Abstract
Dermcidin (DCD) is an antimicrobial peptide which is constitutively expressed in eccrine sweat glands. By postsecretory proteolytic processing in sweat, the DCD protein gives rise to anionic and cationic DCD peptides with a broad spectrum of antimicrobial activity. Many antimicrobial peptides induce membrane permeabilization as part of their killing mechanism, which is accompanied by a loss of the bacterial membrane potential. In this study we show that there is a time-dependent bactericidal activity of anionic and cationic DCD-derived peptides which is followed by bacterial membrane depolarization. However, DCD-derived peptides do not induce pore formation in the membranes of gram-negative and gram-positive bacteria. This is in contrast to the mode of action of the cathelicidin LL-37. Interestingly, LL-37 as well as DCD-derived peptides inhibit bacterial macromolecular synthesis, especially RNA and protein synthesis, without binding to microbial DNA or RNA. Binding studies with components of the cell envelope of gram-positive and gram-negative bacteria and with model membranes indicated that DCD-derived peptides bind to the bacterial envelope but show only a weak binding to lipopolysaccharide (LPS) from gram-negative bacteria or to peptidoglycan, lipoteichoic acid, and wall teichoic acid, isolated from Staphylococcus aureus. In contrast, LL-37 binds strongly in a dose-dependent fashion to these components. Altogether, these data indicate that the mode of action of DCD-derived peptides is different from that of the cathelicidin LL-37 and that components of the bacterial cell envelope play a role in the antimicrobial activity of DCD.
Antimicrobial peptides (AMPs) serve as a first line of innate host defense in many species such as plants, amphibians, insects, and mammals. AMPs show a broad-spectrum antimicrobial activity against a wide range of pathogens including bacteria, fungi, and enveloped viruses (51). Most gene-encoded AMPs are synthesized as proforms, which are subsequently processed into mature peptides of various lengths. A common feature of most of these peptides is that they are cationic and form amphipathic structures (3). The mode of action of most AMPs is incompletely understood. It is believed that most AMPs kill microorganisms by membrane permeation either via pore formation or via membrane disintegration like that induced by the human cathelicidin LL-37. However, membrane disruption may not reflect the complex processes involved in the killing of microorganisms (5, 48). In addition, several AMPs clearly act differently and intracellular target sites have been identified (15). The mode of action has been unraveled for only a few AMPs which act via defined targets, such as the lantibiotic nisin, which specifically binds to bacterial lipid II, a membrane-bound component involved in peptidoglycan (PG) synthesis (7, 46). Similarly, the lantibiotic mersacidin interferes with transglycosylation and PG synthesis in gram-positive bacteria by direct targeting of lipid II (6). Furthermore, buforin II kills microorganisms by disruption of critical intracellular processes such as the inhibition of macromolecular biosynthesis or by interacting with nucleic acids inside the microorganisms (33). For several AMPs it has been demonstrated that the charge and the composition of the bacterial cell envelope determine sensitivity to AMPs (37). Staphylococcus aureus mutants lacking specific modifications in the bacterial envelope are highly susceptible to a variety of cationic AMPs. For example, incorporation of d-alanine into S. aureus teichoic acids by the dltA enzymes or the lysinylation of phosphatidylglycerol by mprF confers resistance to defensins, protegrins, and other AMPs by repulsion of the cationic peptides (36).
Dermcidin (DCD) was identified by our group as a human AMP which is constitutively expressed in eccrine sweat glands and secreted into sweat (40). By postsecretory proteolytic processing in human sweat, the precursor protein gives rise to several truncated DCD peptides varying in length from 25 to 48 amino acids and with net charges between −2 and +2 (2, 10, 38). Several DCD peptides show antimicrobial activity against pathogenic microorganisms such as S. aureus, Escherichia coli, Enterococcus faecalis, Candida albicans, Staphylococcus epidermidis, Pseudomonas putida, and methicillin-resistant S. aureus as well as rifampin- and isoniazid-resistant Mycobacterium tuberculosis (9, 25, 40, 41, 45). We were able to show that DCD-derived peptides are also active under high-salt conditions and in a buffer resembling human sweat (40). Antimicrobially active DCD peptides, namely, the anionic peptides DCD-1L (48-mer) and DCD-1 (47-mer) and the cationic peptides SSL-25 (25-mer) and SSL-23 (23-mer), are derived from the C-terminal region of the precursor protein. Interestingly, these peptides have diverse and overlapping spectra of activity which are independent of the net peptide charge (41). In previous studies we showed that DCD peptides interact with the bacterial cell envelope and kill gram-negative bacteria without forming pores in membranes (41). In this study we investigated the mode of antimicrobial activity of DCD-derived peptides in more detail and studied bacterial factors that govern sensitivity or tolerance to DCD in the model microorganism S. aureus. In our first approach, we tried to identify the bacterial surface molecules to which DCD peptides bind. Second, we analyzed the bacterial response to DCD peptide challenge. Finally, we analyzed bacterial mutants to elucidate the mechanisms determining DCD sensitivity.
MATERIALS AND METHODS
Bacterial strains.
The following bacterial strains were used: S. aureus ATCC 25923, S. aureus 113 (ATCC 35556), and S. aureus Newman. The following defined S. aureus 113 mutants were used: Δatl mutant, deficient in autolysin; Δsrt mutant, deficient in sortase; ΔspA mutant, deficient in protein A; ΔypfP mutant, with a reduced level of lipoteichoic acid (LTA); and ΔtagO mutant, deficient in wall teichoic acid (WTA). Furthermore, S. aureus Newman mutant strains lacking agr, arl, sar, sae, or sigB (kindly provided by Christiane Wolz) were used. All strains were cultured in Luria-Bertani (LB) medium at 37°C and maintained on blood agar plates.
Peptides and chemicals.
Peptides were synthesized utilizing 9-fluorenylmethoxycarbonyl-tertiary butyl chemistry using a multiple peptide synthesizer, Syro II (MultiSynTech). After cleavage, the crude peptide was purified by high-pressure liquid chromatography on a reversed-phase C18 Nucleosil 100-5C column to a purity of >95% using a linear gradient of 5 to 80% acetonitrile in 0.05% trifluoroacetic acid for 45 min. Peptide purity was controlled by matrix-assisted laser desorption ionization-time of flight mass spectroscopy and electrospray ionization. Peptides were dissolved in LiChrosolv H2O (Merck) to a concentration of 1 mg/ml and stored at −20°C. Labeling of the N terminus of DCD-1L, LL-37, and control peptide (GATTTAPSLSGKGNPEEEDVDTSQVLYE) with fluorescein isothiocyanate (FITC) was achieved by elongation of the N-terminal bound peptide with β-alanine and afterwards by FITC. The molecular mass was determined by matrix-assisted laser desorption ionization mass spectroscopy analysis and agreed with the calculated mass. FITC-labeled bovine serum albumin (BSA) was purchased from Invitrogen. All FITC-labeled peptides were prepared in double-distilled water (ddH2O) to a final concentration of 1 mg/ml. Radioisotopes (l-[4,5-3H]leucine, [methyl-3H]thymidine, and [5-3H]uridine) were purchased from Perkin-Elmer and diluted to appropriate concentrations in phosphate-buffered saline (PBS). DiBAC4(3) [bis-(1,3-dibutylbarbituric acid) trimethine oxonol] (Molecular Probes) was resuspended in methanol and diluted in ddH2O to a final concentration of 1 μg/ml. Propidium iodide (PI; Sigma) was dissolved in ddH2O to a final concentration of 10 μg/ml.
Antimicrobial assays.
Antimicrobial assays were performed using the CFU assay as described previously (40, 41). Briefly, overnight bacterial cultures were diluted 1:100 in LB medium and grown to an optical density at 600 nm of 0.4 to 0.8. The bacteria were pelleted, washed two times in sodium phosphate buffer, and then diluted to a concentration of 106 CFU/ml in 10 mM sodium phosphate buffer-10 mM NaCl (pH 7.4). Twenty microliters of the dilutions were incubated at 37°C for 2 h with the respective peptide diluted in water in a total volume of 60 μl in 10 mM sodium phosphate buffer-10 mM NaCl (pH 7.4). Time-dependent antimicrobial assays were performed under the conditions described above. After incubation, cells were diluted 1:100 in 10 mM sodium phosphate buffer-10 mM NaCl (pH 7.4) and 100 μl of the bacterial suspensions was plated in triplicate on blood agar. Bacterial colonies were counted after incubation for 18 to 24 h at 37°C. The antimicrobial activity was calculated as [(cell survival after peptide incubation)/(cell survival in buffer without peptide) × 100]. The IC90 designates the lethal concentration of the current synthetic peptide in μg/ml which leads to 90% reduction of CFU compared to the buffer control.
Flow cytometry.
Flow cytometric analysis of S. aureus was performed as described by Nuding et al. (30) and Mason and Gant (28) on a FACSCalibur flow cytometer (Becton Dickinson). All detectors were used with logarithmic amplification. Bacterial suspensions of 1 × 107 to 1 × 108 CFU/ml were incubated for 2 h with DCD-derived peptides or LL-37 (each at 50 μg/ml) in a total volume of 100 μl under the same conditions used in the antimicrobial assays. Bacteria were pelleted by centrifugation, washed with sterile PBS, and incubated for 5 to 10 min at room temperature with either DiBAC4(3) or PI (mass, 668.4 Da). Afterwards, bacteria were pelleted, washed in PBS, and resuspended in 300 μl PBS. A total of 10,000 bacteria were analyzed in each sample, using Cell Quest software (Becton Dickinson) for data acquisition and analysis.
Flow cytometric analyses of the binding of FITC-labeled DCD-1L, LL-37, and (as control) BSA or control peptide to S. aureus 113 wild type and S. aureus ΔtagO were performed as follows. Bacteria (3 × 108 CFU/ml) in a total volume of 100 μl were incubated with the above-mentioned labeled peptides each at 50 μg/ml for 2 h at room temperature. Afterwards, bacteria were washed two times with PBS and then resuspended in PBS. Bacteria at a final concentration of 1 × 108 CFU/ml were analyzed by flow cytometry using Cell Quest software (Becton Dickinson).
Gel retardation assay.
Gel retardation assays were performed as described by Park et al. (33) by mixing 200 ng pBluescript II plasmid DNA or 2 μg of yeast total RNA in 20 μl of binding buffer (5% glycerol, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1 mM dithiothreitol, 20 mM KCl, and 50 μg/ml BSA) at room temperature with different amounts of DCD peptides. After 60 min of incubation at room temperature, the reaction was stopped and the samples were loaded on a 1% agarose gel and detected by ethidium bromide staining.
Fluorescent electrophoretic mobility shift assay.
The binding of peptides to teichoic acids was measured in a fluorescent mobility shift assay. Therefore, 0.25 mg/ml FITC-labeled peptides were incubated with 0.75 mg/ml of LTA or WTA in a total volume of 16 μl for 1 h at room temperature. After native gel electrophoresis at 150 V for 1 h in a 1% agarose gel, the fluorescent peptides were detected using a fluorescence imager (Storm; GE Healthcare) with blue laser excitation. Binding to LTA or WTA provides additional negative charges to the peptides, resulting in a migration toward the anode.
Incorporation of radioactive metabolites.
The effect of DCD peptides and LL-37 on the synthesis of macromolecules was studied by measuring the incorporation of 3H-labeled precursors (l-[4,5-3H]leucine, [methyl-3H]thymidine, and [5-3H]uridine) as described by Yenugu et al. (49). Briefly, 1 × 106 CFU of mid-log-phase S. aureus ATCC 25923 resuspended in 10 mM sodium phosphate buffer-10 mM NaCl (pH 7.4) were incubated at room temperature for 1 min, 30 min, and 60 min with subinhibitory concentrations of several DCD peptides (10 μg/ml) and LL-37 (1 μg/ml) in the presence of 10 μl/ml of either l-[4,5-3H]leucine (27 Ci/mmol), [methyl-3H]thymidine (25 Ci/mmol), or [5-3H]uridine (26 Ci/mmol) to detect metabolite uptake. Bacterial suspensions were prepared in 96-well plates and harvested (MicroBeta Filter Mate-96 harvester; Perkin-Elmer) after each time point. Samples were afterwards collected onto filters (Filtermat A, GF/C; Perkin-Elmer) using vacuum filtration and washed thoroughly four to five times with ddH2O. The filters were dried for 2 min in a microwave oven and subsequently counted on a liquid scintillation analyzer (MicroBeta; Perkin-Elmer).
PG binding assay.
PG (0.25 mg/ml) isolated from S. aureus 113 wild type was incubated with 30 μl of DCD peptides (DCD-1L and SSL-25, 1 mg/ml) in a total volume of 130 μl. Probes were incubated for 60 min with gentle shaking at room temperature. After centrifugation, the supernatant was separated from the PG-peptide reaction mixture and was collected. The pellet was washed once with PBS, and the supernatant of the washing step was also collected. Pellet was resuspended again in 130 μl PBS, and elution was performed by boiling the reaction mixture for 5 min at 90°C. Samples (20 μl) were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie PageBlue protein staining solution (Fermentas).
LTA and LPS binding assay.
Wells of a flat-bottomed 96-well Microplate (Falcon; BD) were coated with LTA or lipopolysaccharide (LPS) (Sigma) by a method based on the work of Tobias et al. (42) and Yu and Kanost (50). LTA or LPS was suspended at 40 μg/ml in ddH2O and sonicated three times for 15 s each, and 50 μl (2 μg) of LPS or LTA suspension was added to each well. The plate was then incubated overnight until the water evaporated completely. The plates were heated at 60°C for 30 min and then blocked with 200 μl/well of 1 mg/ml BSA in Tris buffer (TB) (50 mM Tris-HCl, 50 mM NaCl, pH 8.0) for 2 h at 37°C. After this, plates were rinsed four times with 200 μl/well of TB. DCD-derived peptides diluted with TB containing 5 mM CaCl2 and 0.1 mg/ml BSA were added at 50 μl/well, and binding was allowed to occur for 3 h at room temperature. The plates were subsequently rinsed four times with 200 μl/well of TB, and afterwards a rabbit polyclonal anti-DCD antiserum or a rabbit polyclonal anti-LL-37 antiserum (diluted 1,000-fold with TB containing 0.1 mg/ml BSA) was added at 100 μl/well. After incubation at 4°C overnight, the wells were rinsed four times with 200 μl/well of TB. Incubation for 2 h at 37°C was followed with biotinylated polyclonal swine anti-rabbit antiserum (Dako) diluted 2,000-fold with TB containing 0.1 mg/ml BSA. Streptavidin-alkaline phosphatase (Roche) diluted 2,000-fold with TB containing 0.1 mg/ml BSA was added at 100 μl/well, and the mixture was incubated for 2 h at 37°C. The wells were washed again as described above, and 50 μl/well of 1 mg/ml p-nitrophenyl phosphate (Sigma) was added and incubated at room temperature for 20 min. Absorbance at 405 nm of each well was determined using a microtiter plate reader (SLT.Spectra).
Preparation of liposomes used for surface acoustic wave (SAW) and Förster resonance energy transfer (FRET) assay.
Rough mutant LPS from E. coli strain WBB01 was extracted by the phenol-chloroform-petroleum ether method (12), purified, lyophilized, and transformed into the triethylamine salt form.
Phospholipid liposomes composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (DOPG), 1:1 (M:M) (Avanti Polar Lipids, Alabaster, AL), and LPS aggregates were prepared as 1 mM aqueous dispersions of the phospholipids or LPS. The lipids were dissolved in chloroform, and the LPS was dissolved in chloroform-methanol (10:1) to a concentration of 1 mg/ml. For the FRET experiments, 1% (mol/mol) of the donor dye nitro-2-1,3-benzoxadiazol-4-yl-phosphatidylethanolamine (NBD-PE) or of the acceptor dye rhodamine-PE (Rh-PE) (both were obtained from Molecular Probes [Eugene, OR]) was added. The solvent was evaporated under a stream of nitrogen; the lipids were resuspended in buffer, mixed thoroughly, and sonicated for 1 min (1-ml solution). Subsequently, the preparation was cooled for 30 min at 4°C, heated for 30 min at 56°C, and cooled to 4°C. Preparations were stored at 4°C overnight prior to measurements.
Lipid binding studies.
The FRET technique was used as a probe dilution assay to obtain information on the intercalation of the peptides into lipid liposomes or LPS aggregates. A preparation of 1 ml of the double-labeled liposomes (10−5 M) at 37°C was excited at 470 nm (λex of NBD-PE), and the intensities of the emission light of the donor NBD-PE (531 nm) and acceptor Rh-PE (593 nm) were measured simultaneously on the SPEX F1T11 fluorescence spectrometer (SPEX Instruments). Five microliters of the peptides DCD-1L, SSL-25, SSL-29, and LL-37 (1 mg/ml) were added to liposomes after 50 s. Since FRET spectroscopy is used here as a probe dilution assay, intercalation of molecules causes an increase in the distance between donor and acceptor in the liposomes and, thus, leads to a reduced energy transfer. This again causes an increase in the donor and decrease in the acceptor intensities. For a qualitative analysis of experiments, the quotient of the intensities of the donor dye and the acceptor dye is plotted against time (designated in the following as the FRET signal). The results shown are representative of three independent experiments. The SAW biosensor system (Biosensor GmbH, Bonn, Germany) was used as described before (1). Briefly, in the SAW biosensor a SAW is propagated along a sensor surface and the modification of its phase and amplitude induced by binding of molecules to the surface is a measure for mass adsorption and change of viscoelastic properties, respectively. Measurements were performed at a continuous buffer flow of 20 μl/min. By injection of 100 μl poly-l-lysine (60 μg/ml; Fluka) and subsequent injection of 100 μl phospholipid liposomes (100 μM) or 100 μl LPS aggregates (100 μM), the bilayer was prepared, and finally 100 μl of the respective peptides (100 μg/ml) was added. All experiments were performed at a temperature of 22°C.
Statistical analysis.
Statistical analyses were performed with a two-tailed unpaired t test. P values < 0.05 were considered to be statistically significant.
RESULTS
Time-dependent killing of S. aureus by DCD-derived peptides is followed by membrane depolarization without pore formation.
Many AMPs induce membrane permeation via pore formation, which is accompanied by a loss of the bacterial membrane potential. Our previous analyses of gram-negative bacteria using membrane permeability assays and efflux assays using liposomes indicated that DCD-derived peptides do not affect membrane permeability (41). In order to analyze the mechanisms determining sensitivity towards anionic and cationic DCD peptides in gram-positive bacteria, we developed a flow cytometric method for the simultaneous measurement of bacterial membrane permeability and membrane potential and compared this with the changes in the number of CFU in a sample over time after treatment with DCD-derived peptides. As a model organism, we used the gram-positive bacterium S. aureus and the anionic DCD peptide DCD-1L and the cationic DCD peptide SSL-25, both antimicrobially active on S. aureus (41). For comparison, we used a pore-formation-inducing AMP, the human cathelicidin LL-37 (Table 1).
TABLE 1.
Amino acid sequences and molecular masses of antimicrobial peptides and control peptides used in this study
| Peptide | Sequence | Molecular mass (Da) | ||||
|---|---|---|---|---|---|---|
| 10 | 20 | 30 | 40 | |||
| DCD-1L | SSLLEKGLDG | AKKAVGGLGK | LGKDAVEDLE | SVGKGAVHDV | KDVLDSVL | 4,818.5 |
| SSL-29 | SSLLEKGLDG | AKKAVGGLGK | LGKDAVEDL | 2,869.9 | ||
| SSL-25 | SSLLEKGLDG | AKKAVGGLGK | LGKDA | 2,412.8 | ||
| SSL-23 | SSLLEKGLDG | AKKAVGGLGK | LGK | 2,666.2 | ||
| LL-37 | LLGDFFRKSK | EKIGKEFKRI | VQRIKDFLRN | LVPRTES | 4,493.3 | |
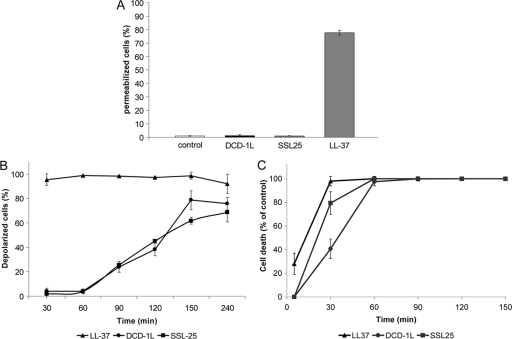
S. aureus bacteria were treated with 50 μg/ml DCD-1L, SSL-25, or LL-37 for 2 hours and then incubated with the DNA-binding dye PI, which binds to DNA only when the bacterial membrane is permeable. Flow cytometric analyses indicated that the antimicrobially active DCD peptides DCD-1L and SSL-25 did not induce pore formation of the bacterial membrane, whereas LL-37 permeabilized the membrane of S. aureus (Fig. 1A).
FIG. 1.
Time-dependent killing of S. aureus by DCD peptides is followed by membrane depolarization without pore formation. (A) Percentage of permeabilized S. aureus bacteria as quantified by uptake of PI by flow cytometric analyses. Quantification of at least three independent experiments. S. aureus ATCC 25923 (1 × 107 CFU/ml) was incubated with DCD-1L or SSL-25 (50 μg/ml) and as a positive control with the AMP LL-37 (50 μg/ml) for 2 h, and damage to the bacterial membrane was measured by uptake of PI. It is evident that DCD-1L and SSL-25 do not form pores in the bacterial membrane (large enough to admit PI), whereas LL-37 induces membrane permeabilization. Lower concentrations of these AMPs (1 to 10 μg/ml) also did not induce pore formation (data not shown). (B) Time-dependent changes in the bacterial membrane potential expressed as the percentage of depolarized S. aureus bacteria after peptide treatment compared to the untreated control. S. aureus ATCC 25923 (1 × 107 CFU/ml) was incubated with DCD-1L, SSL-25, and LL-37 (each at 50 μg/ml). After the indicated time points, bacterial membrane potential was measured after incubation with the membrane-potential-sensitive dye DiBAC4(3). Depolarization of bacterial membranes leads to an uptake of the anionic dye DiBAC4(3) inside the membrane, resulting in an increased fluorescent signal. In contrast to LL-37, which induces an immediate depolarization of the bacterial membrane, DCD-1L and SSL-25 change the bacterial membrane potential over time, starting with approximately 20% depolarized bacteria 90 min after peptide incubation and going up to 80% depolarized S. aureus bacteria 4 h after DCD treatment. (C) Time-dependent killing of S. aureus ATCC 25923 by DCD-1L (circles), SSL-25 (squares), and LL-37 (triangles) (each at 50 μg/ml) measured by the CFU assay. Bacteria in the mid-logarithmic phase of growth were incubated with the peptides for the indicated times (0 to 150 min). The antibacterial activity of these peptides is expressed as percentage of bacterial cell death compared to the buffer control at the respective time points.
In a second step, S. aureus were incubated after peptide incubation with the membrane-potential-sensitive dye DiBAC4(3). Depolarization of bacterial membranes leads to an uptake of the anionic dye DiBAC4(3) inside the cells, resulting in an increased fluorescent signal. As seen in Fig. 1B, DCD-1L and SSL-25 changed the bacterial membrane potential over time starting with approximately 20% depolarized bacteria 90 min after peptide incubation and going up to 80% depolarized S. aureus bacteria 4 h after DCD treatment. In contrast, the pore-forming peptide LL-37 depolarized the membrane of S. aureus within the first 30 min (Fig. 1B). This resembles the time kinetics of antimicrobial activity of these peptides: whereas LL-37 killed S. aureus in the first 30 min after incubation, DCD-1L and SSL-25 showed 100% killing only after 60 min (Fig. 1C). Comparison of the time kinetics of killing and those of membrane depolarization suggests that depolarization of the bacterial membrane may be a secondary event following the lethal hit of the DCD peptides against the bacteria. However, as was pointed out by Jepras et al. (20), whereas the depolarization assay depicts results in real time, colony count data indicate that the lethal hit probably occurred sometime during the overnight incubation.
DCD-derived peptides inhibit bacterial macromolecular synthesis without binding to microbial DNA or RNA.
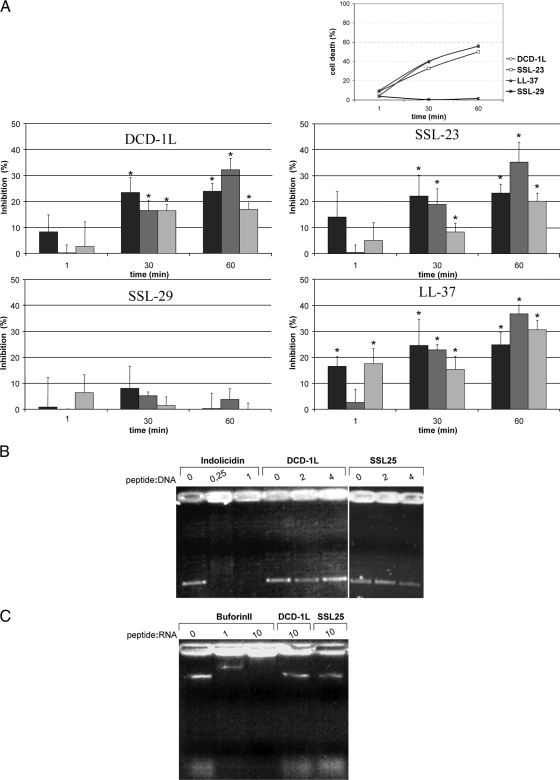
To analyze whether DCD-derived peptides inhibited the macromolecular synthesis in S. aureus, the incorporation of the radioactive precursors l-[4,5-3H]leucine, [methyl-3H]thymidine, and [5-3H]uridine for protein, DNA, and RNA synthesis, respectively, after treatment with subinhibitory concentrations of DCD-1L and SSL-23 (each at 10 μg/ml) and LL-37 (1 μg/ml) was observed over time (1 to 60 min). Interestingly, both DCD-derived peptides inhibited mainly RNA and protein synthesis and to a minor degree DNA synthesis, whereas the nonactive DCD peptide SSL-29 did not significantly affect the metabolic rates in S. aureus (Fig. 2A). The positive controls, ciprofloxacin, rifampin, and tetracycline, caused inhibition of DNA, RNA, and protein synthesis, respectively, over time and were used as controls in the incorporation assays (data not shown).
FIG. 2.
DCD-derived peptides inhibit bacterial macromolecular synthesis without binding to microbial DNA or RNA. (A) Percent inhibition of protein (black bars), RNA (dark gray bars), and DNA (light gray bars) synthesis in S. aureus ATCC 25923 (1 × 106 CFU/ml) incubated for the indicated time points with subinhibitory concentrations of DCD-1L, SSL-23, and SSL-29 (each at 10 μg/ml) or LL-37 (1 μg/ml) compared to control (untreated bacteria). These peptide concentrations kill approximately 50% of the bacteria. Protein, RNA, and DNA synthesis was measured by incorporation of respective radioactive precursors over time. The antimicrobially active DCD peptides DCD-1L and SSL-23 significantly inhibited mainly RNA and protein synthesis and to a minor degree DNA synthesis, whereas the nonactive DCD peptide SSL-29 did not significantly affect the metabolic rates in S. aureus. Significant changes (P < 0.05) are marked with an asterisk. At top the time-dependent not significantly affect the metabolic rates in S. aureus. Significant changes (P < 0.05) are marked with an asterisk. At top the time-dependent antimicrobial activity of subinhibitory concentrations of DCD-1L, SSL-23, and SSL-29 (each at 10 μg/ml) and LL-37 (1 μg/ml) used in the metabolic assays is shown. SSL-23 has antimicrobial activity similar to that of SSL-25. Data represent the means of at least three independent experiments showing percentages of cell death over time. (B) Gel retardation assay with bacterial DNA. Increasing amounts of DCD-1L and SSL-25 (400 ng and 800 ng) and (as a positive control) indolicidin (50 ng and 200 ng) were incubated with 200 ng of pBluescript II plasmid DNA and run on an agarose gel. No shift was detectable for DCD-1L and SSL-25, indicating that they do not bind to plasmid DNA. In contrast, indolicidin, which does bind to DNA, causes retardation of the DNA. (C) Gel retardation assay with yeast RNA. DCD-1L and SSL-25 (20 μg) or (as a positive control) buforin II (2 μg and 20 μg) was incubated with 2 μg yeast RNA and run on an agarose gel. Whereas no shift can be detected for DCD-1L and SSL-25, buforin binds efficiently to yeast RNA.
To analyze whether the inhibition of RNA and protein synthesis was due to binding of DCD-1L and SSL-25 to microbial DNA or RNA, we performed a gel retardation assay with two other AMPs known to bind to DNA or RNA: indolicidin is known to bind to bacterial DNA (19, 27) and buforin II (a histone 2A-derived peptide) is known to bind to yeast RNA (33). As shown in Fig. 2B and C, neither DCD-1L nor SSL-25 was able to bind to microbial DNA or RNA. There is also no binding of LL-37 to microbial DNA or RNA under these conditions (data not shown).
Deletion of global regulatory molecules in S. aureus does not change sensitivity toward DCD-derived peptides.
It has been suggested that DCD induces the expression of the key regulators of virulence factors agr and sarA involved in quorum sensing in S. epidermidis (24). We asked whether in S. aureus the quorum sensing regulation and global regulators play a role in sensitivity to DCD. Therefore, we analyzed the antimicrobial activity of DCD-1L, SSL-25, and LL-37 against mutant S. aureus strains lacking the key regulator agr, arl, sar, sae, or sigB. The sensitivity of S. aureus mutants toward the analyzed peptides did not differ (IC90 for wild-type S. aureus and the mutant strains for DCD-1L and SSL-25 is between 9 and 9.5 μg/ml and that for LL-37 is >5 μg/ml). This indicated that these global regulators do not play a role in the regulation of DCD and LL-37 antimicrobial activity.
DCD-derived peptides in contrast to LL-37 show only a weak binding to components of the cell envelope of gram-negative and gram-positive bacteria.
In our previous studies using immunoelectron microscopy, we showed that anionic and cationic DCD peptides interact with the bacterial cell envelope (41). In order to identify the targets with which DCD peptides mainly interact, we performed binding studies with components of the cell envelope of gram-positive and gram-negative bacteria and with model membranes.
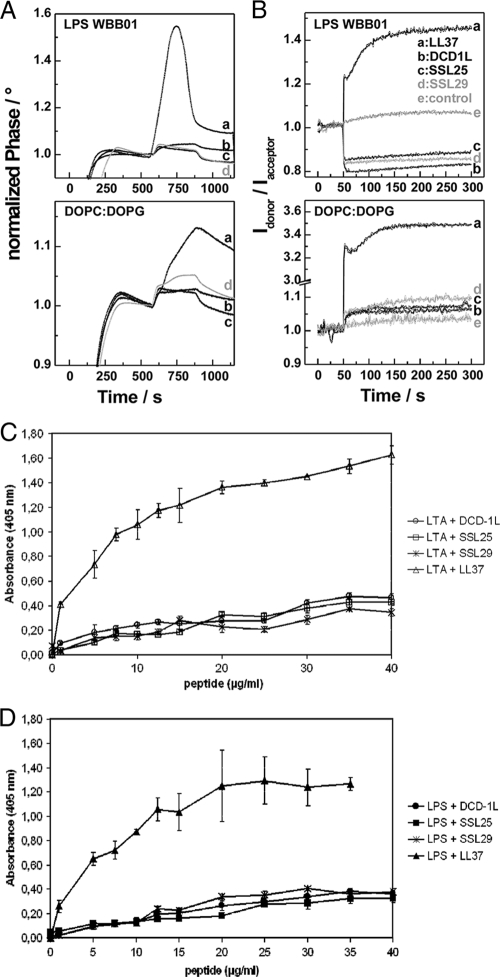
First, we incubated DCD-1L (anionic), SSL-25 (cationic), SSL-29 (neutral), and LL-37 (cationic) with unilamellar liposomes made of a 1:1 molar ratio of the phospholipids phosphatidylcholine (DOPC) and phosphatidylglycerol (DOPG). We measured the interaction or intercalation of the peptides with the model membranes using SAW biosensor analysis and FRET spectroscopy. All analyzed DCD peptides irrespective of charge and antimicrobial activity bound to DOPC-DOPG membranes and could intercalate into the membranes; however, the cathelicidin LL-37 bound much more strongly at the tested concentrations (Fig. 3A and B). Furthermore, binding studies with LPS from gram-negative bacteria indicated that DCD peptides bind only weakly to LPS compared to LL-37 at the tested concentrations (Fig. 3A and B). Addition of lipid II did not have an effect on binding (data not shown).
FIG. 3.
DCD-derived peptides show only weak binding to components of the cell envelope of gram-negative and gram-positive bacteria. (A) Binding of the peptides to LPS from strain WBB01 and DOPC-DOPG membranes determined in a SAW biosensor assay. After the adsorption of the respective lipid (0 to 300 s), 100 μl of each of the peptides (100 μg/ml) was injected at 600 s. The buffer was 10 mM NaCl-10 mM sodium phosphate, pH 7.0; the temperature was 22°C; and the flow rate was 20 μl/min. (B) Intercalation of the peptides into LPS from WBB01 and DOPC-DOPG aggregates. Changes of the FRET signal versus time after addition of 5 μl peptide (1 mg/ml) at 50 s to 1 ml suspensions of aggregates (10 μM) double labeled with NBD-PE and Rh-PE. The buffer was 10 mM NaCl-10 mM sodium phosphate, pH 7.0; the temperature was 37°C. (C and D) Dose-dependent binding of DCD-derived peptides (DCD-1L, SSL-25, and SSL-29) and LL-37 to 2 μg of immobilized LTA (C) or LPS (D) on microtiter plates. Binding assays were performed in triplicate, and the mean values with standard deviations are shown.
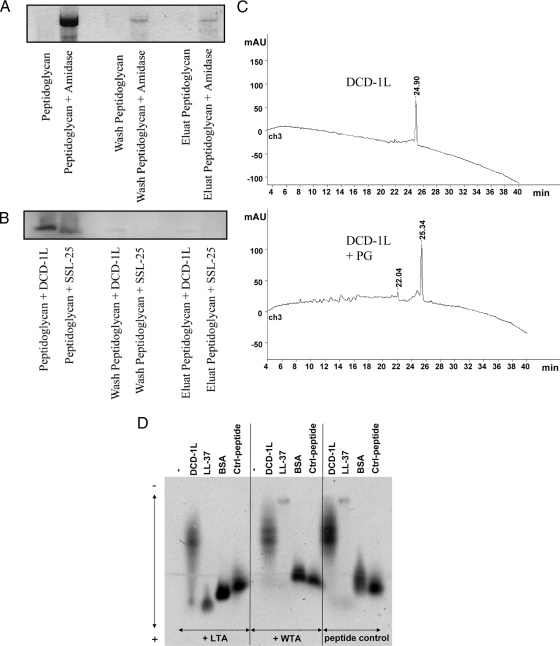
In order to test whether DCD peptides are able to interact with components of gram-positive bacteria, we performed binding studies with LTA, WTA, and PG isolated from S. aureus. Similar to the results from the studies described above, DCD peptides showed only a weak binding to LTA in contrast to LL-37, which bound in a dose-dependent fashion to these components (Fig. 3C and 4D). Furthermore, DCD-1L and SSL-25 showed only low-level binding to PG (Fig. 4B and C), in contrast to S. aureus amidase used as a positive control (Fig. 4A). Interestingly, both DCD-1L and LL-37 did not bind to WTA (Fig. 4D).
FIG. 4.
DCD-derived peptides show only weak binding to PG and teichoic acids of gram-positive bacteria. (A to C) Shown is a PG binding assay. Peptides and amidase as a control were incubated with PG. After incubation, supernatant, wash fraction, and eluate were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie blue. (A) S. aureus amidase can be detected in all fractions (supernatant, wash, and elution), indicating binding to PG. (B) DCD-1L and SSL-25 could be detected only in supernatant; however, there was no visible detection in the wash or the elution fraction. (C) PG binding was observed via high-pressure liquid chromatography by comparing decreases of DCD-1L before and after incubation with PG. The absence of a decrease in DCD-1L reveals that there was no binding to PG. mAU, milliabsorption units over time (in min). (D) Peptide gel shift assay. FITC-labeled peptides (0.25 mg/ml) were incubated with or without the putative binding partners WTA and LTA (0.75 mg/ml). Native agarose gel electrophoresis of the peptides revealed a measurable gel shift toward the anode due to the negative charge of LTA/WTA when there was binding. Whereas no binding was seen with WTA, DCD-1L-FITC (moderate binding) and LL-37-FITC (strong binding) showed moderate and strong binding, respectively, to LTA. BSA-FITC and another control peptide served as controls. Using lower DCD peptide concentrations did not result in different binding behavior.
Antimicrobial activity against S. aureus cell envelope mutants.
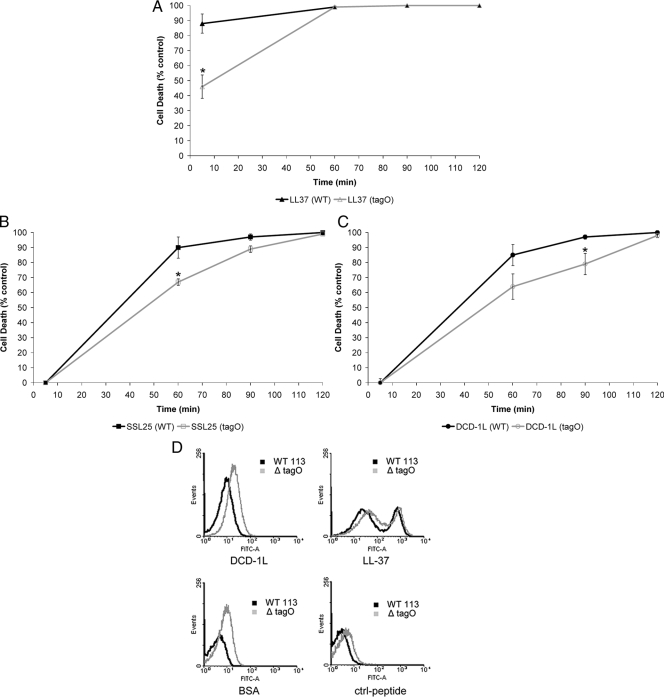
To analyze whether mutants of S. aureus lacking specific cell envelope modifications are more susceptible to cationic and anionic DCD-derived peptides, we performed antimicrobial assays with the S. aureus Δatl mutant, deficient in the major autolysin; the Δsrt mutant, deficient in sortase; the ΔspA mutant, deficient in protein A; the ΔypfP mutant, with a reduced level of LTA; and the ΔtagO mutant, deficient in WTA, and the corresponding wild-type strain 113. As can be seen in Table 2, all mutants showed susceptibilities to DCD-1L, SSL-25, and LL-37 similar to that of the wild-type S. aureus. Interestingly, the S. aureus ΔtagO mutant showed a time kinetic of antimicrobial activity of DCD-1L, SSL-25, and LL-37 different from that of the wild type (Fig. 5A to C). To figure out the reason for the delay of antimicrobial activity in the ΔtagO mutant, we performed binding studies with FITC-labeled DCD-1L and LL-37 as well as an irrelevant control peptide and the control protein BSA with the wild-type strain and the ΔtagO mutant and directly analyzed binding of the FITC-labeled peptides to the bacterial envelope by flow cytometry (Fig. 5D). Interestingly, binding of all peptides including the control peptides to the cell envelope of the ΔtagO mutant was stronger than to the wild-type S. aureus strain. This indicated that the WTA-deficient mutant had the ability to bind peptides in a more unspecific way.
TABLE 2.
Inhibitory concentrations of DCD-1L, SSL-25, and LL-37 on S. aureus 113 wild-type strain and mutants lacking cell envelope modifications
| S. aureus straina | IC90 (μg/ml)b
|
||
|---|---|---|---|
| DCD-1L | SSL-25 | LL-37 | |
| 113 wild type | 12.5 (±0.7) | 12 (±2.8) | 0.6 (±0.2) |
| 113 Δatl | 8 (±2.4) | 8 (±2.8) | 0.5 (±0.1) |
| 113 Δsrt | 11 (±1.4) | 9 (±1.4) | 0.7 (±0.1) |
| 113 ΔspA | 9.5 (±0.7) | 9 (±1) | 1.5 (±0.7) |
| 113 ΔypfP | 12 (±1.7) | 10 (±0.3) | 0.5 (±0.1) |
| 113 ΔtagO | 18 (±1.7) | 15 (±1) | 1.5 (±0.7) |
The following defined S. aureus mutants were used: Δatl mutant, deficient in autolysin; Δsrt mutant, deficient in sortase; ΔspA mutant, deficient in protein A; ΔypfP mutant, reduced level of LTA; ΔtagO mutant, deficient in WTA.
IC90 designates the concentration of peptide in μg/ml able to kill 90% of the microorganisms (1 × 106 CFU/ml) in 10 mM sodium phosphate-10 mM NaCl in a CFU assay compared to the control incubated only in buffer without peptide. The values are means ± standard deviations of at least three independent experiments. There is no significant change of antimicrobial activity for the mutant strains compared to the wild-type S. aureus strain 113 (P > 0.05).
FIG. 5.
WTA deficiency in S. aureus confers delayed activity of DCD-derived peptides and LL-37 by unspecific binding of peptides. Killing characteristics expressed by cell death (percent) over time (min). Comparison of S. aureus wild-type (WT) strain 113 versus the ΔtagO mutant. (A to C) Peptides used in the killing assay were LL-37 (A), SSL-25 (B), and DCD-1L (C) at concentrations of 10 μg/ml each. At least three experiments were performed in triplicate. All three AMPs show delayed activity toward S. aureus ΔtagO. Significant changes are marked with an asterisk (P < 0.05). (D) Flow cytometric analyses of FITC-labeled DCD-1L, BSA, LL-37, and control peptide (50 μg/ml) after incubation for 2 h with S. aureus wild-type strain 113 and ΔtagO mutant (1 × 108 CFU/ml in 10 mM sodium phosphate buffer-10 mM NaCl). Comparison of binding of either BSA, another control peptide (GATTTAPSLSGKGNPEEEDVDTSQVLYE), DCD-1L, or LL-37 to S. aureus wild-type strain 113 and the ΔtagO mutant.
DISCUSSION
In order to elucidate the mechanism of action of DCD-derived peptides, we studied bacterial factors that govern sensitivity or tolerance to DCD in the model microorganism S. aureus. We could show that there is a time-dependent killing by anionic and cationic DCD-derived peptides which is followed by bacterial membrane depolarization without pore formation. Interestingly, DCD-derived peptides inhibit bacterial macromolecular synthesis without binding to microbial DNA or RNA. In our previous studies using immunoelectron microscopy, we could show that anionic and cationic DCD peptides interact with the bacterial cell envelope (41). In order to identify the targets of DCD peptide binding, we performed binding studies with components of the cell envelope of gram-positive and gram-negative bacteria and with model membranes. These studies indicated that DCD-derived peptides show only weak binding to LPS from gram-negative bacteria as well as to PG, LTA, and WTA isolated from S. aureus. In contrast, LL-37 binds strongly to these components in a dose-dependent fashion. This is also reflected by the stronger binding of LL-37 than of DCD-1L to the cell envelope of S. aureus as seen by flow cytometric analyses. These data indicate that the mode of action of DCD-derived peptides is different from that of the cathelicidin LL-37 and probably most other pore-formation-inducing AMPs.
In our previous studies, we showed that DCD peptides interact with the bacterial cell envelope. We showed that the active part of the DCD-1L sequence resides in the first 23 amino acids and that the net charge of the DCD peptides is not essential for the antimicrobial function. Cationic (SSL-25) and anionic (DCD-1L) DCD peptides exhibit similar spectra of activity against several gram-positive and gram-negative bacteria including methicillin-resistant S. aureus (41). In this study we extended these studies and showed that the anionic DCD-1L and the cationic SSL-25 behave similarly in respect to inhibition of macromolecular synthesis, depolarization of the bacterial membrane, and binding to components of the bacterial cell envelope in S. aureus. Furthermore, our data indicate that the charge of the DCD peptide is not essential for binding to the bacterial membrane or for structure formation and that the first 23 amino acids of DCD-1L are crucial for binding of the peptide to the bacterial cell envelope.
Disruption of the dltA or mprF gene in S. aureus increases the susceptibility to several cationic AMPs like defensins, protegrins, or amphibian magainin (35, 36). In previous studies we showed that in a similar way the mprF and dltA bacterial envelope mutants lacking cell envelope modifications are more sensitive to the activity of DCD peptides irrespective of the charge of the latter (41). In this study we found a similar susceptibility toward DCD-1L, SSL-25, and LL-37 of several mutants of S. aureus lacking specific cell envelope modifications compared to the wild-type S. aureus strain. Interestingly, it was recently shown that WTA deficiency in S. aureus confers a selective increase in bacterial resistance to the AMPs mammalian group IIA phospholipase A2 and human β-defensin 3 (22). These findings suggested a selective role of WTA in the antistaphylococcal actions of certain AMPs and indicate an important and general role of components of the staphylococcal cell envelope in the sensitivity to several AMPs including DCD peptides. In this study we show that binding of DCD peptides and the control peptides to the cell envelope of the WTA-deficient mutant is stronger than to the wild-type S. aureus strain. This may be due to the distinct morphology of the ΔtagO mutant, which has a rough surface with many surface protrusions, in contrast to wild-type S. aureus cells, which are round and have a smooth surface (22). This suggests that the increase in resistance to several AMPs in this mutant may be partly due to the fact that AMPs are trapped in an unspecific way on the rough and enlarged cell envelope and because of this can find their specific targets only in a less efficient way.
Cell death due to cationic AMPs may begin as quickly as 2 to 3 min after initial exposure (4, 16, 26, 43). It has been primarily attributed to membrane perturbation due to pore formation, membrane permeabilization, or depolarization of the bacterial membrane, which leads to loss of ions and metabolites, cessation of essential vital functions, and ultimately to cell death (5, 17, 20, 48). In gram-negative bacteria human defensins sequentially permeabilize the outer and the inner membrane (13, 26). In gram-positive cells, exposure to some AMPs results in an immediate increase in water and ion flow, an efflux of ions, swelling, and osmotic dysregulation (21, 29). Time kinetics indicated that the killing of bacteria by DCD peptides is a rather slow process, taking at least 1 hour in vitro. This is in contrast to LL-37, which is able to kill the bacteria by membrane permeabilization after the first few minutes. Interestingly, DCD peptides show a time-dependent inhibition pattern of macromolecular synthesis similar to that of LL-37, although the two differ with respect to the ability to induce pore formation and binding to components of the bacterial cell envelope. This suggests that inhibition of RNA and protein synthesis by these AMPs is only a secondary effect and that the mechanism of antimicrobial activity of these peptides has still to be elucidated. Since DCD peptides are antimicrobially active against gram-negative and gram-positive microorganisms and Candida albicans, DCD might use a general mechanism of action which is similar in different types of microorganisms.
It has been suggested that a given AMP may use more than one antimicrobial mechanism (37). In this respect DCD and LL-37 are two examples with different major but also overlapping antimicrobial mechanisms. It may be that DCD peptides act similarly to the non-membrane-permeabilizing AMPs already described (5, 31, 48). Several AMPs are suggested to target bacterial components other than membranes such as nucleic acids. In recent years it became evident that a series of cationic antibacterial peptides can enter bacterial cells by passive transport (11). Buforin II and its truncated analogs penetrate the cell membrane but do not permeabilize it. Buforin II kills bacteria by binding to nucleic acids (33). Furthermore, several AMPs are able to kill microorganisms by inhibition of macromolecular biosynthesis (18, 33, 44) or by interacting with specific vital components inside the microorganisms (39). Similarly to buforin II, thrombin-induced platelet microbicidal protein 1 and human neutrophil defensin 1 kill bacteria slowly by inhibiting protein and DNA synthesis (47). The frog AMP dermaseptin inhibits RNA synthesis. It translocates across lipid bilayers but does not permeabilize membranes (34). The proline-rich peptide apidaecin binds nonspecifically to outer membrane components, followed by invasion of periplasmic space and by a specific and irreversible engagement with a membrane-bound receptor molecule. Finally, the peptide is translocated into the cell, where it meets its ultimate target (8). Pyrrhocoricin and drosocin are proline-rich AMPs and kill bacteria by inactivating ATPase activity of the bacterial heat shock protein DnaK and inhibiting chaperone-assisted protein folding (23, 32). Apparently many AMPs interfere with bacterial enzymatic processes (31). One hypothesis is that in addition to membrane activities, cationic peptides and proteins might render bacteria nonviable by activating their autolytic wall enzymes, muramidases, resulting in bacteriolysis (14). In this respect cationic AMPs mimic the bactericidal effects exerted by β-lactam antibiotics.
Analysis of the bacterial response to DCD-1L peptide challenge by transcriptome analysis in S. epidermidis indicated that DCD induces a general stress response with upregulated expression of the global regulatory systems agr, sarA, and saeRS (24). However, this stress response seems to be a secondary effect and not essential for the antimicrobial function of DCD peptides, since S. aureus mutants lacking each of these global regulators show the same sensitivity toward DCD-1L and SSL-25.
In conclusion, we suggest a model in which negatively or positively charged DCD peptides bind to the bacterial cell envelope by electrostatic interaction and by binding to several defined targets (not yet identified) without causing massive permeabilization. This may impair vital functions to such an extent that the system gets highly stressed and gives the bacterium the signal to reduce bacterial RNA and protein synthesis. Still, the mode of antimicrobial activity of DCD-derived peptides has to be elucidated and the targets in the bacterial envelope to which DCD peptides bind have to be identified.
Acknowledgments
We thank Ines Wanke and Jasmin Schnaut for expert technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft SCHI 510/3-3, SFB 766, and SFB 617 (T.G.).
Footnotes
Published ahead of print on 3 April 2009.
REFERENCES
- 1.Andra, J., A. Bohling, T. M. Gronewold, U. Schlecht, M. Perpeet, and T. Gutsmann. 2008. Surface acoustic wave biosensor as a tool to study the interaction of antimicrobial peptides with phospholipid and lipopolysaccharide model membranes. Langmuir 24:9148-9153. [DOI] [PubMed] [Google Scholar]
- 2.Baechle, D., T. Flad, A. Cansier, H. Steffen, B. Schittek, J. Tolson, T. Herrmann, H. Dihazi, A. Beck, G. A. Mueller, M. Mueller, S. Stevanovic, C. Garbe, C. A. Mueller, and H. Kalbacher. 2006. Cathepsin D is present in human eccrine sweat and involved in the postsecretory processing of the antimicrobial peptide DCD-1L. J. Biol. Chem. 281:5406-5415. [DOI] [PubMed] [Google Scholar]
- 3.Bals, R. 2000. Epithelial antimicrobial peptides in host defense against infection. Respir. Res. 1:141-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondelle, S. E., K. Lohner, and M. Aguilar. 1999. Lipid-induced conformation and lipid-binding properties of cytolytic and antimicrobial peptides: determination and biological specificity. Biochim. Biophys. Acta 1462:89-108. [DOI] [PubMed] [Google Scholar]
- 5.Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. [DOI] [PubMed] [Google Scholar]
- 6.Brotz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H. G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brotz, H., M. Josten, I. Wiedemann, U. Schneider, F. Gotz, G. Bierbaum, and H. G. Sahl. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317-327. [DOI] [PubMed] [Google Scholar]
- 8.Castle, M., A. Nazarian, S. S. Yi, and P. Tempst. 1999. Lethal effects of apidaecin on Escherichia coli involve sequential molecular interactions with diverse targets. J. Biol. Chem. 274:32555-32564. [DOI] [PubMed] [Google Scholar]
- 9.Cipakova, I., J. Gasperik, and E. Hostinova. 2006. Expression and purification of human antimicrobial peptide, dermcidin, in Escherichia coli. Protein Expr. Purif. 45:269-274. [DOI] [PubMed] [Google Scholar]
- 10.Flad, T., R. Bogumil, J. Tolson, B. Schittek, C. Garbe, M. Deeg, C. A. Mueller, and H. Kalbacher. 2002. Detection of dermcidin-derived peptides in sweat by ProteinChip technology. J. Immunol. Methods 270:53-62. [DOI] [PubMed] [Google Scholar]
- 11.Futaki, S., S. Goto, T. Suzuki, I. Nakase, and Y. Sugiura. 2003. Structural variety of membrane permeable peptides. Curr. Protein Pept. Sci. 4:87-96. [DOI] [PubMed] [Google Scholar]
- 12.Galanos, C., O. Luderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9:245-249. [DOI] [PubMed] [Google Scholar]
- 13.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 14.Ginsburg, I. 2004. Bactericidal cationic peptides can also function as bacteriolysis-inducing agents mimicking beta-lactam antibiotics; it is enigmatic why this concept is consistently disregarded. Med. Hypotheses 62:367-374. [DOI] [PubMed] [Google Scholar]
- 15.Hale, J. D., and R. E. Hancock. 2007. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti Infect. Ther. 5:951-959. [DOI] [PubMed] [Google Scholar]
- 16.Hancock, R. E., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 18.Haukland, H. H., H. Ulvatne, K. Sandvik, and L. H. Vorland. 2001. The antimicrobial peptides lactoferricin B and magainin 2 cross over the bacterial cytoplasmic membrane and reside in the cytoplasm. FEBS Lett. 508:389-393. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, C. H., C. Chen, M. L. Jou, A. Y. Lee, Y. C. Lin, Y. P. Yu, W. T. Huang, and S. H. Wu. 2005. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res. 33:4053-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jepras, R. I., F. E. Paul, S. C. Pearson, and M. J. Wilkinson. 1997. Rapid assessment of antibiotic effects on Escherichia coli by bis-(1,3-dibutylbarbituric acid) trimethine oxonol and flow cytometry. Antimicrob. Agents Chemother. 41:2001-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juretic, D., H. C. Chen, J. H. Brown, J. L. Morell, R. W. Hendler, and H. V. Westerhoff. 1989. Magainin 2 amide and analogues. Antimicrobial activity, membrane depolarization and susceptibility to proteolysis. FEBS Lett. 249:219-223. [DOI] [PubMed] [Google Scholar]
- 22.Koprivnjak, T., C. Weidenmaier, A. Peschel, and J. P. Weiss. 2008. Wall teichoic acid deficiency in Staphylococcus aureus confers selective resistance to mammalian group IIA phospholipase A2 and human β-defensin 3. Infect. Immun. 76:2169-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kragol, G., S. Lovas, G. Varadi, B. A. Condie, R. Hoffmann, and L. Otvos, Jr. 2001. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40:3016-3026. [DOI] [PubMed] [Google Scholar]
- 24.Lai, Y., A. E. Villaruz, M. Li, D. J. Cha, D. E. Sturdevant, and M. Otto. 2007. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol. Microbiol. 63:497-506. [DOI] [PubMed] [Google Scholar]
- 25.Lai, Y. P., Y. F. Peng, Y. Zuo, J. Li, J. Huang, L. F. Wang, and Z. R. Wu. 2005. Functional and structural characterization of recombinant dermcidin-1L, a human antimicrobial peptide. Biochem. Biophys. Res. Commun. 328:243-250. [DOI] [PubMed] [Google Scholar]
- 26.Lehrer, R. I., A. Barton, K. A. Daher, S. S. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchand, C., K. Krajewski, H. F. Lee, S. Antony, A. A. Johnson, R. Amin, P. Roller, M. Kvaratskhelia, and Y. Pommier. 2006. Covalent binding of the natural antimicrobial peptide indolicidin to DNA abasic sites. Nucleic Acids Res. 34:5157-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mason, D. J., and V. A. Gant. 1995. The application of flow cytometry to the estimation of bacterial antibiotic susceptibility. J. Antimicrob. Chemother. 36:441-443. [DOI] [PubMed] [Google Scholar]
- 29.Matsuzaki, T., M. Irahara, and T. Aono. 1997. Physiology and action of prolactin. Nippon Rinsho 55:2871-2875. (In Japanese.) [PubMed] [Google Scholar]
- 30.Nuding, S., K. Fellermann, J. Wehkamp, H. A. Mueller, and E. F. Stange. 2006. A flow cytometric assay to monitor antimicrobial activity of defensins and cationic tissue extracts. J. Microbiol. Methods 65:335-345. [DOI] [PubMed] [Google Scholar]
- 31.Otvos, L., Jr. 2005. Antibacterial peptides and proteins with multiple cellular targets. J. Pept. Sci. 11:697-706. [DOI] [PubMed] [Google Scholar]
- 32.Otvos, L., Jr., O. Insug, M. E. Rogers, P. J. Consolvo, B. A. Condie, S. Lovas, P. Bulet, and M. Blaszczyk-Thurin. 2000. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 39:14150-14159. [DOI] [PubMed] [Google Scholar]
- 33.Park, C. B., H. S. Kim, and S. C. Kim. 1998. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 244:253-257. [DOI] [PubMed] [Google Scholar]
- 34.Patrzykat, A., C. L. Friedrich, L. Zhang, V. Mendoza, and R. E. Hancock. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46:605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 37.Peschel, A., and H. G. Sahl. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529-536. [DOI] [PubMed] [Google Scholar]
- 38.Rieg, S., S. Seeber, H. Steffen, A. Humeny, H. Kalbacher, S. Stevanovic, A. Kimura, C. Garbe, and B. Schittek. 2006. Generation of multiple stable dermcidin-derived antimicrobial peptides in sweat of different body sites. J. Investig. Dermatol. 126:354-365. [DOI] [PubMed] [Google Scholar]
- 39.Ruissen, A. L., J. Groenink, E. J. Helmerhorst, E. Walgreen-Weterings, W. Van't Hof, E. C. Veerman, and A. V. Nieuw Amerongen. 2001. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 356:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schittek, B., R. Hipfel, B. Sauer, J. Bauer, H. Kalbacher, S. Stevanovic, M. Schirle, K. Schroeder, N. Blin, F. Meier, G. Rassner, and C. Garbe. 2001. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat. Immunol. 2:1133-1137. [DOI] [PubMed] [Google Scholar]
- 41.Steffen, H., S. Rieg, I. Wiedemann, H. Kalbacher, M. Deeg, H. G. Sahl, A. Peschel, F. Gotz, C. Garbe, and B. Schittek. 2006. Naturally processed dermcidin-derived peptides do not permeabilize bacterial membranes and kill microorganisms irrespective of their charge. Antimicrob. Agents Chemother. 50:2608-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tobias, P. S., K. Soldau, and R. J. Ulevitch. 1989. Identification of a lipid A binding site in the acute phase reactant lipopolysaccharide binding protein. J. Biol. Chem. 264:10867-10871. [PubMed] [Google Scholar]
- 43.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 44.Ulvatne, H., O. Samuelsen, H. H. Haukland, M. Kramer, and L. H. Vorland. 2004. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Lett. 237:377-384. [DOI] [PubMed] [Google Scholar]
- 45.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 46.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H. G. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 47.Xiong, Y. Q., M. R. Yeaman, and A. S. Bayer. 1999. In vitro antibacterial activities of platelet microbicidal protein and neutrophil defensin against Staphylococcus aureus are influenced by antibiotics differing in mechanism of action. Antimicrob. Agents Chemother. 43:1111-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]
- 49.Yenugu, S., K. G. Hamil, F. S. French, and S. H. Hall. 2004. Antimicrobial actions of the human epididymis 2 (HE2) protein isoforms, HE2alpha, HE2beta1 and HE2beta2. Reprod. Biol. Endocrinol. 2:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, X. Q., and M. R. Kanost. 2000. Immulectin-2, a lipopolysaccharide-specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J. Biol. Chem. 275:37373-37381. [DOI] [PubMed] [Google Scholar]
- 51.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]