Abstract
Escherichia coli is refractory to elevated doses of antibiotics when it is growing in a biofilm, and this is potentially due to high numbers of multidrug-tolerant persister cells in the surface-adherent population. Previously, the chromosomal toxin-antitoxin loci hipBA and relBE have been linked to the frequency at which persister cells occur in E. coli populations. In the present study, we focused on the dinJ-yafQ-encoded toxin-antitoxin system and hypothesized that deletion of the toxin gene yafQ might influence cell survival in antibiotic-exposed biofilms. By using confocal laser scanning microscopy and viable cell counting, it was determined that a ΔyafQ mutant produced biofilms with a structure and a cell density equivalent to those of the parental strain. In-depth susceptibility testing identified that relative to wild-type E. coli, the ΔyafQ strain had up to a ∼2,400-fold decrease in cell survival after the biofilms were exposed to bactericidal concentrations of cefazolin or tobramycin. Corresponding to these data, controlled overexpression of yafQ from a high-copy-number plasmid resulted in up to a ∼10,000-fold increase in the number of biofilm cells surviving exposure to these bactericidal drugs. In contrast, neither the inactivation nor the overexpression of yafQ affected the tolerance of biofilms to doxycycline or rifampin (rifampicin). Furthermore, deletion of yafQ did not affect the tolerance of stationary-phase planktonic cells to any of the antibacterials tested. These results suggest that yafQ mediates the tolerance of E. coli biofilms to multiple but specific antibiotics; moreover, our data imply that this cellular pathway for persistence is likely different from that of multidrug-tolerant cells in stationary-phase planktonic cell cultures.
Biofilms, which are assemblages of microorganisms encased in a self-produced matrix of extracellular polymers, are a prevalent mode of microbial life that is thought to be responsible for many chronic, recalcitrant infections (8, 13, 30, 31). The bacteria in biofilms have decreased sensitivities to antibiotics and other antimicrobial agents, and this decreased sensitivity occurs independently of the presence of resistance determinants on mobile genetic elements (7). Recent findings suggest that growth in a biofilm might be a population-based strategy for bacterial survival, as multidrug resistance and tolerance can be linked to an ongoing process of phenotypic diversification among the surface-adherent cells (16, 39). One type of phenotypic variant produced by single-species bacterial populations is the persister cell, a metabolically quiescent cell that neither grows nor dies when it is exposed to bactericidal concentrations of antibiotics (2, 3, 19, 36, 38). Although subpopulations of these physiological variants represent 0.1% or less of logarithmically growing planktonic cell cultures (29, 36), the proportion of persister cells in biofilms might be as high as 10% (36). Therefore, it is reasonable to hypothesize that persister cells contribute to the multidrug tolerance of bacterial biofilms (20, 36).
Although bacterial persistence was first reported in the literature during the 1940s (3), the molecular mechanisms involved in the formation of persister cells continue to be a subject of controversy (23, 26, 32, 41, 42). An association exists between toxin-antitoxin (TA) genes and persistence on the basis of empirical observations, but a mechanistic understanding of this phenomenon remains unclear. For instance, there are well-established genetic links between the frequencies of persister cells in Escherichia coli populations and the chromosomal TA gene modules hipBA and relBE (4, 5, 9, 20, 25, 27-29, 34). However, it is likely that different toxins and antitoxins have distinct physiological roles, as there is experimental evidence suggesting that TA systems may be involved in a diverse range of behaviors, such as antibiotic-mediated programmed cell death, cellular stasis, and biofilm dispersion (11, 12, 21, 24, 32). Adding to the complexity of this problem are reports that drug-tolerant cells may also be generated through different biochemical pathways that apparently do not depend on chromosomal TA gene pairs (23). Nonetheless, Keren et al. (20) have previously described that stationary-phase cultures of an E. coli strain with double mutations in hipBA have decreased survival after exposure to antibiotics. Moreover, transcriptional profiling of persisters isolated from planktonic cell cultures indicates that expression of the TA genes mazE, yefM-yoeB, relE, and dinJ-yafQ might be upregulated (20, 35). Of the TA gene modules, dinJ-yafQ has been characterized the least, and so we investigated the chromosomal toxin gene yafQ to determine whether it has a role in multidrug tolerance.
Since the vast majority of work examining links between multidrug tolerance and TA gene modules has been done with stationary-phase planktonic cells, we postulated that antibiotic susceptibility testing with biofilms might yield a phenotype missed by commonly used broth microdilution assays. In the study described in this report, we have found that a single, in-frame, markerless deletion of yafQ results in decreased bacterial survival after the exposure of E. coli biofilms but not stationary-phase planktonic cells to specific bactericidal antibiotics. Moreover, we show that the controlled overexpression of yafQ can induce a multidrug-tolerant state for the cells in a biofilm.
MATERIALS AND METHODS
Strains and growth media.
The E. coli strains and plasmids used in this study were obtained from the publically available Keio and ASKA Collections (1) (Table 1). All strains were stored at −70°C in Luria-Bertani (LB) broth with 8% dimethyl sulfoxide and were routinely cultured on LB plates that contained 1.5% (wt/vol) agar (EMD Chemicals Inc.). The agar plates were incubated at 37°C for up to 24 h. All serial dilutions were carried out with 0.9% NaCl.
TABLE 1.
Escherichia coli strains and plasmids used in this study
| Strain or plasmid | Genotype | Relevant characteristic or deleted gene | Source |
|---|---|---|---|
| Escherichia coli strains | |||
| AG1 | recA1 endA1 gyrA96 thi-1 hsdR17 (rK−mK+) supE44 relA1 | Cloning host strain | 22 |
| K-12 BW25113 | MG1655 F− λ−rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 | Wild type | 1 |
| JW0215 | K-12 BW25113 ΔyafQ | Toxin, relBE family, inhibits translation through mRNA cleavage | 1 |
| Plasmids | |||
| pCA24N ptac | pCA24N pT5-lac lacIq Cmr | High copy no., IPTG-inducible expression vector | 22 |
| pCA24N ptax::yafQ | pCA24N pT5-lac::yafQ lacIq Cmr | pCA24N containing the yafQ gene from E. coli K-12 | 22 |
Antibiotics.
All antibiotics were purchased from Sigma Chemical Company (St. Louis, MO). Stock solutions of cefazolin, doxycycline, and tobramycin were made up in double-distilled water at a concentration of 5,120 μg ml−1. Rifampin (rifampicin) was dissolved in 50% methanol. All stock solutions were split into 0.5-ml aliquots and were stored at −70°C until used.
Biofilm cultivation.
Biofilms were grown in a Calgary biofilm device (commercially available as the MBEC physiology and genetics assay [Innovotech Inc., Edmonton, AB, Canada]), as originally described by Ceri et al. (7). The Calgary biofilm device consists of a polystyrene lid with 96 pegs that can be fit into a standard 96-well microtiter plate. Starting with cryogenic stocks, the desired bacterial strain was streaked out twice on LB agar, and an inoculum was prepared by suspending colonies from the second agar subculture in 0.9% NaCl to match a 1.0 McFarland standard. This standard inoculum was diluted 30-fold in growth medium to get a starting viable cell count of roughly 1.0 × 107 CFU/ml. A total of 150 μl of this inoculum was transferred into each well of a 96-well microtiter plate, and the sterile peg lid of the Calgary biofilm device was inserted into this plate. The inoculated device was then placed on a gyratory shaker at 125 rpm and incubated for 24 h at 37°C and 95% relative humidity.
Following this initial period of incubation, the biofilms were rinsed once with 0.9% saline (by placing the lid in a microtiter plate containing 200 μl of 0.9% NaCl in each well) to remove loosely adherent planktonic cells. Biofilm formation was evaluated by breaking off four pegs from each device after it had been rinsed. The biofilms from these pegs were disrupted in 200 μl of 0.9% NaCl by using an ultrasonic cleaner on the high setting for 5 min (Aquasonic model 250 HT; VWR Scientific, Mississauga, ON, Canada), as described previously (7). The disrupted biofilms were serially diluted and plated onto agar for counting of the viable cells. Biofilms cultivated in this fashion were then used for susceptibility testing (see below).
It was additionally ascertained that the strains used in this study formed equivalent biofilms on the pegs of the Calgary biofilm device. To ascertain this, we repeated the cultivation protocol and then disrupted all of the biofilms from the lid of the Calgary biofilm device in a microtiter plate containing 200 μl of 0.9% NaCl in each well. Of these biofilms, cells recovered from 48 biofilms in one half of the device (pegs 1 to 6 from rows A to H) were then serially diluted and plated onto LB agar for counting of the viable cells. These agar plates were incubated for 24 h at 37°C, and then the colonies were enumerated. The viable cell counts were grouped by row of the Calgary biofilm device, and these values were compared by one-way analysis of variance (ANOVA).
Biofilm susceptibility testing.
After the biofilms that had been grown on the lids of the Calgary biofilm device were rinsed, the peg lids were inserted into microtiter plates containing serial, twofold dilutions of the desired antibiotics. Following antimicrobial exposure, the biofilms were rinsed twice more and then placed in a microtiter recovery plate, each well of which contained 200 μl LB medium and 1% Tween 20. Bacterial cells were immediately recovered from the biofilms at room temperature by disrupting the biofilms in the recovery medium by using an ultrasonic cleaner (as described above). Tenfold serial dilutions of 20-μl aliquots from the wells of the recovery plates were made in 0.9% NaCl, and then these diluted cultures were plated onto LB agar. To maximize the recovery of viable but potentially slowly growing bacteria surviving antibiotic exposure, at least 24 h of incubation at 37°C was allowed before the growth on these agar plates was scored.
Confocal laser scanning microscopy (CLSM).
Pegs were broken from the lid of the Calgary biofilm device with needle nose pliers. Staining of the E. coli biofilms was carried out by immersing the biofilm pegs in 200 μl of 0.05% acridine orange for 5 min. The fluorescently labeled biofilms were then placed in 2 drops of 0.9% saline on the surface of a glass coverslip. These pegs were examined with a DM IRE2 spectral confocal and multiphoton microscope (Leica Microsystems, Richmond Hill, ON, Canada) with a TCS SP2 acoustic optical beam splitter (Leica Microsystems), as described previously (17, 18). Samples were scanned by using 476-nm excitation, and emission was monitored in the green region of the spectrum. A ×63 water-immersion objective was used in all imaging experiments. The capture and assembly of the slices of the biofilm image were performed with Leica Confocal software (Leica Microsystems), a software package provided with the instrument. Microscope and program settings were user defined and were adjusted to maintain data integrity, as previously described by Harrison et al. (18). Images were adjusted for contrast and brightness by using Photoshop CS3 software (Adobe Systems Inc., San Jose, CA).
MIC determinations.
MICs were determined by broth microdilution according to the routinely used Clinical and Laboratory Standards Institute protocols.
Cultivation and susceptibility testing of stationary-phase planktonic cells.
To obtain stationary-phase planktonic cells for susceptibility testing, 25 ml of LB medium was inoculated with a single colony that was picked with a sterile cotton swab from a first agar subculture. The E. coli cells in these cultures were grown to early stationary phase by overnight (18 to 20 h) incubation on a gyratory shaker set at 225 rpm and 37°C. Stationary-phase cells were collected by centrifugation (5,000 × g for 10 min), after which the spent medium was discarded. The cell pellets were suspended in 12.5 ml of fresh growth medium, thereby concentrating the cells into half of the original volume. In contrast to the susceptibility determinations for biofilms, the antibiotic challenge plates were prepared with agents at twice the desired concentration but in 100-μl volumes, as opposed to 200 μl, in the wells of the microtiter plates. To these challenge plates, 100 μl of the stationary-phase cells concentrated two times was added to each well. The challenge and recovery media, neutralizing agents, serial dilutions, incubation conditions, and viable cell counting protocols were identical to those used for susceptibility testing with the Calgary biofilm device. In all cases, the starting numbers of cells in these suspensions were determined by counting the viable cells.
Overexpression assays.
Standard molecular methods were carried out according to the protocols of Sambrook and Russell (33). Isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible pCA24N expression vectors were obtained from the ASKA Collection in host strain E. coli AG1 (22). The plasmids were isolated from these strains with a QIAprep spin miniprep kit (Qiagen) and were transformed into E. coli K-12 BW25113. Cells containing these constructs were grown in LB medium with 30 μg/ml chloramphenicol and were prepared for susceptibility testing by using the protocols described above, except that transformants were induced with IPTG prior to antibiotic exposure. In this case, induction was achieved by inserting the biofilms into microtiter plates containing 150 μl of LB, 30 μg/ml chloramphenicol, and 5 μM IPTG in each well and incubating the plates for 4 h at 37°C on a gyratory shaker set at 125 rpm.
Statistical tests and data analysis.
The log killing of the mutant populations relative to that of the wild-type population was considered significantly different if (i) there was a minimum 50-fold (log10 1.7) change in the mean log killing of the mutant population compared to the mean log killing of the wild-type population and (ii) there was no overlap in the standard deviations of these two measurements. These criteria have been set by using an empirical approach to assess the normal variation in high-throughput cell viability data generated with the Calgary biofilm device (J. J. Harrison and H. Ceri, unpublished data). When appropriate, statistical tests, such as one-way ANOVA of the viable cell counts, were performed with the Minitab program (release 14; Minitab Inc., State College, PA) to analyze log10-transformed raw data. Alternate hypotheses were tested at the 95% level of confidence. All other calculations were performed with the Excel 2003 program (Microsoft Corporation, Redmond, WA). The data were plotted as required by using the Prism (version 4.0) program (GraphPad Software Inc., La Jolla, CA).
RESULTS AND DISCUSSION
Biofilm formation by wild-type E. coli K-12 BW25113 and its isogenic ΔyafQ mutant is similar.
Recent evidence shows that an E. coli strain bearing multiple deletions at five chromosomal TA gene loci has an altered pattern of biofilm development (21); therefore, we examined E. coli K-12 BW25113 and its isogenic ΔyafQ mutant for differences in surface-adherent growth. On the polystyrene surface of the Calgary biofilm device, both of these strains produced biofilms with similar cell densities (Fig. 1). To additionally validate this finding, we examined these strains for equivalent growth in different regions of the Calgary biofilm device, and when they were grouped by rows of pegs, the biofilms of the wild-type and the ΔyafQ strains had statistically equivalent mean viable cell counts (by one-way ANOVA, P ≥ 0.05; data not shown). The biofilms were also fluorescently stained with acridine orange and were examined by CLSM. In that analysis, both strains produced robust biofilms at the air-liquid-surface interface of the peg surfaces (Fig. 1). The fluid dynamics in the Calgary biofilm device are complex, and thus, biofilms may have subtle variations in structural conformations on the same peg and from one peg to the next. However, no gross morphological differences in the three-dimensional biofilm structures could be distinguished for the wild-type and the ΔyafQ strains (Fig. 1), especially when the differences between the wild-type and the ΔyafQ strains were compared to the differences that we have previously reported for other microbial strains of various genetic backgrounds grown under different culture conditions (10, 17, 18). Since the deletion of yafQ did not influence E. coli biofilm development, we directly compared the antibiotic-mediated killing of cells in the wild-type biofilms to that in the mutant biofilms.
FIG. 1.
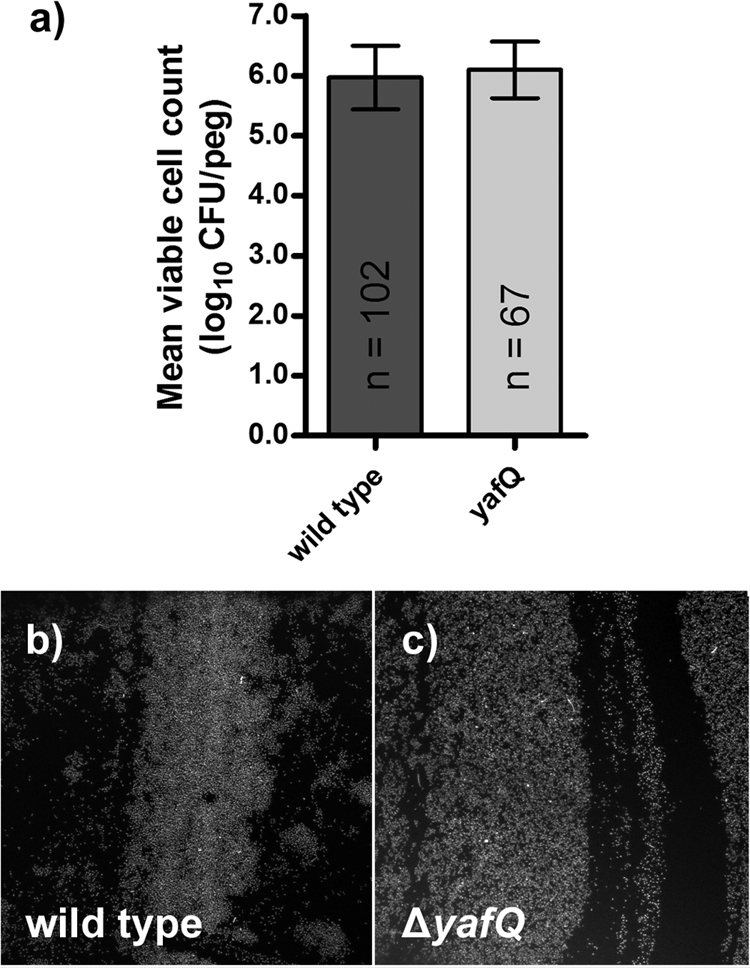
Biofilm formation by wild-type E. coli K-12 BW25113 is similar to that of its isogenic ΔyafQ mutant. (a) Biofilm mean viable cell counts and standard deviations were comparable for the two strains, and this assessment was based on the indicated number of control measurements (n). When their structures were examined by CLSM, the wild-type (b) and mutant (c) strains also had similar structures at the air-liquid-surface interface of polystyrene pegs. Each microscopy panel is a representative of four independent replicates and captures an area of 238 by 238 μm.
yafQ influences cell survival in E. coli biofilms exposed to specific antibiotics.
At the population level, persister cells give rise to biphasic dynamics of population survival that are either time dependent (2, 15, 40) or concentration dependent (6). In the present study, biofilms grown in the Calgary biofilm device were assessed for their concentration-dependent sensitivities to representative antibiotics from four different functional classes: cefazolin (a β-lactam and cell wall synthesis inhibitor), tobramycin (an aminoglycoside and protein synthesis inhibitor), doxycycline (a tetracycline and protein synthesis inhibitor), and rifampin (a RNA synthesis inhibitor). Ciprofloxacin (a DNA synthesis inhibitor) was also tested; however, the high variation in population survival precluded interpretation of the results of assays with this drug (data not shown). The latter observation was consistent with previously described difficulties in the high-throughput screening of the Keio Collection of isolates for their ofloxacin sensitivities, and here, these results were likely dependent on the growth conditions (14).
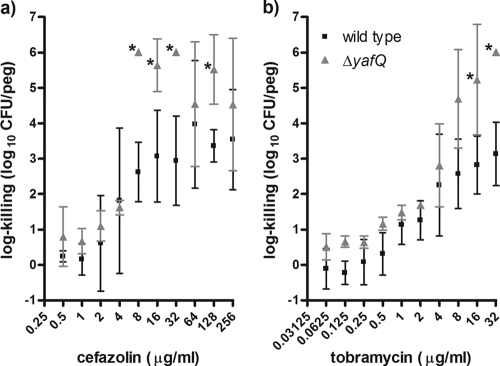
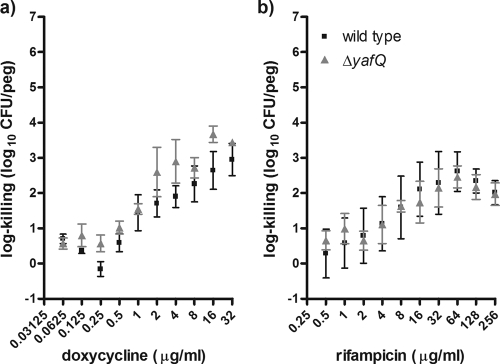
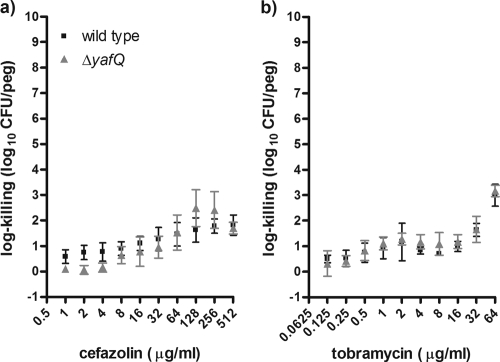
Nonetheless, our process of biofilm susceptibility testing indicated that the ΔyafQ mutant had significantly higher levels of cell death than the wild-type strain when the strains were exposed to bactericidal concentrations of cefazolin or tobramycin (Fig. 2). The log killings of the ΔyafQ biofilm populations by cefazolin and tobramycin were maximally increased ∼2,400- and ∼760-fold, respectively, relative to the killing of the wild-type strain (Fig. 2). By contrast, there were no differences in cell death in biofilms exposed to doxycycline or rifampin (Fig. 3). The lack of a rifampin-sensitive phenotype for the ΔyafQ mutant fits with the findings presented in a previous report of a study in which a role for TA gene systems in rifampin tolerance failed to be established (41). Overall, these data suggest that the chromosomal toxin gene yafQ mediates the tolerance of E. coli biofilms to specific antibiotics. To validate these findings, we investigated whether the overexpression yafQ might increase the antibiotic tolerance of biofilms.
FIG. 2.
Biofilm populations of the ΔyafQ strain have decreased numbers of cells surviving exposure to cefazolin and tobramycin compared to the numbers of parental E. coli K-12 BW25113 cells. Mean log killing was calculated after the biofilms had been exposed to cefazolin (a) and tobramycin (b) for 24 h. Each datum point is based on 4 to 16 independent replicates. Asterisks, significant difference in log killing of the mutant population compared to that of the wild-type population by using the criteria indicated in Materials and Methods. The mean viable cell counts for the biofilms not exposed to antibiotics were 6.19 ± 0.31 and 5.99 ± 0.19 log10 CFU/peg for the wild-type and ΔyafQ strains, respectively.
FIG. 3.
Biofilm populations of the ΔyafQ strain and isogenic parental strain E. coli K-12 BW25113 had similar numbers of cells surviving exposure to doxycycline and rifampin. Mean log killing was calculated from the viable cell counts after the biofilms had been exposed to doxycycline (a) and rifampin (b) for 24 h. Each datum point is based on 4 to 20 independent replicates. The mean viable cell counts for the biofilms not exposed to antibiotics were 5.84 ± 0.58 and 5.85 ± 0.29 log10 CFU/peg for the wild-type and ΔyafQ strains, respectively.
Controlled overexpression of yafQ increases the number of multidrug-tolerant cells in E. coli biofilms.
An approach that has been used to identify genes involved in multidrug tolerance relies on the overexpression of the genes of interest prior to the exposure of bacterial cells to antibiotics (14, 37). In the present study, we used this approach to look for changes in the survival of E. coli K-12 BW25113 biofilm cells bearing an IPTG-inducible yafQ expression vector, pCA24N ptac::yafQ, relative to the survival of cells of the same strain bearing the vector without the gene of interest, pCA24N ptac. Corresponding to the findings described in a previous report (21), we found that high levels of yafQ induction, which were achieved by adding ≥100 μM IPTG to the growth medium, greatly reduced the number of viable cells in the biofilms (data not shown). However, lower levels of yafQ induction, achieved by adding ≤5.0 μM IPTG to the growth medium, had little effect on the mean viable cell counts in the biofilm. Therefore, 5.0 μM IPTG was used to induce pCA24N ptac::yafQ and to look for changes in the antibiotic susceptibilities of the biofilm populations.
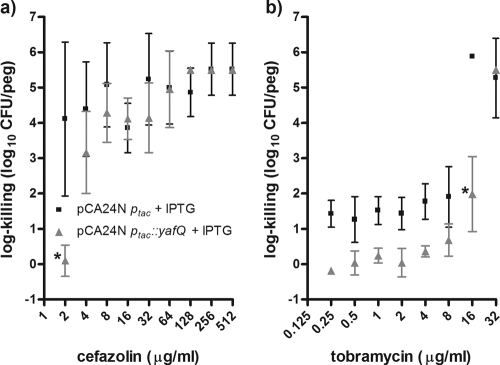
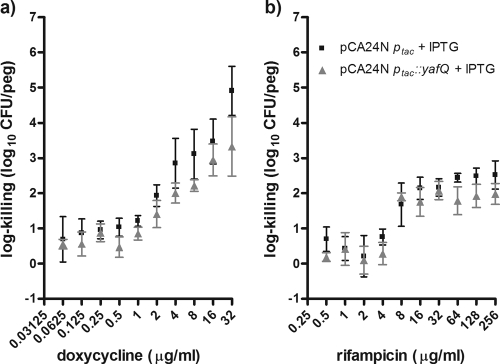
When pCA24N ptac::yafQ was induced with 5 μM IPTG for 4 h prior to antibiotic exposure, pCA24N ptac::yafQ protected the biofilm cells from killing by cefazolin and tobramycin (Fig. 4). The induction of yafQ maximally resulted in ∼10,000- and ∼3,300-fold increases in cell survival in biofilms exposed to these two drugs, respectively (Fig. 4). It is worth noting that although the changes in survival were below our empirically set threshold for significance (see Materials and Methods), there was an observable trend for yafQ induction to result in greater cell survival for E. coli biofilms over a broad range of tobramycin concentrations (Fig. 4). In no instances did the overexpression of yafQ protect the biofilms against killing by doxycycline or rifampin (Fig. 5). Collectively, these data validate the hypothesis that the chromosomal toxin gene yafQ functions as a multidrug tolerance determinant for E. coli biofilms but indicate that this protection is limited to specific antibiotics. A logical next step was to repeat some of this work by using protocols for the antibiotic susceptibility testing of planktonic cells.
FIG. 4.
Overexpression of yafQ from a high-copy-number plasmid increased the number of cells in E. coli K-12 BW25113 biofilm populations surviving exposure to bactericidal concentrations of cefazolin and tobramycin. Mean log killing was calculated from the viable cell counts after the biofilms had been exposed to cefazolin (a) and tobramycin (b) for 24 h. The biofilms were incubated with 5 μM IPTG for 4 h before antibiotic exposure. All assays were conducted with 30 μg/ml chloramphenicol in the growth medium to select for the pCA24N expression vector. Each datum point is based on four to eight independent replicates. Asterisks, significant difference in log killing of the mutant population compared to that of the wild-type population by using the criteria indicated in Materials and Methods. The mean viable cell counts for the biofilms unexposed to antibiotics were 5.34 ± 0.80 and 5.44 ± 0.15 log10 CFU/peg for the wild-type and ΔyafQ strains, respectively.
FIG. 5.
Overexpression of yafQ from a high-copy-number plasmid had no effect on the number of cells in E. coli K-12 BW25113 biofilm populations surviving exposure to doxycycline or rifampin. Mean log killing was calculated from the viable cell counts after the biofilms had been exposed to doxycycline (a) and rifampin (b) for 24 h. The biofilms were incubated with 5 μM IPTG for 4 h before antibiotic exposure. All assays were conducted with 30 μg/ml chloramphenicol in the growth medium to select for the pCA24N expression vector. Each datum point is based on four to eight independent replicates. The mean viable cell counts for the biofilms not exposed to antibiotics were 5.38 ± 0.09 and 5.65 ± 0.22 log10 CFU/peg for the wild-type E. coli K-12 BW25113 strain bearing pCA24Nptac and pCA24Nptac yafQ, respectively.
Deletion of yafQ has no effect on the antibiotic resistance or tolerance of planktonic cells.
Antibiotic MICs were determined for wild-type E. coli K-12 BW25113 and its isogenic ΔyafQ mutant by using the standard Clinical and Laboratory Standards Institute protocol. There were no significant differences in MICs between these strains for any of the antibiotics tested (Table 2). A lack of a difference in MICs for planktonic cells is a defining characteristic of multidrug tolerance determinants. We also examined wild-type and ΔyafQ E. coli strains grown in stationary-phase planktonic cell cultures, growth conditions that reportedly result in high levels of persister cells in the population (36), for hypersensitivity to cefazolin and tobramycin. In no instance was there increased killing of the ΔyafQ mutant compared to the level of killing of the wild-type strain (Fig. 6). These data imply that yafQ functions as multidrug tolerance determinant for biofilms but not for stationary-phase planktonic cells.
TABLE 2.
Antibiotic MICs for wild-type E. coli K-12 BW25113 and the ΔyafQ strain
| Antibiotic | Median (range) MIC (μg/ml)
|
|
|---|---|---|
| Wild type | ΔyafQ | |
| Cefazolin | 4 | 4 |
| Doxycycline | 4 | 3 (2-4) |
| Rifampin | 16 (16-32) | 16 |
| Tobramycin | 1.5 (1-2) | 2 (2-4) |
FIG. 6.
Stationary-phase planktonic cell populations of ΔyafQ and isogenic parental strain E. coli K-12 BW25113 had similar numbers of cells surviving exposure to cefazolin and tobramycin. Mean log killing was calculated after the biofilms had been exposed to cefazolin (a) and tobramycin (b) for 24 h. Each datum point is based on four to eight independent replicates. The mean viable cell counts for the stationary-phase planktonic populations not exposed to antibiotics were 8.74 ± 0.35 and 8.99 ± 0.06 log10 CFU/ml for the wild-type and ΔyafQ strains, respectively.
Concluding remarks.
The E. coli K-12 chromosome encodes at least seven TA gene systems: mazEF (or chpAR), relBE, yefM-yoeB, chpB-chpS, dinJ-yafQ, hipBA, and hicAB. To the best of our knowledge, there is no evidence in the literature that E. coli strains bearing a single inactivated toxin or antitoxin gene have drug-sensitive phenotypes, even when survival has been quantitatively determined for stationary-phase planktonic cell cultures (26). Although screening strategies involving stationary-phase planktonic cell cultures have served as a surrogate for biofilm susceptibility testing, the data in this report indicate that these strategies may miss certain chromosomal determinants affecting biofilm susceptibility to antibiotics. Conceptually, this implies that the mechanisms of bacterial persistence might be different for stationary-phase planktonic cells and biofilms.
Our research group is currently reexamining TA gene loci for their physiological roles specific to the biofilm mode of growth. So far, no effects have been discerned for single deletions of hipA, relE, or mazF on the antibiotic tolerance of E. coli K-12 BW25113 biofilms (J. J. Harrison, W. D. Wade, R. J. Turner, and H. Ceri, unpublished data). The results presented in this report as well as information in the literature suggest that yafQ might be hitherto distinct among chromosomal TA genes for its isolated, physiological role in biofilm multidrug tolerance.
Although the number of cells surviving exposure to cefazolin and tobramycin was greatly diminished in the ΔyafQ strain, the survival of small numbers of cells in biofilm populations indicates that additional mechanisms of multidrug tolerance are likely at work. This belief is additionally supported by evidence that the deletion of yafQ has no effect on biofilm tolerance to doxycycline or rifampin. While it is reasonable to hypothesize that yafQ may play a role in the formation of biofilm persister cells, it is possible both that there is more than one physiologically distinct subpopulation of multidrug-tolerant cells in biofilms and that more than one cellular pathway requisite for the tolerance of a single subpopulation of cells to multiple antibiotics exists.
Acknowledgments
This work has been supported through discovery grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada to R.J.T. and H.C. The NSERC has also provided a Canada graduate scholarship-doctoral award to J.J.H., who was additionally supported by a Ph.D. studentship from the Alberta Heritage Foundation for Medical Research. CLSM was made possible through a Canadian Foundation for Innovation Bone and Joint Disease Network grant to H.C.
Footnotes
Published ahead of print on 23 March 2009.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Kirill, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 3.Bigger, J. W. 1944. Treatment of staphylococcal infections with penicillin. Lancet ii:497-500. [Google Scholar]
- 4.Black, D. S., B. Irwin, and H. S. Moyed. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. W. Morck, and A. G. Buret. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities in bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Davidson, C. J., and M. G. Surette. 2008. Individuality in bacteria. Annu. Rev. Genet. 42:253-268. [DOI] [PubMed] [Google Scholar]
- 10.Davies, J. A., J. J. Harrison, L. L. R. Marques, G. R. Foglia, C. A. Stremick, D. G. Storey, R. J. Turner, M. E. Olson, and H. Ceri. 2007. The GacS sensor kinase controls phenotypic reversion of small colony variants isolated from biofilms of Pseudomonas aeruginosa PA14. FEMS Microbiol. Ecol. 59:32-46. [DOI] [PubMed] [Google Scholar]
- 11.Engelberg-Kulka, H., S. Amitai, I. Kolodkin-Gal, and R. Hazan. 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLOS Genet. 2:1518-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 13.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, S., K. Lewis, and M. Vulić. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52:2718-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison, J. J., H. Ceri, N. J. Roper, E. A. Badry, K. M. Sproule, and R. J. Turner. 2005. Persister cells mediate tolerance to metal oxyanions in Escherichia coli. Microbiology 151:3181-3195. [DOI] [PubMed] [Google Scholar]
- 16.Harrison, J. J., H. Ceri, and R. J. Turner. 2007. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 5:928-938. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, J. J., H. Ceri, J. Yerly, M. Rabiei, Y. Hu, R. Martinuzzi, and R. J. Turner. 2007. Metal ions may suppress or enhance cellular differentiation in Candida albicans and Candida tropicalis biofilms. Appl. Environ. Microbiol. 73:4940-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison, J. J., H. Ceri, J. Yerly, C. A. Stremick, Y. Hu, R. Martinuzzi, and R. J. Turner. 2006. The use of microscopy and three-dimensional visualization to evaluate the structure of microbial biofilms cultivated in the Calgary biofilm device. Biol. Proceed. Online 8:194-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison, J. J., R. J. Turner, and H. Ceri. 2005. Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Environ. Microbiol. 7:981-994. [DOI] [PubMed] [Google Scholar]
- 20.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, Y., X. Wang, Q. Ma, X. S. Zhang, and T. K. Wood. 2009. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191:1258-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitigawa, M., T. Ara, M. Arifuzzaman, T. Ioka-Nakamichi, E. Inamoto, H. Toyonaga, and H. Mori. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291-299. [DOI] [PubMed] [Google Scholar]
- 23.Klapper, I., P. Gilbert, B. P. Ayati, J. Dockery, and P. S. Stewart. 2007. Senescence can explain microbial persistence. Microbiology 153:3623-3630. [DOI] [PubMed] [Google Scholar]
- 24.Kolodkin-Gal, I., and H. Engelber-Kulka. 2008. The extracellular death factor: physiological and genetic factors influencing its production and response in Escherichia coli. J. Bacteriol. 190:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., and Y. Zhang. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moyed, H., and S. Broderick. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 31.Potera, C. 1998. Forging a link between biofilms and disease. Science 283:1837-1839. [DOI] [PubMed] [Google Scholar]
- 32.Rice, K. C., and K. W. Bayles. 2008. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 72:85-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170:3321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah, D., Z. Zhang, A. Khodursky, N. Kaldalu, K. Kurg, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spoering, A., and K. Lewis. 2001. Biofilm and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spoering, A. L., M. Vulic, and K. Lewis. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stewart, P. S. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107-113. [DOI] [PubMed] [Google Scholar]
- 39.Stewart, P. S., and J. W. Costerton. 2001. The antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 40.Sufya, N., D. G. Allison, and P. Gilbert. 2003. Clonal variation in maximum specific growth rate and susceptibility towards antimicrobials. J. Appl. Microbiol. 95:1261-1267. [DOI] [PubMed] [Google Scholar]
- 41.Tsilibaris, V., G. Maenhaut-Michel, N. Mine, and L. Van Melderen. 2007. What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J. Bacteriol. 189:6101-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vásquez-Laslop, N., H. Lee, and A. A. Neyfakh. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 188:3494-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]