Abstract
We report here the characterization of a novel aminoglycoside resistance gene, aac(6′)-Iaf, present in two multidrug-resistant (MDR) Pseudomonas aeruginosa clinical isolates. These isolates, IMCJ798 and IMCJ799, were independently obtained from two patients, one with a urinary tract infection and the other with a decubitus ulcer, in a hospital located in the western part of Japan. Although the antibiotic resistance profiles of IMCJ798 and IMCJ799 were similar to that of MDR P. aeruginosa IMCJ2.S1, which caused outbreaks in the eastern part of Japan, the pulsed-field gel electrophoresis patterns for these isolates were different from that for IMCJ2.S1. Both IMCJ798 and IMCJ799 were found to contain a novel chromosomal class 1 integron, In123, which included aac(6′)-Iaf as the first cassette gene. The encoded protein, AAC(6′)-Iaf, was found to consist of 183 amino acids, with 91 and 87% identity to AAC(6′)-Iq and AAC(6′)-Im, respectively. IMCJ798, IMCJ799, and Escherichia coli transformants carrying a plasmid containing the aac(6′)-Iaf gene and its upstream region were highly resistant to amikacin, dibekacin, and kanamycin but not to gentamicin. The production of AAC(6′)-Iaf in these strains was confirmed by Western blot analysis. Thin-layer chromatography indicated that AAC(6′)-Iaf is a functional acetyltransferase that specifically modifies the amino groups at the 6′ positions of aminoglycosides. Collectively, these findings indicate that AAC(6′)-Iaf contributes to aminoglycoside resistance.
Pseudomonas aeruginosa is a nosocomial pathogen that exhibits a remarkable ability to acquire resistance to several antibiotics. The most serious problem has been the emergence of multidrug-resistant (MDR) P. aeruginosa strains with resistance to all β-lactams, aminoglycosides, and quinolones (39, 40). In Japan, MDR P. aeruginosa is defined as having resistance to carbapenem (MIC ≥ 16 μg/ml), amikacin (AMK; MIC ≥ 32 μg/ml), and fluoroquinolone (MIC ≥ 4 μg/ml).
Bacterial resistance to aminoglycosides can result from three causes (44): decreased membrane permeability (13), the modification of 16S RNA (14, 16, 17, 49) or ribosomal proteins (13), and the enzymatic modification of aminoglycosides. In P. aeruginosa isolates, resistance to aminoglycosides is due primarily to the production of aminoglycoside-modifying enzymes (4, 47). The aminoglycoside acetyltransferases (AACs) are aminoglycoside-modifying enzymes that transfer acetyl groups to the amino groups of aminoglycosides. The AACs can be grouped into four classes, AAC(1), AAC(2′), AAC(3′), and AAC(6′), based on the acetylation sites of the aminoglycosides (22, 44). N-acetylation at the 6′ position catalyzed by AAC(6′) is one of the most prevalent forms of modification of aminoglycosides (32). AAC(6′)-I confers resistance to AMK but not to gentamicin (GEM) (41). To date, at least 27 AAC(6′)-I enzymes, designated AAC(6′)-Ia to AAC(6′)-Iae, have been identified and characterized (15, 22, 38, 44). In contrast, only two AAC(6′)-II enzymes, which confer resistance to GEM but not to AMK, have been identified (41). The aac genes are often found in class 1 integrons (21). These integrons possess two conserved segments at each end, separated by a variable region that includes integrated antibiotic resistance gene cassettes (19, 20). The 5′-conserved segment (5′-CS) contains the intI gene, and the 3′-conserved segment (3′-CS) contains the qacEΔ1 and sul1 genes (19).
We previously described a nosocomial outbreak of catheter-associated urinary tract infection with an MDR P. aeruginosa strain, IMCJ2.S1, in a hospital in the Tohoku region in the eastern part of Japan (39). IMCJ2.S1 was found to harbor an aminoglycoside 6′-N-acetyltransferase gene, aac(6′)-Iae, in a chromosomal integron. We developed kits to detect the aac(6′)-Iae gene and the AAC(6′)-Iae protein and used these kits to survey MDR P. aeruginosa strains in hospitals throughout Japan (27, 39). During surveillance in the western part of Japan, two MDR P. aeruginosa clinical isolates negative for aac(6′)-Iae were identified. Each of these isolates contained a novel aminoglycoside 6′-N-acetyltransferase gene, aac(6′)-Iaf. We report here the structure of this gene and the properties of its product.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Two P. aeruginosa clinical isolates, IMCJ798 and IMCJ799, were individually obtained from two patients, one with a urinary tract infection and the other with a decubitus ulcer. P. aeruginosa ATCC 27853 was obtained from the American Type Culture Collection (Manassas, VA) and used as a reference strain for antibiotic susceptibility testing. Escherichia coli strains DH5α (Takara Bio, Shiga, Japan) and JM109 (Stratagene, La Jolla, CA) were used as hosts for recombinant plasmids. E. coli BL21(DE3)(pLysS) (Invitrogen, Carlsbad, CA) was used for the expression of recombinant aac(6′)-Iaf. A rifampin-resistant mutant of P. aeruginosa, ATCC 27853 Rfpr, was used for conjugation. P. aeruginosa GN17203, carrying plasmid pMS350 containing blaIMP-1 (46), was kindly provided by S. Iyobe (Kitasato University, Sagamihara, Japan).
Antimicrobial agents.
Amikacin (AMK) and imipenem (IPM) were obtained from Banyu Pharmaceutical Co. (Tokyo, Japan), arbekacin (ABK) and dibekacin (DIB) were purchased from Meiji Seika Kaisha, Ltd. (Tokyo, Japan), aztreonam (ATM) was obtained from Eizai (Tokyo, Japan), ceftazidime (CAZ) was acquired from GlaxoSmithKline K.K. (Tokyo, Japan), gentamicin (GEM) and neomycin B and C mixtures (NEO) were obtained from Nacalai Tesque, Inc. (Kyoto, Japan), isepamicin (ISP), netilmicin (NET), and sisomicin (SIS) were from Schering-Plough K.K. (Osaka, Japan), kanamycin A (KAN) and polymyxin B (PMB) were purchased from Sigma-Aldrich (St. Louis, MO), meropenem (MEM) was obtained from Sumitomo Pharmaceutical Co., Ltd. (Osaka, Japan), ofloxacin (OFX) was acquired from Daiichi Pharmaceutical Co., Ltd. (Tokyo, Japan), piperacillin (PIP) and piperacillin-tazobactam (TZP) were obtained from Tomiyama Pure Chemical Industries, Ltd. (Tokyo, Japan), and tobramycin (TOB) was purchased from Towa Pharmaceutical Co., Ltd. (Osaka, Japan).
In vitro susceptibility tests.
MICs were determined using a microdilution method according to the protocols recommended by the Clinical and Laboratory Standards Institute (10).
Serotyping.
The O serotypes of isolates were determined with a slide agglutination test kit (Denka Seiken Co., Tokyo, Japan).
Detection of MDR P. aeruginosa using a LAMP method and an agglutination test.
The aac(6′)-Iae gene was assessed using a loop-mediated isothermal amplification (LAMP) method, and the AAC(6′)-Iae protein was evaluated using an agglutination test, as described previously (39).
PFGE.
DNA plugs were prepared as described previously (18) and digested overnight at 37°C with SpeI and XbaI (Takara Bio). Pulsed-field gel electrophoresis (PFGE) analysis was performed as described previously (38).
PCR amplification of class 1 integrons.
Genomic DNA was extracted as described previously (36) and used as PCR templates. Class 1 integrons were detected by PCR using 5′-CS and 3′-CS primers as described previously (11, 29) and genetically mapped using the primers listed in Table 1. An Expand high-fidelity PCR system (Roche Diagnostics GmbH, Penzberg, Germany) was used for all PCR amplifications. All PCR products were sequenced to identify genes and their orders in the integrons.
TABLE 1.
PCR primers used in this study
| Primer | Sequencea (5′ to 3′) | Label in Fig. 2 | Description | Reference |
|---|---|---|---|---|
| 5′-CS | GGCATCCAAGCAGCAAG | B | 5′-End common segment of class 1 integrons | 29 |
| 3′-CS | AAGCAGACTTGACCTGA | F | 3′-End common segment of class 1 integrons | 29 |
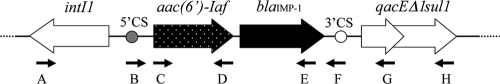
| intI-R | TGCGTGTAAATCATCGTCGT | A | Positions 196-177 in intI1 | 38 |
| qacEdelta-R | GCAATTATGAGCCCCATACC | G | Positions 287-268 in qacEΔ1 | 38 |
| sul1-R | GGGTTTCCGAGAAGGTGATT | H | Positions 787-768 in sul1 | 38 |
| aac(6′)Iaf-F | TTGGACTATTCAATATGCGA | C | Positions 1-20 in aac(6′)-Iaf | This study |
| aac(6′)Iaf-R | CTAGCTAATATCTTTCCACA | D | Positions 552-533 in aac(6′)-Iaf | This study |
| blaIMP-1-F | GAAGTTAACGGGTGGGGCG | Positions 124-142 in blaIMP-1 | This study | |
| blaIMP-1-R | CTTTAACCGCCTGCTCTAAT | E | Positions 700-681 in blaIMP-1 | This study |
| 16S-rRNA-F | ATGCAAGTCGAGCGGATGAAGGGAG | Positions 55-79 in 16S rRNA gene | This study | |
| 16S-rRNA-R | TAGTCGACATCGTTTACGGCGTGGA | Positions 822-798 in 16S rRNA gene | This study | |
| 23S-rRNA-F | CGAGGACAGTGTATGGTGGGCAGT | Positions 2207-2231 in 23S rRNA gene | This study | |
| 23S-rRNA-R | CTCAACGCCTCACAACGCTTACACA | Positions 2856-2832 in 23S rRNA gene | This study | |
| PstI-aac-F | aactgcagGGCTTGTTATGACTGTTTTT | Sequence in the 185- to 166-bp upstream region of aac(6′)-Iaf with PstI site | This study | |
| EcoRI-aac-R | ggaattcCTAGCTAATATCTTTCCACA | Positions 552-533 in aac(6′)-Iaf with EcoRI site | This study | |
| SphI-aac-F | aaagcatgcgATGGACTATTCAATATGCGA | Positions 1-20 in aac(6′)-Iaf with SphIb | This study | |
| PstI-aac-R | aactgcagCTAGCTAATATCTTTCCACA | Positions 552-533 in aac(6′)-Iaf with PstI site | This study |
Lowercase letters represent restriction enzyme recognition sites attached on the 5′ ends of primers.
The initiation codon TTG in aac(6′)-Iaf was replaced with ATG.
DNA sequencing.
DNA sequences were determined using an ABI PRISM 3100 sequencer (Applied Biosystems). Homology searches of nucleotide and translated protein sequences were performed using BLAST (2, 3). Multiple-sequence alignments and searches for open reading frames (ORFs) were performed using the Clustal W2 program (28) and GENETYX software (Genetyx, Tokyo, Japan). The dendrogram for AACs was determined with the Clustal W2 program (28).
Plasmid extraction.
The methods of Kado and Liu (25) and Casse et al. (8), modified as follows, were used to extract plasmid DNA from P. aeruginosa. The bacterial pellet was lysed by the addition of 2 ml of lysis buffer (50 mM Tris-Cl, 20 mM EDTA, 4% sodium dodecyl sulfate [SDS], pH 12.6), followed by gentle shaking for 30 min at 37°C. The lysate was neutralized by adding 400 μl of 1 M Tris-Cl (pH 7.5), and the proteins were precipitated by adding 250 μl of 5 M NaCl. The solution was extracted with an equal volume of phenol-chloroform solution (1:1, vol/vol). The plasmid DNA in the aqueous phase was precipitated by adding a twofold volume of 100% ethanol. The DNA pellet was collected. Plasmid DNA preparations were analyzed by electrophoresis on 0.7% agarose gels in 0.5× Tris-borate-EDTA buffer at 4°C.
Transformation using plasmid preparations from P. aeruginosa IMCJ798 and IMCJ799.
Plasmid preparations from P. aeruginosa strains were used to transform E. coli DH5α and P. aeruginosa PAO1 by electroporation using a Gene Pulser Xcell system (Bio-Rad Laboratories, Hercules, CA). The transformants were cultured on Luria-Bertani (LB) agar plates containing 20 μg/ml AMK for 24 h at 37°C.
Transfer of aminoglycoside resistance.
Drug resistance was transferred from P. aeruginosa clinical isolates to a rifampin-resistant mutant of P. aeruginosa, ATCC 27853 Rfpr, using the broth mating method (26). The transconjugants were selected on Mueller-Hinton agar plates containing rifampin (200 μg/ml) and IPM (16 μg/ml) or AMK (20 μg/ml).
Genome typing by I-CeuI digestion and Southern blot hybridization.
DNA plugs containing total genomic DNA from isolates were digested overnight with I-CeuI. DNA fragments were separated by PFGE. Southern hybridization was performed using an enhanced chemiluminescence direct nucleic acid-labeling and detection system according to the instructions of the manufacturer (GE Healthcare, Tokyo, Japan), as described previously (24, 30, 34), to determine whether the novel class 1 integron identified in the P. aeruginosa isolates, designated In123, has a chromosomal location. Probes for aac(6′)-Iaf, blaIMP-1, 16S rRNA, and 23S rRNA genes from IMCJ798 were amplified by PCR using the primer sets aac(6′)Iaf-F/aac(6′)Iaf-R, blaIMP-1-F/blaIMP-1-R, 16S-rRNA-F/16S-rRNA-R, and 23S-rRNA-F/23S-rRNA-R, respectively (Table 1).
Cloning of aac(6′)-Iaf gene.
The ORF of aac(6′)-Iaf and 185 bp of the upstream region of the gene, which includes the promoter, were PCR amplified from P. aeruginosa IMCJ798 by using the primer set PstI-aac-F and EcoRI-aac-R (Table 1). The PCR products were digested with EcoRI and PstI and ligated into the PstI and EcoRI sites of pSTV28, at a polarity opposite the transcriptional direction of the promoter on the vector. The plasmids were used to transform DH5α, and transformants were selected on LB agar containing 30 μg/ml of chloramphenicol. The resulting plasmid was designated pSTV-aacWT. To determine MICs, E. coli JM109 was transformed with pSTV-aacWT, which represses transcription driven by the promoter on the pSTV28 vector.
Site-directed mutagenesis.
The putative initiation codon on pSTV-aacWT, TTG, was replaced by ATG by using a QuikChange site-directed mutagenesis kit (Stratagene). The resulting plasmid was designated pSTV-aac(TTG→ATG). To determine MICs, E. coli JM109 was transformed with this plasmid.
Construction of AAC(6′)-Iaf-overexpressing strains.
The aac(6′)-Iaf gene from P. aeruginosa IMCJ798 was PCR amplified using the primer set SphI-aac-F and PstI-aac-R (Table 1), and the product was digested with SphI and PstI and ligated into pQE2 (Invitrogen), which had been digested with the same restriction enzymes. The plasmid was used to transform DH5α, and the transformants were selected on LB agar containing 100 μg/ml of ampicillin. The resulting plasmid, pQE-aac(6′)-Iaf, was used to transform E. coli BL21(DE3)(pLys), which was used for recombinant protein purification.
Purification of recombinant AAC(6′)-Iaf.
E. coli BL21(DE3)(pLysS) carrying plasmid pQE2-aac(6′)-Iaf was grown in LB medium containing 200 μg/ml ampicillin at 37°C until the A600 reached 0.3. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a concentration of 0.1 mM to induce the expression of AAC(6′)-Iaf, and the culture was incubated for 4 h at 37°C. The hexahistidine-tagged AAC(6′)-Iaf was purified from the soluble fraction using Ni-nitrilotriacetic acid agarose according to the instructions of the manufacturer (Qiagen, Tokyo, Japan). The final concentration of protein was determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL).
Purification of native AAC(6′)-Iaf from P. aeruginosa.
Rabbits were immunized with recombinant AAC(6′)-Iaf protein emulsified in Freund's adjuvant. The animal experiments were approved by the ethical committee for animal experiments at the Research Institute of the International Medical Center of Japan. Anti-AAC(6′)-Iaf immunoglobulin G (IgG), purified from the rabbit sera on protein G-Sepharose (GE Healthcare), was coupled to NHS-activated Sepharose according to the instructions of the manufacturer (GE Healthcare). Bacterial cells from overnight cultures of P. aeruginosa IMCJ798 were disrupted by sonication, and the cleared lysate was applied to the IgG-coupled Sepharose column. After the column was washed with phosphate-buffered saline containing 0.05% Tween 20, protein was eluted with 0.1 M glycine-HCl (pH 2.5). The purified protein was dialyzed in Tris-buffered saline (50 mM Tris, 150 mM NaCl, pH 8.0) and separated by SDS-15% polyacrylamide gel electrophoresis (SDS-15% PAGE), and the N-terminal sequence was analyzed by a commercial service (Nippi, Inc., Tokyo, Japan).
Western blotting.
E. coli JM109 bacteria carrying pSTV28, pSTV-aacWT, or pSTV-aac(TTG→ATG) were cultivated for 16 h at 37°C in LB broth containing 30 μg/ml chloramphenicol. P. aeruginosa isolates IMCJ798 and IMCJ799 were cultivated for 16 h at 37°C in LB broth containing 20 μg/ml AMK. One milliliter of each culture was collected by centrifugation, and whole-cell lysates in 200 μl of SDS-PAGE sample buffer were prepared. A 5-μl aliquot of each cell lysate was separated on an SDS-15% PAGE gel, and the proteins were transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% skim milk in a mixture of 20 mM Tris (pH 8.0), 150 mM NaCl, and 0.05% Tween 20 and incubated with rabbit polyclonal anti-AAC(6′)-Iaf antibodies, obtained by immunization with His-AAC(6′)-Iaf. After the incubation of the membranes with secondary horseradish peroxidase-linked anti-rabbit IgG (GE Healthcare), bands were detected by chemiluminescence. The intensity of each band was quantified using Quantity One software (Bio-Rad Laboratories).
TLC analysis of acetylated aminoglycosides.
Mixtures containing 2 mM aminoglycoside, 2 mM acetyl coenzyme A (acetyl-CoA), and 50 μg/ml AAC(6′)-Iaf in 20 μl of phosphate buffer (pH 7.4) were incubated for 16 h at 37°C, 3 μl of each aminoglycoside mixture was spotted onto the surface of a silica gel 60 thin-layer chromatography (TLC) plate containing a fluorescence indicator with a 254-nm excitation wavelength (Merck Ltd., Japan), and the results were developed with 5% phosphate potassium solution. The aminoglycosides and their acetylated products were detected with 0.5% ninhydrin in acetone (50).
Nucleotide sequence accession number.
The nucleotide sequence of In123 determined in this study has been deposited in the EMBL and GenBank databases and the DDBJ and assigned accession number AB462903.
RESULTS AND DISCUSSION
Characterization of P. aeruginosa IMCJ798 and IMCJ799.
We obtained two P. aeruginosa clinical isolates, IMCJ798 and IMCJ799, from two patients, one with a urinary tract infection and the other with a decubitus ulcer, from a hospital in the Chugoku region of western Japan in 2007. After the implementation of various infection control measures, no other patients with such infections were detected.
The genotypic and phenotypic properties of IMCJ798 and IMCJ799 were compared with those of the MDR P. aeruginosa strain IMCJ2.S1, which had been reported previously to be found in Japan (38). The MICs for P. aeruginosa IMCJ798, IMCJ799, IMCJ2.S1, and ATCC 27853 are shown in Table 2. Multidrug resistance phenotypes were observed in IMCJ798 and IMCJ799. These isolates were resistant to all antibiotics except for GEM. In particular, they showed high levels of resistance to β-lactams, AMK, and OFX. These results were similar to those for IMCJ2.S1, except for ABK and GEM (Table 2). IMCJ798, IMCJ799, and IMCJ2.S1 also had the same serotype, O:11. Although IMCJ798 and IMCJ799 seemed to be derived from IMCJ2.S1, both were negative for the aac(6′)-Iae gene by the LAMP method (data not shown) and for AAC(6′)-Iae protein by the agglutination test (data not shown), whereas IMCJ2.S1 was positive for aac(6′)-Iae (39). The PFGE patterns of SpeI- and XbaI-digested fragments from the IMCJ798 and IMCJ799 isolates were identical but differed from those of fragments from IMCJ2.S1 (Fig. 1). The PFGE patterns for IMCJ798 and IMCJ799 showed similarities of 56.4% (SpeI) and 70.5% (XbaI), respectively, to that for IMCJ2.S1. Thus, the genotypic properties of IMCJ798 and IMCJ799 differed from those of IMCJ2.S1, although these strains had similar phenotypes. Further nationwide, hospital-based surveillance of MDR P. aeruginosa is required.
TABLE 2.
Antimicrobial susceptibility parameters of IMCJ798, IMCJ799, IMCJ2.S1, and ATCC 27853 for various antibioticsa
| Isolate name | MIC (μg/ml) of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | CAZ | IPM | MEM | ATM | AMK | ABK | GEM | OFX | PMB | |
| IMCJ798 | 256 | 256 | 512 | 128 | >512 | 64 | 128 | 8 | 4 | >128 | 4 |
| IMCJ799 | 256 | 256 | 512 | 128 | >512 | 64 | 128 | 16 | 2 | >128 | 4 |
| IMCJ2.S1 | 256 | 256 | 512 | 128 | 512 | 128 | 128 | 2 | 16 | 128 | 2 |
| ATCC 27853 | <4 | 4 | <1 | 4 | 1 | 2 | 2 | <0.5 | <1 | <0.5 | 2 |
PIP, piperacillin; TZP, piperacillin-tazobactam; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; ATM, aztreonam; AMK, amikacin; ABK, arbekacin; GEM, gentamicin; OFX, ofloxacin; PMB, polymyxin B.
FIG. 1.
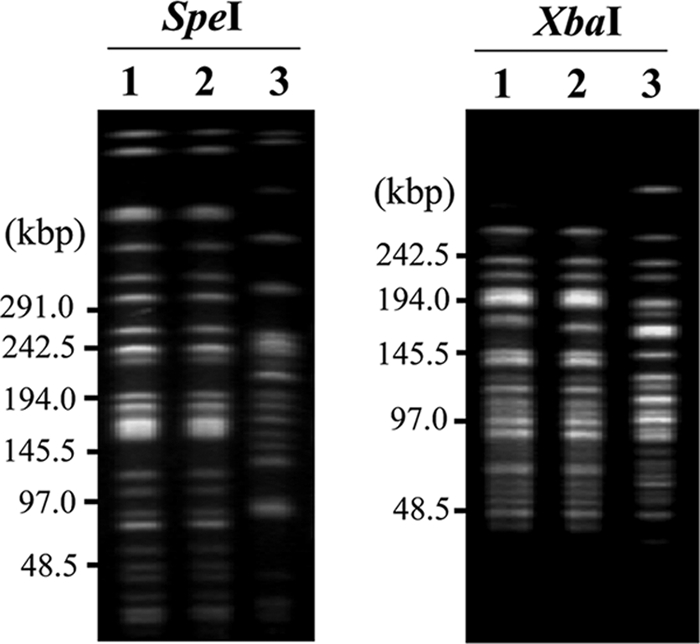
PFGE patterns for SpeI- and XbaI-digested genomic DNA from MDR P. aeruginosa strains IMCJ798 and IMCJ799. The DNA fragments were detected by ethidium bromide staining. Results for IMCJ798 (lanes 1), IMCJ799 (lanes 2), and IMCJ2.S1 (lanes 3) are shown.
aac(6′)-Iaf in the class 1 integron.
To identify the drug resistance genes of IMCJ798 and IMCJ799, the variable regions of class 1 integrons were amplified with primers 5′-CS and 3′-CS (Table 1). Amplicons of 1.7 kbp generated from both strains were found to be identical by DNA sequencing. Sequence analysis revealed a variable region containing two cassettes, one carrying a novel aac(6′) gene and the other carrying a blaIMP-1 metallo-β-lactamase gene (Fig. 2). The novel aac(6′) gene comprised an ORF of 552 bp, starting with a TTG codon, and its sequence showed 94 and 91% identity to those of aac(6′)-Iq from Klebsiella pneumoniae (9) and aac(6′)-Im from Citrobacter freundii (23). Based on the standard nomenclature (45), we named this ORF aac(6′)-Iaf.
FIG. 2.
Genetic structure of In123. Primers labeled A, B, C, D, E, F, G, and H are described in Table 1. Arrows indicate primer locations and directions.
The 5′ CS and 3′ CS of the integron were further mapped with PCR cartography using external primers (Table 1; Fig. 2). Typical 59-base elements (42) were observed in both cassettes. These results supported the idea that the aac(6′)-Iaf gene in P. aeruginosa IMCJ798 and IMCJ799 is localized within the class 1 integron. The sequence of the integron was not found in any database; we therefore named the integron In123.
In addition, the aac(6′)-Iaf gene has a G+C content of 34.4%; in contrast, the average G+C contents of the P. aeruginosa PAO1 and K. pneumoniae MGH78578 genomes are 66.6 and 57.1%, respectively (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi?view=1). These findings suggested that aac(6′)-Iaf may be derived from species with intrinsically low G+C contents, not from Pseudomonas or Klebsiella species.
Location of In123.
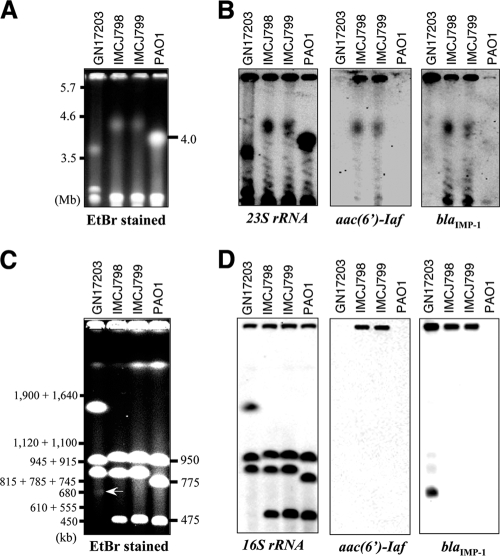
Class 1 integrons are frequently located on plasmids, and they can be transferred among bacteria (5). Plasmid preparation, transformation, conjugation, and Southern hybridization using genomic DNA digested by I-CeuI were carried out to determine the locations and transmission ability of In123 in IMCJ798 and IMCJ799. P. aeruginosa GN17203, which harbors pMS350 containing blaIMP-1, was used as the positive control (46). Initially, we prepared plasmid DNA as described in Materials and Methods. No plasmid in IMCJ798 or IMCJ799 was detected by electrophoresis, whereas pMS350 was detected in P. aeruginosa GN17203 (data not shown). E. coli DH5α and P. aeruginosa PAO1 were transformed with the plasmid DNA preparations by electroporation. No transformants were obtained on LB agar plates containing AMK. In conjugation tests using P. aeruginosa ATCC 27853 Rfpr as a recipient strain, the AMK resistance was not transferred from IMCJ798 and IMCJ799 to P. aeruginosa ATCC 27853 Rfpr whereas carbapenem resistance was transferred from P. aeruginosa GN17203 to ATCC 27853 Rfpr. In order to confirm that In123 is located on the chromosome, PFGE analysis and Southern hybridizations using P. aeruginosa genomic DNA digested by I-CeuI were performed. In all strains, four chromosomal fragments of various sizes (PAO1, 4,063, 950, 775, and 475 kb; GN17203, ca. 3,600, 1,500, 945, and 900 kb; and IMCJ798 and IMCJ799, ca. 4,500, 950, 900, and 480 kb) were detected by the rRNA gene probes (Fig. 3A and C and left panels in B and D). The aac(6′)-Iaf probe detected the 4,500-kb fragments from IMCJ798 and IMCJ799. The band hybridized by the aac(6′)-Iaf probe was also recognized by the rRNA gene probe. The blaIMP-1 probe detected the same fragments in the IMCJ798 and IMCJ799 clinical isolates as the aac(6′)-Iaf and rRNA gene probes. Additionally, the blaIMP-1 probe detected a 700-kbp extrachromosomal fragment, which may correspond to pMS350 in GN17203 (46), that was not detected by the rRNA gene probe. Another smaller blaIMP-1 probe-specific band detected in the IMCJ798 and IMCJ799 clinical isolates, as shown in Fig. 3B, was not observed in the analysis presented in Fig. 3D, probably due to differences in electrophoretic conditions (see the legend to Fig. 3). It is likely that this band was not resolved under the conditions used in the analyses presented in Fig. 3C and D.
FIG. 3.
PFGE patterns (A and C) and Southern hybridization analyses (B and D) of P. aeruginosa genomic DNA digested by I-CeuI. PFGE analysis of P. aeruginosa genomic DNA digested with I-CeuI was done under the following two different sets of conditions: condition set 1 for the separation of the largest, 4.0-Mb fragment of PAO1 (A) consisted of a 106° angle, 0.8% agarose, and linear switching times of 20 to 30 min for 48 h with a voltage gradient of 2 V/cm, and condition set 2 for the separation of the 950-, 775-, and 475-kb fragments of PAO1 (C) consisted of a 120° angle, 1% agarose, and nonlinear switching times of 5.3 to 120 s for 19.5 h with a voltage gradient of 6 V/cm. The molecular standards were Schizosaccharomyces pombe (A) and Saccharomyces cerevisiae YPH80 (C). An arrow in panel C indicates the location of an extrachromosomal band that may correspond to pMS350. DNA fragments for which results are shown in panels A and C were transferred onto membranes and were used for the hybridization analyses presented in panels B and D, respectively. Southern hybridization was performed with probes for rRNA genes, aac(6′)-Iaf, and blaIMP-1, as shown in panels B and D. EtBr, ethidium bromide.
Collectively, these results strongly suggest that In123, which carries aac(6′)-Iaf, is located on a chromosome, not on a plasmid, in P. aeruginosa IMCJ798 and IMCJ799. This arrangement is similar to those for other class 1 integrons, including aac(3)-Ib, aac(3)-Ic, and aac(6′)-Iae integrons in P. aeruginosa (35, 37) and the aac(3)-Id integron in Vibrio fluvialis (1).
Comparison of AAC(6′)-Iaf with other AAC(6′)-I enzymes.
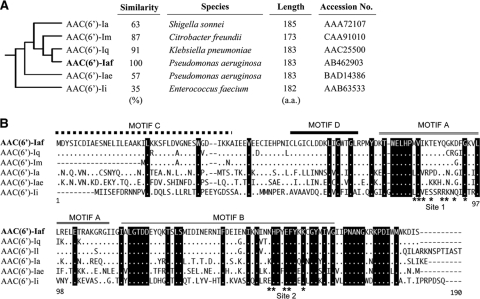
AAC(6′)-Iaf, encoded by the first cassette gene in In123, consists of 183 amino acids. The amino acid sequence of AAC(6′)-Iaf was compared to those of other AAC(6′)-I enzymes. The deduced molecular phylogeny of these sequences suggests that all the AAC(6′)-I enzymes can be classified into three subfamilies (44), the first containing AAC(6′)-Ib and AAC(6′)-Ie, the second containing AAC(6′)-Ic, AAC(6′)-Id, and AAC(6′)-Ih, and the third containing AAC(6′)-Ia, AAC(6′)-Iae, and AAC(6′)-Iq. It was found that AAC(6′)-Iaf belonged to the third subfamily, whose members show considerable phylogenetic distance from those of the other two subfamilies, which include AAC(6′)-Ib or AAC(6′)-Iad (15, 38, 44). Using multiple-sequence alignments, AAC(6′)-Iaf was found to have 91, 87, 63, 57, and 35% identity to AAC(6′)-Iq from K. pneumoniae (9), AAC(6′)-Im from C. freundii (23), AAC(6′)-Ia from Shigella sonnei (43), AAC(6′)-Iae from P. aeruginosa (38), and AAC(6′)-Ii from Enterococcus faecium (12), respectively (Fig. 4A). Moreover, four motifs (C, D, A, and B) of GCN5-related N-acetyltransferases (33) were also observed in AAC(6′)-Iaf, as well as most other AAC(6′)-I enzymes (Fig. 4B). Additionally, the crystal structure of AAC(6′)-Ii, which also belongs to the third subfamily, has been resolved, and two acetyl-CoA binding sites have been reported (6, 7). Putative sites required for acetyl-CoA binding, sites 1 and 2, were also found in AAC(6′)-Iaf (Fig. 4B).
FIG. 4.
Phylogenic tree and amino acid sequence alignments for the AAC(6′)-I subfamily. (A) Phylogenic relationships were determined with the Clustal W2 program. aa, amino acids. (B) Dots indicate amino acids identical to those of AAC(6′)-Iaf. Black-highlighted amino acids are conserved among this AAC(6′)-I subfamily. Dashes represent gaps introduced to optimize similarity.
Effects of aac(6′)-Iaf on aminoglycoside resistance.
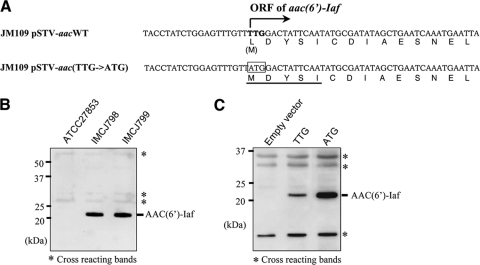
Both P. aeruginosa IMCJ798 and IMCJ799 were resistant to AMK, DIB, ISP, KAN, NET, and TOB but were sensitive to GEM (Table 3). To determine whether aac(6′)-Iaf mediates aminoglycoside resistance, the MICs of various aminoglycosides were assessed using E. coli JM109 transformants carrying pSTV-aacWT or pSTV-aac(TTG→ATG). pSTV-aacWT includes both aac(6′)-Iaf and its 185-bp upstream region from IMCJ798. By using E. coli JM109, the aac(6′)-Iaf gene was expressed utilizing its native promoter and initiation codon. In pSTV-aac(TTG→ATG), the putative initiation codon TTG was replaced with ATG, as shown in the nucleotide sequences in Fig. 5A, to solve the problem of low-level expression in E. coli caused by a rare initiation codon, as described previously (9, 31). As shown in Table 3, the aminoglycoside resistance profile for E. coli JM109 carrying pSTV-aacWT or pSTV-aac(TTG→ATG) was correlated with those for IMCJ798 and IMCJ799. For the E. coli transformants, effective increases of MICs of the same aminoglycosides to which IMCJ798 and IMCJ799 exhibited resistance, AMK, DIB, ISP, KAN, NET, and TOB, were observed. Furthermore, MICs of five of the six above-listed aminoglycosides increased more than twofold for JM109 carrying pSTV-aac(TTG→ATG) compared those for JM109 carrying pSTV-aacWT. To confirm whether the initiation codon of aac(6′)-Iaf is TTG, N-terminal sequencing of the native AAC(6′)-Iaf protein was performed. The native protein was purified from IMCJ798 by using an affinity column with polyclonal anti-AAC(6′)-Iaf rabbit IgG coupled to Sepharose. The molecular mass of purified native AAC(6′)-Iaf, as determined by 15% PAGE, corresponded to the estimated size of approximately 21 kDa. The N-terminal sequence of AAC(6′)-Iaf was found to be MDYSI (Fig. 5A). Additionally, AAC(6′)-Iaf production in IMCJ798, IMCJ799, and E. coli JM109 transformants was confirmed by Western blot analysis using polyclonal anti-AAC(6′)-Iaf IgG (Fig. 5B and C). The production level of AAC(6′)-Iaf in JM109 carrying pSTV-aac(TTG→ATG) increased 2.4-fold compared to that in JM109 carrying pSTV-aacWT, whereas the intensities of cross-reacting protein bands were unchanged (Fig. 5B). This increase was similar to results observed previously for the 6-hydroxy-d-nicotine oxidase gene carrying TTG as an initiation codon (31). These results demonstrate that the ORF of aac(6′)-Iaf initiates from TTG and that aac(6′)-Iaf plays a crucial role in aminoglycoside resistance in P. aeruginosa IMCJ798 and IMCJ799.
TABLE 3.
MICs of various aminoglycosides for P. aeruginosa strains and E. coli strains transformed with aac(6′)-Iaf
| Straina | MICb (μg/ml) of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMK | ABK | DIB | GEM | ISP | KAN | NET | SIS | TOB | NEO | |
| P. aeruginosa IMCJ798 | 128 | 8 | >128 | 4 | >128 | >128 | >128 | 32 | 32 | 8 |
| P. aeruginosa IMCJ799 | 128 | 16 | >128 | 2 | >128 | >128 | >128 | 32 | 32 | 8 |
| E. coli JM109/pSTV28 | 1 | 2 | 1 | 0.5 | 1 | 2 | 0.5 | 0.5 | 0.125 | 2 |
| E. coli JM109/pSTV-aacWT | 16 | 4 | 16 | 1 | 4 | 64 | 8 | 1 | 4 | 4 |
| E. coli JM109/pSTV-aac(TTG→ATG) | 32 | 4 | 32 | 1 | 16 | 128 | 8 | 2 | 4 | 4 |
The MICs for E. coli strains were determined with Mueller-Hinton broth preparations containing chloramphenicol (30 μg/ml) and individual aminoglycosides.
AMK, amikacin; ABK, arbekacin; DIB, dibekacin; GEM, gentamicin; ISP, isopamicin; KAN, kanamycin; NET, netilmicin; SIS, sisomicin; TOB, tobramycin; NEO, neomycin.
FIG. 5.
Production of AAC(6′)-Iaf in P. aeruginosa clinical isolates and E. coli transformants used in aminoglycoside susceptibility tests. Each lysate was separated by SDS-15% PAGE and analyzed by Western blotting using polyclonal anti-AAC(6′)-Iaf antibodies. The positions of nonspecifically reacting protein bands serving as loading controls are indicated by asterisks. (A) Partial sequences of the aac(6′)-Iaf gene and its promoter region on pSTV-aacWT and pSTV-aac(TTG→ATG) and of the corresponding peptides. The putative wild-type initiation codon is shown in boldface type. The initiation codon altered to ATG is boxed. The N-terminal sequence of native AAC(6′)-Iaf, as determined by the Edman degradation method, is underlined. (B) Expression of AAC(6′)-Iaf in P. aeruginosa clinical isolates IMCJ798 and IMCJ799. (C) Expression of AAC(6′)-Iaf in E. coli JM109 carrying pSTV28 (lane 1; empty vector), pSTV-aacWT (lane 2; TTG), and pSTV-aac(TTG→ATG) (lane 3; ATG).
Acetylation activity of AAC(6′)-Iaf.
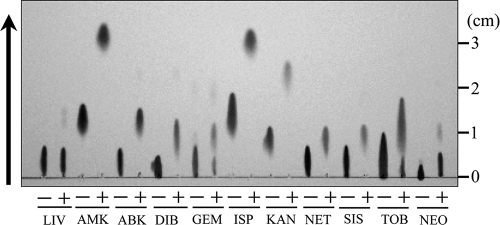
To examine the biochemical properties of AAC(6′)-Iaf in aminoglycoside resistance, acetylation activities against various aminoglycosides were assayed by TLC using native AAC(6′)-Iaf. TLC analyses showed that all aminoglycosides with an amino group at the 6′ position were acetylated by native AAC(6′)-Iaf in the presence of acetyl-CoA (Fig. 6). Commercially available GEM is a mixture of derivatives of GEM, such as GEM C1, C1a, C2, and C2b (41, 44). NEO also consists of derivatives of NEO B and C. In these two reagent mixtures, partially acetylated reagents were observed. Surprisingly, lividomycin A, which has a hydroxyl group at the 6′ position, was also a substrate for AAC(6′)-Iaf, although only an extremely small amount of AAC activity was detected. This partial acetylation of lividomycin A suggests that AAC(6′)-Iaf may have acetylation activities for an alternate amino group in the aminoglycoside molecule. These findings indicate that aac(6′)-Iaf encodes a functional aminoglycoside 6′-N-acetyltransferase that effectively modifies the amino groups at the 6′ positions of aminoglycosides in vitro. However, E. coli JM109 carrying pSTV-aacWT, expressing exogenous AAC(6′)-Iaf, did not show reduced susceptibility to ABK, GEM, or NEO (Table 3). E. faecium producing AAC(6′)-Ii is susceptible to NEO even though AAC(6′)-Ii acetylates NEO (48). ABK and NEO were shown previously to retain their antibiotic effects on an ABK-resistant actinomycete strain, even after they were acetylated at the 6′ positions by AAC(6′) enzymes (50). These results suggest that the acetylation of ABK and NEO at the 6′ positions does not affect the antimicrobial activities of these drugs. The antimicrobial activity retained after treatment with AAC(6′)-Iaf may be due to residual unacetylated ABK or NEO. GEM derivatives C1 and C2b carry methyl groups at the 6′ positions, and they may be refractory to AAC(6′)-I enzymes. Further work is needed to determine the detailed biochemical properties of AAC(6′)-Iaf.
FIG. 6.
Analysis of acetylated aminoglycosides by TLC. Native AAC(6′)-Iaf and various aminoglycosides were incubated in the absence (−) or presence (+) of acetyl-CoA. LIV, lividomycin A. The arrow indicates the direction of development.
Acknowledgments
We are grateful to Kayo Shimada and Tomoko Kuwahara for molecular epidemiological analyses of clinical isolates. We thank Shizuko Iyobe (Kitasato University, Sagamihara, Japan) for providing the P. aeruginosa GN17203 strain. We also thank Michael S. Patrick for revising the manuscript carefully.
This study was supported in part by the Ministry of Health, Labor and Welfare of Japan (grants H18-Shinko-011 and H21-Shinko-008 for genotyping, phenotyping, and functional analyses and H19-shinko-001 for molecular epidemiology).
Footnotes
Published ahead of print on 6 April 2009.
REFERENCES
- 1.Ahmed, A. M., T. Nakagawa, E. Arakawa, T. Ramamurthy, S. Shinoda, and T. Shimamoto. 2004. New aminoglycoside acetyltransferase gene, aac(3)-Id, in a class 1 integron from a multiresistant strain of Vibrio fluvialis isolated from an infant aged 6 months. J. Antimicrob. Chemother. 53:947-951. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., J. C. Wootton, E. M. Gertz, R. Agarwala, A. Morgulis, A. A. Schaffer, and Y. K. Yu. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272:5101-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azucena, E., and S. Mobashery. 2001. Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resist. Updat. 4:106-117. [DOI] [PubMed] [Google Scholar]
- 5.Bissonnette, L., and P. H. Roy. 1992. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J. Bacteriol. 174:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burk, D. L., N. Ghuman, L. E. Wybenga-Groot, and A. M. Berghuis. 2003. X-ray structure of the AAC(6′)-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 12:426-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burk, D. L., B. Xiong, C. Breitbach, and A. M. Berghuis. 2005. Structures of aminoglycoside acetyltransferase AAC(6′)-Ii in a novel crystal form: structural and normal-mode analyses. Acta Crystallogr. D 61:1273-1279. [DOI] [PubMed] [Google Scholar]
- 8.Casse, F., C. Boucher, J. S. Julliot, M. Michel, and J. Denarie. 1979. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J. Gen. Microbiol. 113:229-242. [Google Scholar]
- 9.Centron, D., and P. H. Roy. 1998. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Iq from the integron of a natural multiresistance plasmid. Antimicrob. Agents Chemother. 42:1506-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M07-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Collis, C. M., and R. M. Hall. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa, Y., M. Galimand, R. Leclercq, J. Duval, and P. Courvalin. 1993. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37:1896-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, B. D. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doi, Y., and Y. Arakawa. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88-94. [DOI] [PubMed] [Google Scholar]
- 15.Doi, Y., J. Wachino, K. Yamane, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Spread of novel aminoglycoside resistance gene aac(6′)-Iad among Acinetobacter clinical isolates in Japan. Antimicrob. Agents Chemother. 48:2075-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galimand, M., P. Courvalin, and T. Lambert. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundmann, H., C. Schneider, D. Hartung, F. D. Daschner, and T. L. Pitt. 1995. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J. Clin. Microbiol. 33:528-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 21.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. 1:109-119. [DOI] [PubMed] [Google Scholar]
- 22.Hamano, Y., Y. Hoshino, S. Nakamori, and H. Takagi. 2004. Overexpression and characterization of an aminoglycoside 6′-N-acetyltransferase with broad specificity from an epsilon-poly-l-lysine producer, Streptomyces albulus IFO14147. J. Biochem. 136:517-524. [DOI] [PubMed] [Google Scholar]
- 23.Hannecart-Pokorni, E., F. Depuydt, L. De Wit, E. van Bossuyt, J. Content, and R. Vanhoof. 1997. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Il [sic] associated with a sulI-type integron. Antimicrob. Agents Chemother. 41:314-318. (Author's correction, 42:485, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Head, N. E., and H. Yu. 2004. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect. Immun. 72:133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, T., Y. Sato, S. Iyobe, and S. Mitsuhashi. 1982. Plasmid-mediated gentamicin resistance of Pseudomonas aeruginosa and its lack of expression in Escherichia coli. Antimicrob. Agents Chemother. 22:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirikae, T., Y. Mizuguchi, and Y. Arakawa. 2008. Investigation of isolation rates of Pseudomonas aeruginosa with and without multidrug resistance in medical facilities and clinical laboratories in Japan. J. Antimicrob. Chemother. 61:612-615. [DOI] [PubMed] [Google Scholar]
- 28.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 29.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mauch, L., V. Bichler, and R. Brandsch. 1990. Functional analysis of the 5′ regulatory region and the UUG translation initiation codon of the Arthrobacter oxidans 6-hydroxy-d-nicotine oxidase gene. Mol. Gen. Genet. 221:427-434. [DOI] [PubMed] [Google Scholar]
- 32.Miller, G. H., F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu, K. J. Shaw, et al. 1997. The most frequent aminoglycoside resistance mechanisms—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clin. Infect. Dis. 24(Suppl. 1):S46-S62. [DOI] [PubMed] [Google Scholar]
- 33.Neuwald, A. F., and D. Landsman. 1997. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22:154-155. [DOI] [PubMed] [Google Scholar]
- 34.Poirel, L., O. Menuteau, N. Agoli, C. Cattoen, and P. Nordmann. 2003. Outbreak of extended-spectrum β-lactamase VEB-1-producing isolates of Acinetobacter baumannii in a French hospital. J. Clin. Microbiol. 41:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riccio, M. L., J. D. Docquier, E. Dell'Amico, F. Luzzaro, G. Amicosante, and G. M. Rossolini. 2003. Novel 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ic, from a Pseudomonas aeruginosa integron. Antimicrob. Agents Chemother. 47:1746-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Schwocho, L. R., C. P. Schaffner, G. H. Miller, R. S. Hare, and K. J. Shaw. 1995. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekiguchi, J., T. Asagi, T. Miyoshi-Akiyama, T. Fujino, I. Kobayashi, K. Morita, Y. Kikuchi, T. Kuratsuji, and T. Kirikae. 2005. Multidrug-resistant Pseudomonas aeruginosa strain that caused an outbreak in a neurosurgery ward and its aac(6′)-Iae gene cassette encoding a novel aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 49:3734-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekiguchi, J., T. Asagi, T. Miyoshi-Akiyama, A. Kasai, Y. Mizuguchi, M. Araake, T. Fujino, H. Kikuchi, S. Sasaki, H. Watari, T. Kojima, H. Miki, K. Kanemitsu, H. Kunishima, Y. Kikuchi, M. Kaku, H. Yoshikura, T. Kuratsuji, and T. Kirikae. 2007. Outbreaks of multidrug-resistant Pseudomonas aeruginosa in community hospitals in Japan. J. Clin. Microbiol. 45:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekiguchi, J., K. Teruya, K. Horii, E. Kuroda, H. Konosaki, Y. Mizuguchi, M. Araake, A. Kawana, H. Yoshikura, T. Kuratsuji, H. Miyazaki, and T. Kirikae. 2007. Molecular epidemiology of outbreaks and containment of drug-resistant Pseudomonas aeruginosa in a Tokyo hospital. J. Infect. Chemother. 13:418-422. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 43.Tenover, F. C., D. Filpula, K. L. Phillips, and J. J. Plorde. 1988. Cloning and sequencing of a gene encoding an aminoglycoside 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J. Bacteriol. 170:471-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vakulenko, S. B., and S. Mobashery. 2003. Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16:430-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanhoof, R., E. Hannecart-Pokorni, and J. Content. 1998. Nomenclature of genes encoding aminoglycoside-modifying enzymes. Antimicrob. Agents Chemother. 42:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright, G. D. 1999. Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 2:499-503. [DOI] [PubMed] [Google Scholar]
- 48.Wright, G. D., and P. Ladak. 1997. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 41:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yokoyama, K., Y. Doi, K. Yamane, H. Kurokawa, N. Shibata, K. Shibayama, T. Yagi, H. Kato, and Y. Arakawa. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362:1888-1893. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, C. B., A. Sunada, J. Ishikawa, Y. Ikeda, S. Kondo, and K. Hotta. 1999. Role of aminoglycoside 6′-acetyltransferase in a novel multiple aminoglycoside resistance of an actinomycete strain #8: inactivation of aminoglycosides with 6′-amino group except arbekacin and neomycin. J. Antibiot. (Tokyo) 52:889-894. [DOI] [PubMed] [Google Scholar]