Abstract
The cfiA gene is clustered in a bicistronic operon encoding an N-acetyltransferase and an O-acetyltransferase related to resistance markers. This genetic context, exclusively found in strains of Bacteroides fragilis division II, has been highly rearranged by the successive integration of two new mobile sequences, a miniature element and ISBf9. Besides that, among the DNA polymorphisms detected in the cfiA locus, only the integration of IS942 at its promoter was a determinant for expression of carbapenemase activity.
The cfiA gene, encoding the unique carbapenemase enzyme found in Bacteroides, is restricted to the division II group of Bacteroides fragilis strains (5). Bacteria belonging to this group were isolated in previous studies of human (3) or animal (unpublished data) infections, where identification of the cfiA genes was performed by PCR with primers P1 and P4 (Table 1) and confirmed by Southern hybridization on genomic DNA digestions (not shown).
TABLE 1.
Primers used in this work
| Primer | Sequence (5′-3′) | Positionb | Usage of primer pair |
|---|---|---|---|
| P1a | AAAGAATAAAATGAAAACAGT | 1855 | PCR of cfiA, probe for Southern hybridization, detection of protein polymorphisms |
| P4a | GATAAAAGTTTTCGCCTCTTC | 2260 | |
| P5 | AAGCAGGGATCTGCTTAC | 164 | PCR of orf1-orf3 intergenic sequence: detection of sequence polymorphisms |
| P6 | ATGACCGTATTCAGATAG | 1109 | |
| R1 | TGCGACAGGGAAAAGCATGGAGA | 1867 | iPCR of cfiA upstream region: detection of sequence polymorphisms. |
| R2 | CCCTCGCCGAAATCGAAGGATG | 1990 |
Imipenemase activity.
Division II strains were cultivated under anaerobic conditions (3), and enzyme activity was determined at 37°C with 0.1 mM imipenem and 50 mM phosphate buffer (pH 7.0) by using an ɛ299 (extinction coefficient at 299 nm) value of 9,670 M−1 cm−1. Only strains MN11 and MN32 expressed imipenemase activity and were resistant to imipenem (Fig. 1).
FIG. 1.
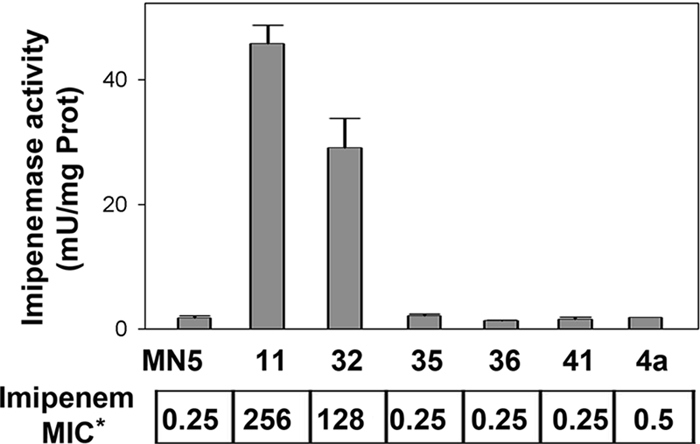
Imipenemase activities and imipenem resistance of the B. fragilis strains. MICs (μg/ml) for some strains were reported previously (3).
Allelic polymorphism.
The sequences of cfiA genes revealed that they encode previously described protein variants (4). The allele from strain MN5 (cfiA11) corresponds to protein polymorphisms in residues 79 (T), 85 (T), 113 (K), 188 (T), and 225 (N), referred to as TTKTN; alleles from strains MN11 (cfiA12), MN32 (cfiA13), and 4a (cfiA14) encode the protein variant MTRAD; and those from strains MN36 (cfiA15) and MN41 (cfiA16) presented the polymorphisms TAKAD and TTKAD, respectively. Only MTRAD alleles were presented in the two strains, MN11 and MN32, expressing imipenemase activity (Fig. 1). However, the occurrence of a particular protein variant does not correlate with the metallolactamase phenotype, since the 4a strain (allele cfiA14; protein variant MTRAD) is sensitive to carbapenems. Accordingly, a previous work reported TAKTN and TAKAD alleles in strains sensitive to carbapenem, while TAKAD, TTKTN, and TAKTD alleles were found in resistant strains, where the expression of imipenemase activity was determined by the integration of IS elements near the promoter of the gene (4).
Upstream sequences.
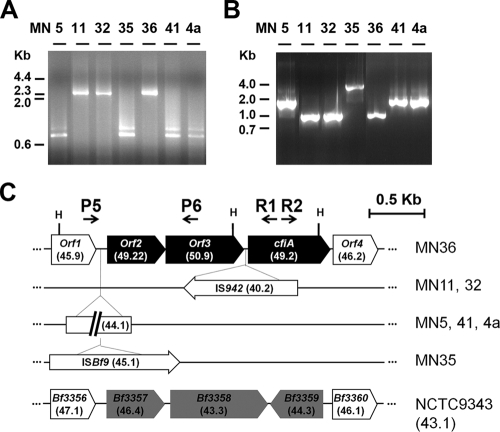
Inverse PCR (iPCR) was performed to isolate the DNA regions upstream from the cfiA genes. Genomic DNAs were digested by HaeIII, and after ligation (3), the R1 and R2 primers (Table 1) were used in a PCR of 35 cycles with annealing/extension at 68°C. A fragment of 0.7 kb was expected from the HaeIII restriction site placed 157 bp upstream of the cfiA start codon (Fig. 2C), but the display of iPCR fragments revealed the polymorphism of that region (Fig. 2A). Thus, strains MN5, MN35, MN41, and 4a presented fragments of the predictable size, while PCR products of 2.4 kb were detected in strains MN11, MN32, and MN36. Sequencing of iPCR fragments showed that the cfiA upstream sequences from strains MN11 and MN32 share the insertion, 7 bp upstream of the gene, of an IS942-like mobile element. As described previously (4), this element might determinate the imipenem resistance phenotype (Fig. 1). In contrast, the DNA fragment from strain MN36 is produced accidentally, since a sequence polymorphism abolished the proximal restriction site. This situation makes available for research the DNA region upstream of the cfiA gene.
FIG. 2.
Structures of cfiA upstream sequences. (A) iPCR fragments amplified with the R1/R2 primer pair and HaeIII (H) digestions. (B) orf1-orf3 intergenic polymorphisms displayed by PCR with the P5/P6 primer pair. (C) Genomic map of the cfiA gene cluster and surrounding regions. Numbers in parentheses indicate %GC of sequence elements. See the text for accession numbers.
Gene context.
Three coding sequences were detected upstream of the cfiA gene from strain MN36 (Fig. 2C). The first sequence, orf1, is closely related to genes Bf3550 and Bf3356 from the genomes of B. fragilis YCH46 and NCTC9343, respectively, where they are annotated as hypothetical membrane proteins. The genes orf2 and orf3, which are minimally overlapped (1 bp), encode nonrelated proteins belonging to the Gcn5-related N-acetyltransferase (8) and streptogramin A acetyltransferase (7) families, respectively.
All DNA fragments spanning further than 151 bp upstream of the cfiA gene, including sequences available in public databases, share the sequence of orf3. The primers P5 and P6 were used in a PCR of 30 cycles, with an annealing temperature at 54°C, which showed the exclusive detection of the orf1-orf2-orf3 gene cluster in division II strains (5) and that the intergenic sequence between orf1 and orf2 is highly polymorphic (Fig. 2B). Sequencing of the amplified fragments revealed that the orf2 upstream sequence is the target for integration of two new mobile sequences (Fig. 2C), a putative miniature element (MITEBf1) and a member of the ISAs1 family, named ISBf9, following the standard nomenclature (6).
Two cfiA gene sequences that exist in public databases (GenBank), under accession numbers M34831 and AY372696, extend downstream from the cfiA gene and span a sequence closely related to Bf3360 from B. fragilis NCTC9343, encoding a member of the small multidrug resistance protein family (1). Thus, the DNA region between Bf3356 and Bf3360 in B. fragilis NCTC9343 could have been exchanged in the strains carrying the metallolactamase gene, a hypothesis supported by the GC content bias (roughly 5%) existing between the orf2-orf3-cfiA gene cluster and its genomic environment (Fig. 2C). The presence of cfiA in a region of genomic plasticity, where sequences related to resistance markers or mobilization functions are detected, might condition the potential for spreading carbapenem resistance among Bacteroides and related bacteria, a clinically relevant gene context that paradoxically remained unknown.
Nucleotide sequence accession numbers.
Accession numbers for cfiA coding and upstream sequences determined in this work are FM200784 to FM200794.
Acknowledgments
This work was supported by grants SCSS0459, SCSS0506, and SCSS0629 (Junta de Extremadura) and grant AGL2005-02416 (Ministerio de Educación y Ciencia, Spain).
Footnotes
Published ahead of print on 13 April 2009.
REFERENCES
- 1.Chung, Y. J., and M. H. Saier, Jr. 2001. SMR-type multidrug resistance pumps. Curr. Opin. Drug Discov. Dev. 4:237-245. [PubMed] [Google Scholar]
- 2.Fang, H., M. Hedberg, C. Edlund, and C. E. Nord. 1999. Identification of the metallo-lactamase gene from clinical isolates of Bacteroides fragilis. Anaerobe 5:431-434. [Google Scholar]
- 3.García, N., G. Gutiérrez, M. Lorenzo, J. E. García, S. Píriz, and A. Quesada. 2008. Genetic determinants for cfxA expression in Bacteroides strains isolated from human infections. J. Antimicrob. Chemother. 62:942-947. [DOI] [PubMed] [Google Scholar]
- 4.Kato, N., K. Yamazoe, C. G. Han, and E. Ohtsubo. 2003. New insertion sequence elements in the upstream region of cfiA in imipenem-resistant Bacteroides fragilis strains. Antimicrob. Agents Chemother. 47:979-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruimy, R., I. Podglajen, J. Breuil, R. Christen, and E. Collatz. 1996. A recent fixation of cfiA genes in a monophyletic cluster of Bacteroides fragilis is correlated with the presence of multiple insertion elements. J. Bacteriol. 178:1914-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siguier, P., J. Perochon, L. Lestrade, J. Mahillon, and M. Chandler. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32-D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugantino, M., and S. L. Roderick. 2002. Crystal structure of Vat(D): an acetyltransferase that inactivates streptogramin group A antibiotics. Biochemistry 41:2209-2216. [DOI] [PubMed] [Google Scholar]
- 8.Vetting, M. W., S. L. P. de Carvalho, M. Yu, S. S. Hegde, S. Magnet, S. L. Roderick, and J. S. Blanchard. 2005. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433:212-226. [DOI] [PubMed] [Google Scholar]