee, D. F., R. W. Sidwell, D. Kefauver, M. Bray, and J. W. Huggins. 2002. Characterization of wild-type and cidofovir-resistant strains of camelpox, cowpox, monkeypox, and vaccinia viruses. Antimicrob. Agents Chemother. 46:1329-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song, E., S. K. Lee, D. M. Dykxhoorn, C. Novina, D. Zhang, K. Crawford, J. Cerny, P. A. Sharp, J. Lieberman, N. Manjunath, and P. Shankar. 2003. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 77:7174-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szajner, P., H. Jaffe, A. S. Weisberg, and B. Moss. 2003. Vaccinia virus G7L protein interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 77:3418-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tattersall, M. H., and K. R. Harrap. 1973. Combination chemotherapy: the antagonism of methotrexate and cytosine arabinoside. Eur. J. Cancer 9:229-232. [DOI] [PubMed] [Google Scholar]
- 66.Tompkins, S. M., C. Y. Lo, T. M. Tumpey, and S. L. Epstein. 2004. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl. Acad. Sci. USA 101:8682-8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valderrama, F., J. V. Cordeiro, S. Schleich, F. Frischknecht, and M. Way. 2006. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science 311:377-381. [DOI] [PubMed] [Google Scholar]
- 68.Vigne, S., R. Germi, S. Duraffour, S. Larrat, G. Andrei, R. Snoeck, D. Garin, and J. M. Crance. 2008. Specific inhibition of orthopoxvirus replication by a small interfering RNA targeting the D5R gene. Antivir. Ther. 13:357-368. [PubMed] [Google Scholar]
- 69.Vigne, S., S. Duraffour, G. Andrei, R. Snoeck, D. Garin, and J. M. Crance. 2008. Anti-orthopoxviral potency of siRNAs targeting three genes of vaccinia virus, abstr. P8.107. Abstr. 17th Int. Poxvirus Iridovirus Conf.
- 70.White, R. L., D. S. Burgess, M. Manduru, and J. A. Bosso. 1996. Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob. Agents Chemother. 40:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiebe, M. S., and P. Traktman. 2007. Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe 1:187-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang, G., D. C. Pevear, M. H. Davies, M. S. Collett, T. Bailey, S. Rippen, L. Barone, C. Burns, G. Rhodes, S. Tohan, J. W. Huggins, R. O. Baker, R. L. Buller, E. Touchette, K. Waller, J. Schriewer, J. Neyts, E. DeClercq, K. Jones, D. Hruby, and R. Jordan. 2005. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J. Virol. 79:13139-13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ying, R. S., C. Zhu, X. G. Fan, N. Li, X. F. Tian, H. B. Liu, and B. X. Zhang. 2007. Hepatitis B virus is inhibited by RNA interference in cell culture and in mice. Antivir. Res. 73:24-30. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, W., H. Yang, X. Kong, S. Mohapatra, H. San Juan-Vergara, G. Hellermann, S. Behera, R. Singam, R. F. Lockey, and S. S. Mohapatra. 2005. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11:56-62. [DOI] [PubMed] [Google Scholar]