Abstract
Farnesol, a precursor in the isoprenoid/sterol pathway, was recently identified as a quorum-sensing molecule produced by the fungal pathogen Candida albicans. Farnesol is involved in the inhibition of germination and biofilm formation by C. albicans and can be cytotoxic at certain concentrations. In addition, we have shown that farnesol can trigger apoptosis in mammalian cells via the classical apoptotic pathways. In order to elucidate the mechanism behind farnesol cytotoxicity in C. albicans, the response to farnesol was investigated, using proteomic analysis. Global protein expression profiles demonstrated significant changes in protein expression resulting from farnesol exposure. Among the downregulated proteins were those involved in metabolism, glycolysis, protein synthesis, and mitochondrial electron transport and the respiratory chain, whereas proteins involved in folding, protection against environmental and oxidative stress, actin cytoskeleton reorganization, and apoptosis were upregulated. Cellular changes that accompany apoptosis (regulated cell death) were further analyzed using fluorescent microscopy and gene expression analysis. The results indicated reactive oxygen species accumulation, mitochondrial degradation, and positive terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) in the farnesol-exposed cells concurrent with increased expression of antioxidant-encoding and drug response genes. More importantly, the results demonstrated farnesol-induced upregulation of the caspase gene MCA1 and the intracellular presence of activated caspases. In conclusion, this study demonstrated that farnesol promotes apoptosis in C. albicans through caspase activation, implying an important physiological role for farnesol in the fungal cell life cycle with important implications for adaptation and survival.
Candida albicans is the most important human fungal pathogen, causing various diseases from superficial mucosal infections to life-threatening systemic disorders (7, 27). As a polymorphic species, C. albicans is capable of switching morphologies among yeast, hyphal, and pseudohyphal forms, and the transitions are central to its pathogenesis and biofilm formation (1, 15, 30, 32). Farnesol was recently described as a quorum-sensing molecule secreted by C. albicans that is able to prevent yeast-to-hyphal conversion (12, 26). In C. albicans, farnesol is endogenously generated in the cell by enzymatic dephosphorylation of farnesyl diphosphate, a precursor for the synthesis of sterols in the sterol biosynthesis pathway (5, 13, 31).
We have previously shown that in addition to inhibiting germination in C. albicans, at high concentrations farnesol also induces cell death (14). Farnesol has been shown to induce apoptosis or programmed cell death in a number of fungal species, such as Saccharomyces cerevisiae and Aspergillus nidulans (6, 34). In C. albicans, however, farnesol-induced apoptosis has not yet been described. More recently, through analyses of typical markers of apoptosis and proteomic studies, we demonstrated that farnesol triggers apoptosis in human oral squamous carcinoma cells through the classical mammalian intrinsic and extrinsic apoptotic signaling pathways (33). More interestingly, the study also demonstrated for the first time the ability of the farnesol produced by C. albicans to induce a similar apoptotic effect on the tumor cells via the same pathways (33).
Apoptosis is characterized by biochemical processes that are largely conserved throughout evolution. In mammalian cells, apoptosis is a tightly regulated process for controlling the proliferation of undesired cells (3, 10). Apoptosis is triggered by various extracellular and intracellular stimuli that result in the activation of caspases, a class of cysteine proteases that play a crucial role in the process of apoptosis by conveying the apoptotic signal in a proteolytic cascade (3, 10, 35, 39). This ultimately leads to cell disassembly, mitochondrial degradation, and growth inhibition, which are major cellular responses characteristic of apoptosis (3, 10, 21, 33, 39).
Mitochondria have a central role in apoptosis, and many important aspects of the apoptotic process converge in this organelle. Mitochondria are the richest source of reactive oxygen species (ROS) in the cell, and the inhibition of the mitochondrial electron transport chain, which results in the subsequent release of ROS, is an early event in apoptotic cell death (37, 38). ROS can damage almost every essential cellular component, and in order to protect against the damaging effects of ROS, fungal cells have evolved specific defense mechanisms to neutralize ROS. The primary enzymatic antioxidants involved in protection against ROS are catalase and the superoxide dismutases (9, 16, 21, 23, 28).
In C. albicans, physiological concentrations of farnesol were reported to target the mitochondria, inducing oxidative stress, and a putative caspase (a mammalian caspase homologue, MCA1) has been identified in the C. albicans genome project (orf19.5995) (4, 28, 29, 37). Furthermore, Phillips et al. (28) recently reported that C. albicans undergoes programmed cell death under certain conditions. However, in these studies, activation of the Ras1-cyclic AMP pathway was determined to be involved in the process in which the deletion of RAS1 resulted in increased survival and decreased apoptosis (28, 29). In contrast, farnesol was shown to result in lower concentrations of cyclic AMP, indicating that farnesol-induced apoptosis likely involves a different pathway (31).
The exact mechanism of farnesol cytotoxicity and whether it is a part of an apoptotic pathway analogous to the caspase-mediated process of apoptosis in mammalian cells remain unclear. Therefore, to elucidate the mechanisms underlying farnesol cytotoxicity and its possible involvement in an apoptotic process in C. albicans, a global two-dimensional proteomic approach combined with a detailed assessment of standard apoptotic markers in farnesol-exposed C. albicans cells was undertaken.
MATERIALS AND METHODS
Organisms, growth conditions, and chemicals.
Farnesol (Sigma-Aldrich Chemical, St. Louis, MO) was obtained as a 3-M stock solution and diluted to a 30-mM solution in 100% methanol. For each of three independent experiments, an aliquot of glycerol stock of C. albicans strain SC5314 was grown and maintained on Sabouraud dextrose agar (BBL, Cockeysville, MD). Cultures were grown overnight in yeast nitrogen base (pH 7.0) with 50 mM glucose (BBL, Sparks, MD) in an orbital shaker (120 rpm) at 30°C under aerobic conditions. Cells were harvested, washed twice in sterile phosphate-buffered saline (PBS), and resuspended to an optical density of 0.1 at 600 nm for all experiments. All experiments were performed on at least three separate occasions. Cells were grown in the absence or presence of 40, 100, or 200 μM farnesol. Farnesol has been shown to be increasingly produced as the culture ages, at a reported estimated concentration of 35 to 40 μM; therefore, 40 μM of farnesol was chosen for its physiological relevance, and higher farnesol concentrations were used in order to demonstrate a dose-dependent effect of farnesol (36). For all experiments, control cultures with no farnesol included methanol at the maximum concentrations incorporated in the farnesol experiments. In addition, experiments were performed using hydrogen peroxide as a positive control.
MTS viability assay.
The proliferation of farnesol-treated C. albicans cells was assessed using an MTS tetrazolium-based viability assay (Promega, Madison, WI). Farnesol was added to C. albicans suspensions (100 μl) with cell densities of 1 × 107 cells/ml at final concentrations of 0, 10, 40, 100, 200, and 300 μM in the wells of 96-well microtiter plates. The plates were incubated for 24 h at 30°C with shaking. Following incubation, 20 μl of the MTS reagent was added to each well and the plates were incubated at 37°C for 4 h or until color fully developed. Following color development, each sample was transferred to a fresh plate and the colorimetric change at 490 nm (A490) was measured with a microtiter plate reader (Multiskan MCC1340; Titertek).
Proteome analysis.
Two-dimensional gel electrophoresis and mass spectrometry were used to compare the proteomes of treated cells with those of untreated cells as previously described (33). Briefly, cytosolic proteins were extracted, subjected to isoelectric focusing, and separated by polyacrylamide gel electrophoresis. Gels nondestructively stained with silver were scanned (resolution, 300 dots per inch), and gel images were analyzed with PDQuest version 7.0 (Bio-Rad Laboratories). Differentially expressed proteins were selected for identification. Spots which changed consistently and significantly (more than 1.5-fold) were excised and subjected to trypsinization. Peptides were extracted and analyzed using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) with an Applied Biosystems Voyager DE-STR MALDI-TOF mass spectrometer. For protein identification, we used the ProFound search engine with Genomic Solution's Knexus software (version 2004.03.15). The database used was the latest NCBI nonredundant database obtained from NIH.
Statistical analysis.
Experiments were performed in duplicate on three separate occasions. Student's t test was employed to assess the statistical significance of treated versus untreated samples along with the standard deviation. Differences were considered statistically significant at P values of less than 0.05. Protein spots were considered to represent differentially expressed proteins if they were up- or downregulated at least 1.5-fold in three independent experiments.
Confocal scanning laser microscopy.
Samples processed for caspase activation, ROS accumulation, and mitochondrial degradation were observed with a Zeiss Axiovert 100 confocal microscope (with a video capture system, automatic camera, and image analysis hardware software), using 20×, 40×, and 100× oil immersion objectives. Imaging of stained cells was accomplished by using a Cy2/Cy3 multitrack filter set. Images were processed for display by using AxioVision 3.x software (Zeiss). For all microscopy experiments, methanol was incorporated in the control experiments and hydrogen peroxide was used as a positive control.
ROS accumulation.
Intracellular ROS accumulation was examined using a mixture of 100 μl of the fluorescent probe dichlorodihydrofluorescein diacetate (5 mM in ethyl alcohol; Molecular Probes), 100 μl propidium iodide (1 mg/ml; Sigma-Aldrich, St. Louis, MO), and 100 μl calcofluor white (1 mg/ml; Sigma-Aldrich). Cells were examined for ROS accumulation following a 3-h exposure to increasing concentrations of farnesol in order to establish a timeline for the effect of farnesol on the fungal cell. The mixture (4.5 μl) was added to washed cell suspensions and incubated for 30 min in the dark. After being incubated, cells were collected by centrifugation (5 min at 4000 × g), washed once with 1 ml PBS, and resuspended in 50 μl PBS. Fluorescence was analyzed by confocal laser microscopy as described above. Quantification of ROS was performed by scoring the number of green fluorescent cells (ROS) relative to all cells. In addition, the percentage of red fluorescent cells (necrotic cells) was scored. Apoptotic cells fluoresce green, whereas nonapoptotic cells appear unstained.
Mitochondrial degradation.
Mitochondrial degradation was evaluated by using a MitoPT 924 mitochondrial permeability transition detection kit (Immunochemistry Technologies, LLC, Bloomington, MN) according to the manufacturer's recommendations. MitoPT dye concentrates in the mitochondria of healthy cells, creating red fluorescent regions within the cell. However, the dye becomes dispersed in apoptotic cells, which appear green.
In vivo detection of caspase activation.
Activated caspases in C. albicans cells were detected microscopically using FLICA apoptosis detection kits (Immunochemistry Technologies, LLC) according to the manufacturer's recommendation. Cells with intracellular active caspases fluoresce green, whereas nonapoptotic cells appear unstained.
TUNEL assay.
A terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed in order to confirm the occurrence of a farnesol-induced apoptosis process. Following a 24-h incubation with farnesol, C. albicans cells were washed twice with PBS and fixed with a fixation solution of 4% paraformaldehyde in PBS (pH 7.4) for 1 h at 20°C. Cells were rinsed twice with PBS and then incubated with permeabilization solution for 2 min on ice. The cells were rinsed in PBS and labeled, using 50 μl of a 9:1 solution of the label and enzyme solutions from an in situ cell death detection kit, fluorescein (Roche Applied Sciences, Mannheim, Germany), with appropriate controls labeled only with the label solution. The cells were incubated for 1 h at 37°C in a humidified atmosphere in the dark, rinsed in PBS, and examined with a fluorescence microscope with a detection range of 515 to 565 nm. In addition, the number of cells determined to be positive by the TUNEL assay was quantified using flow cytometry. Negative (no enzyme added) and positive (DNase-treated) controls were used to set the region of fluorescence intensity corresponding to cells positive by the TUNEL assay and quantified as percentages of total gated cells (30,000 cells for all samples).
Quantitative PCR (qPCR) analysis of MCA1 expression.
Total RNA was isolated from cultures, using an Invisorb spin cell total RNA minikit (Invitek, Berlin, Germany) after cells were ground in liquid nitrogen. DNA was removed by treatment with DNase I (New England Biolabs) followed by cleaning up using the RNeasy protocol (Qiagen Benelux B.V., Venlo, The Netherlands). The RNA concentration was determined, using a NanoDrop UV-VIS spectrophotometer (Isogen Biosolutions, Maarsen, The Netherlands). The absence of genomic DNA was confirmed by performing PCR using an ACT1 primer pair (Table 1) prior to the reverse transcription step. Reverse transcription was performed on 1 μg of total RNA, using an iScript cDNA synthesis kit (Bio-Rad, Veenendaal, The Netherlands) as specified. Synthesized cDNA was diluted 1:20 in diethyl pyrocarbonate-treated water and stored at −80°C until needed. Sample preparation was performed using a CAS-1200 pipetting robot (Corbett Life Science, Sydney, Australia). Real-time PCR was performed on a MyCycler real-time PCR machine (Bio-Rad, Veenendaal, The Netherlands), using an Absolute QPCR SYBR green mix (Abgene, Epsom, United Kingdom) and the primer pairs listed in Table 1 (Isogen Life Science, Maarsen, The Netherlands), designed using PerlPrimer (22). Amplification was achieved using the following cycle settings: 15 min at 95°C, followed by 35 cycles at 95°C for 1 min, 58°C for 30 s, and 72°C for 30 s. After amplification, a melt curve was analyzed to ensure the absence of primer dimers. Expression of MCA1, which encodes a mammalian homologue of caspase, was calculated using the 2−ΔΔCT method, using ACT1 and RPP2B as the reference genes, as suggested previously (19, 25).
TABLE 1.
Primer sequences used in qPCRa
| Primer | Sequence (5′→3′) | Fragment size (bp) |
|---|---|---|
| ACT1 f1 | ATCAAGGTATCATGGTTGGTATGG | 100 |
| ACT1 r2 | TGTTCAATTGGGTATCTCAAGGTC | 100 |
| RPP2B f3 | ACTTAGCTGCTTACTTATTGTTAG | 143 |
| RPP2B r4 | GTCTTTACCTTCCAAATCTTTCA | 143 |
| MCAI f3 | AGGGTATAACCAAGGTTACG | 200 |
| MCAI r4 | CAGAAGGTCTATTGTATTGTCC | 200 |
All primers were optimized at 58°C and showed efficiencies between 90 and 110%.
RESULTS
For all experiments performed, methanol did not have any effect at the concentrations tested.
MTS viability assay.
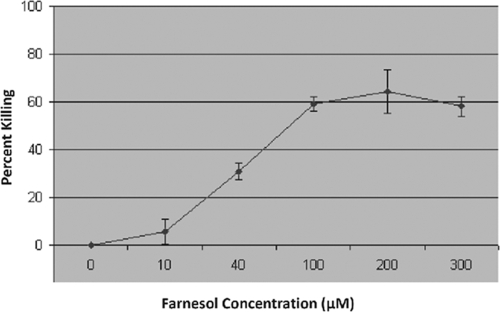
The proliferation of C. albicans cells exposed to increasing concentrations (10, 40, 100, 200, and 300 μM) of farnesol was assessed using the MTS assay. Following incubation, color development was assessed by measuring the A490. Results indicated a minimal killing effect (<10%) for 10 μM farnesol on C. albicans; however, the percentage of killed cells increased significantly in proportion to the farnesol concentration and reached a maximum of around 60% (Fig. 1).
FIG. 1.
Percentage of killed C. albicans cells in total of 2 × 107 cells/ml following 24-h exposure to farnesol (0 to 300 μM), as determined by MTS metabolic assay. Error bars indicate standard errors of the means.
Proteome analysis.
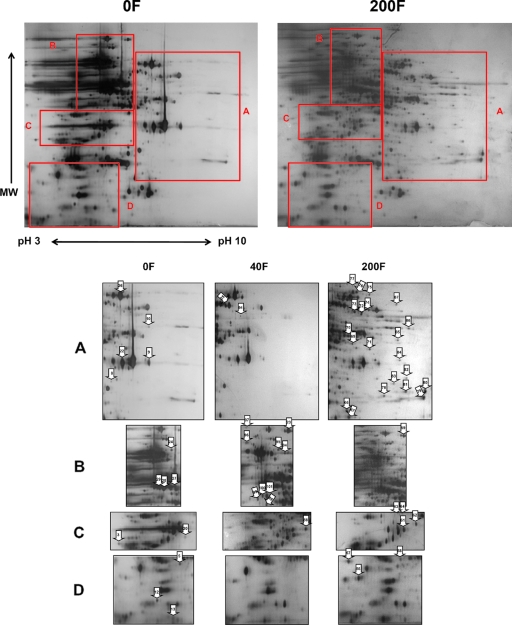
Three independent proteomic analyses showed high reproducibility in demonstrating a significant number of proteins to be differentially regulated upon a 24-h treatment with 40 or 200 μM farnesol (Fig. 2). Proteomic analysis of the farnesol-exposed cells compared to unexposed cells revealed a significant number of proteins to be differentially expressed. Among the 67 altered proteins excised and analyzed by MALDI-TOF MS, 42 proteins were positively identified, including those which function in diverse biological processes such as protein binding and folding, metabolism, and oxidative stress (Table 2). Among those, 22 were downregulated and 26 were upregulated by farnesol treatment (Table 2).
FIG. 2.
Representative two-dimensional gels of extracted proteins from C. albicans grown in the presence of 0 (0F), 40 (40F), or 200 (200F) μM farnesol, demonstrating differential protein expression. Forty-five protein spots (marked with numbered arrows) displayed consistent alterations upon farnesol treatment. Panels A to D represent the corresponding sections marked on the two-dimensional gels. Information on the identities and functions of these spots are listed in Table 2. MW, molecular weight.
TABLE 2.
Proteins down- or upregulated with farnesola
| Protein regulation and amount of farnesol (μM) | Protein spot | Protein (gene) | Function | MW | pI | No. of peptide matches | % Protein score/ confidence interval (%) | NCBI nr database accession no. |
|---|---|---|---|---|---|---|---|---|
| Downregulation | ||||||||
| 40 | 2a | Putative stress response protein (DDR48) | Damage response | 22602 | 4.27 | 11 | 100 | 68483600 |
| 2b | Translational elongation factor 1β (TEF1) | Protein synthesis | 2494267 | 4.25 | 3 | 100 | 2494267 | |
| 5 | Riboflavin synthase; hypothetical Ca019-11507 (RIB5) | Involved in riboflavin synthesis | 30917.3 | 5.8 | 8 | 100 | 68467869 | |
| 6 | Phosphoglycerate mutase; hypothetical CaO19.8522 (GPM1-2) | Glycolysis; changes with drug exposure | 27437.5 | 5.79 | 15 | 100 | 68469783 | |
| 7 | G-beta-like protein; hypothetical Ca019-9606 (ASC1) | Unknown | 23622.1 | 6.3 | 4 | 100 | 68487301 | |
| 8 | Similar to mammalian aldo/keto reductase; hypothetical CaO19.6757 (GRE3) | Involved in stress response | 33095.2 | 6.17 | 7 | 100 | 68472117 | |
| 9 | Glyceraldehyde-3-phosphate dehydrogenase (TDH1) | Glycolysis; changes with drug exposure | 35926.7 | 6.61 | 15 | 100 | 68472227 | |
| 10 | Enolase; hypothetical Ca019-8025 (ENO) | Glycolysis | 47202.5 | 5.54 | 13 | 100 | 68488457 | |
| 200 | 17 | ATP synthase subunit D; hypothetical CaO19.10301 (ATP7) | Mitochondrial; catalyzes ATP synthesis | 19364.1 | 6.19 | 8 | 100 | 68488805 |
| 18 | NADH:quinone oxidoreductase; hypothetical CaO19.11095 (CQR2) | Enzyme complex of the respiratory chain | 21714.2 | 6.51 | 5 | 100 | 68483141 | |
| 19 | Heat shock protein 60 (HSP60) | Mitochondrial; involved in proteins imported into the mitochondrion | 60378.6 | 5.22 | 16 | 100 | 68485963 | |
| 20 | Orthologous to carboxymethylenebutenolidase; hypothetical CaO19.2966 (YDL086W) | Hydrolase enzyme | 28828.4 | 5.84 | 4 | 100 | 68468813 | |
| 22 | Glyceraldehyde-3-phosphate dehydrogenase (TDH1-3) | Glycolysis | 35926.7 | 6.61 | 18 | 100 | 68472227 | |
| 24 | Dihydroxy acid dehydratase (ILV3) | Mitochondrion; involved in branched amino acid synthesis | 63462.9 | 6.2 | 12 | 100 | 68467901 | |
| 26 | Pyruvate kinase (PYK1) | Glycolysis | 55757.7 | 6.54 | 23 | 100 | 68482226 | |
| 27 | S-adenosylmethionine synthase; hypothetical CaO19.8272 (SAM2) | Methionine metabolism | 42465.8 | 5.64 | 16 | 100 | 68484437 | |
| Upregulation | ||||||||
| 40 | 23 | Putative mitochondrial aconitate hydratase (ACO1) | Tricarboxylic acid cycle | 84632.7 | 5.96 | 24 | 100 | 68479387 |
| 88 | Coproporphyrinogen III oxidase (HEM13) | Mitochondrion; involved in heme biosynthesis | 37089.4 | 5.66 | 15 | 100 | 68490312 | |
| 89 | Branched chain amino acid transaminase; hypothetical CaO19.6994 (BAT1) | Involved in branched amino acid synthesis | 40894 | 5.89 | 11 | 100 | 68482781 | |
| 90a | Alcohol dehydrogenase; hypothetical CaO19.3997 (ADH3) | Functions in formaldehyde metabolism | 46499.6 | 8.26 | 19 | 100 | 68468132 | |
| 91 | Catalase A (CTA1) | Reactive oxygen metabolism | 54945.2 | 6.18 | 16 | 100 | 68474218 | |
| 92 | 6-Phosphogluconate dehydrogenase; hypothetical CaO19.12491 (GND1) | Plays a critical role in protecting cells from oxidative stress | 57164.1 | 6.14 | 19 | 100 | 68467359 | |
| 93 | Pyruvate decarboxylase (CDC19/PYK1) | Glycolysis | 62750 | 5.39 | 18 | 100 | 68480872 | |
| 96 | Heat shock protein 70; hypothetical CaO19.12447 (SSA1) | Molecular chaperone; involved in response to environmental stress | 70445 | 5.06 | 24 | 100 | 68467277 | |
| 97 | Alanine/arginine aminopeptidase; hypothetical CaO19.12664 (APE2) | Protein degradation | 107584.4 | 5.63 | 37 | 100 | 68491573 | |
| 98 | Heat shock protein 90 homologue; hypothetical CaO19.6515 (HSP90) | Molecular chaperone; involved in response to environmental stress | 80773.2 | 4.81 | 28 | 100 | 68469132 | |
| 99a | Aryl-alcohol dehydrogenase; hypothetical CaO19.1048 (CAD4) | Involved in drug resistance | 39539.7 | 6.84 | 16 | 100 | 6325169 | |
| 200 | 57 | Heat shock protein 90 (HSP90) | Molecular chaperone; involved in response to environmental stress | 23899.4 | 4.33 | 1 | 63 | 68475757 |
| 58 | Putative Rho protein GDP dissociation factor (RDI1) | Involved in reorganization of actin cytoskeleton | 22947.9 | 5.15 | 7 | 100 | 68465635 | |
| 59 | Putative mitochondrial aconitate dehydrogenase (ACO1) | Required for tricarboxylic acid cycle and mitochondrial genome maintenance | 84632.7 | 5.96 | 28 | 100 | 68479387 | |
| 60 | Heat shock protein 60 (HSP60) | Mitochondria; oxidative stress response | 60378.6 | 5.22 | 1 | 50 | 68485963 | |
| 61 | Aldose reductase (GRE3) | Induced by oxidative stress | 42602.9 | 7.05 | 17 | 100 | 68470494 | |
| 62 | Coproporphyrinogen III oxidase (HEM13) | Mitochondria; involved in heme biosynthesis | 37089.4 | 5.66 | 10 | 100 | 68490312 | |
| 66 | GTP binding protein; homologue to human Ranp (GSP2) | Involved in the maintenance of nuclear organization, RNA processing | 25148.7 | 6.22 | 6 | 100 | 6324759 | |
| 67 | Manganese superoxide dismutase | Involved in defense against ROS | 14782.7 | 8.04 | 2 | 46 | 60760051 | |
| 69 | Mitochondrial 2-enoyl thioester reductase | Required for respiration and maintenance of mitochondrial function | 38645.3 | 6.36 | 13 | 99.99 | 68486065 | |
| 70 | Putative aspartate aminotransferase (AAT21) | Aspartate metabolism | 49035.3 | 8.61 | 17 | 100 | 68482311 | |
| 72 | Glutathione reductase (GLR1) | Oxidative stress | 57190.1 | 7.18 | 10 | 100 | 55294642 | |
| 76 | Acetyl-CoA hydrolase/transferase (ACH1) | Involved in acetyl-CoA pathway | 58172.5 | 6.42 | 8 | 98.53 | 68482646 | |
| 78 | Phosphoglycerate mutase; hypothetical CaO19.8669 (GPM1) | Glycolysis | 30678.1 | 7.19 | 5 | 99.98 | 68484809 | |
| 80 | Outer mitochondrial membrane porin; hypothetical CaO19.1042 (POR1) | Required for the maintenance of mitochondrial osmotic stability and membrane permeability | 29748.5 | 8.5 | 12 | 100 | 68484582 | |
| 81 | NADH-cytochrome b5 reductase (MCR1) | Involved in resistance against oxidative stress | 33555.6 | 8.54 | 1 | 48 | 68490698 |
Proteins identified by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and MALDI-TOF MS as being differentially expressed in cells exposed to increasing concentrations of farnesol and unexposed cells. Acetyl-CoA, acetyl coenzyme A.
Downregulated proteins.
Most notable among the downregulated proteins as a result of farnesol exposure were those involved in glycolysis (GPM1-2, TDH1, ENO, and PYK1), protein synthesis (TEF1 and ILV3), and metabolism. In addition, S-adenosylmethionine synthetase (SAM2), an enzyme suggested to be morphology specific, was downregulated. With 200 μM farnesol, mitochondrial enzymes involved in ATP synthesis (ATP7), mitochondrial electron transport, and the respiratory chain (CQR2, HSP60, and ILV3) were downregulated (Table 2).
Upregulated proteins.
Among the 29 proteins displaying significant overexpression, 11 were upregulated with 40 μM farnesol and 18 with 200 μM farnesol. The stress protein CTA1 and the heat shock proteins HSP90 and HSP70 (SSA1) were upregulated with 40 and 200 μM farnesol. With 200 μM farnesol, proteins involved in protein folding and protection against environmental and oxidative stress (HSP90, HSP60, GLR1, and SOD2) were significantly upregulated. The Rho protein GDP dissociation inhibitor (RDI1) was also found to be upregulated with 200 μM farnesol (Table 2).
ROS accumulation.
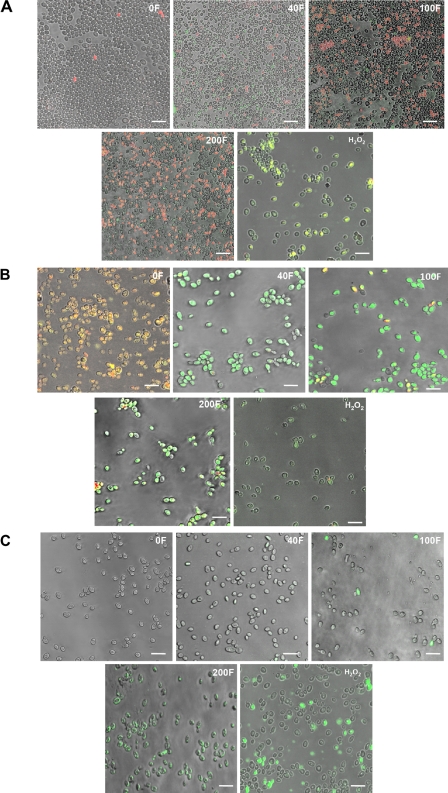
Consistent with the results demonstrating that farnesol had a damaging effect on the mitochondria, ROS accumulation was assessed using a cocktail of the dichlorodihydrofluorescein diacetate fluorescent probe, which demonstrates the presence of ROS as green fluorescence, propidium iodide, which stains necrotic cells red, and the chitin dye calcofluor white, which allows visualization of healthy cells. Results demonstrated no green or red fluorescence in the unexposed cells. However, an increasing number of cells appeared green, orange, and red, indicating an accumulation of ROS in the cells, followed by necrosis and death, proportional to the farnesol concentration (Fig. 3A; Table 3). ROS accumulation occurred within 3 h of exposure to farnesol and hydrogen peroxide (Fig. 3A).
FIG. 3.
Representative confocal scanning laser fluorescence images of farnesol-treated and untreated C. albicans cells, revealing the presence of (A) ROS accumulation, indicated by green fluorescence, and necrotic or dead cells, indicated by red fluorescence; (B) mitochondrial degradation in the farnesol-exposed cells, indicated by green fluorescence, with healthy mitochondria appearing as red aggregates; and (C) activation of intracellular caspases in the farnesol-exposed cells, indicated by green fluorescence. Minimal patchy fluorescence was observed in the cells treated with 40 μM farnesol, with increasing fluorescence seen with increasing concentrations of farnesol. The amount of farnesol is indicated in each panel by a number and the letter “F.” The bar represents 20 μm.
TABLE 3.
Percentages of cells demonstrating mitochondrial degradation, ROS accumulation, or caspase activationa
| Farnesol conc (μM) | Mitochondrial degradation | ROS accumulation | Caspase activity | % Dead cells |
|---|---|---|---|---|
| 0 | <1 | 0 | <1 | 0 |
| 40 | 90 | 26 | 5 | 13 |
| 100 | 93 | 80 | 13 | 20 |
| 200 | 95 | 65 | 84 | 35 |
Indicate occurrences of apoptosis followed by death upon exposure to increasing concentrations of farnesol.
Mitochondrial degradation.
Treatment of farnesol-exposed cells with the MitoPT reagent resulted in the appearance of apoptotic cells with green fluorescence, whereas the cells unexposed to farnesol appeared nonapoptotic, or red (Fig. 3B; Table 3). The ratios of red aggregates and green fluorescence varied, indicating cells in various stages of apoptosis. Minimal patchy green fluorescence was observed in the cells treated with 40 μM farnesol, and increasing fluorescence was observed in proportion to the increase in farnesol concentration. Mitochondrial degradation was also observed following exposure to hydrogen peroxide (Fig. 3B).
In vivo detection of caspase activation.
Caspases are activated upon induction of apoptosis. In order to determine the presence of activated caspases, which is indicative of an apoptotic process, a FLICA apoptosis detection kit was used. Images revealed the presence of green fluorescence in the farnesol-exposed cells, indicating the activation of intracellular caspases, whereas minimal or no fluorescence was observed in the unexposed cells (Fig. 3C; Table 3). The fraction of cells containing activated caspases increased with increasing farnesol concentrations.
TUNEL assay.
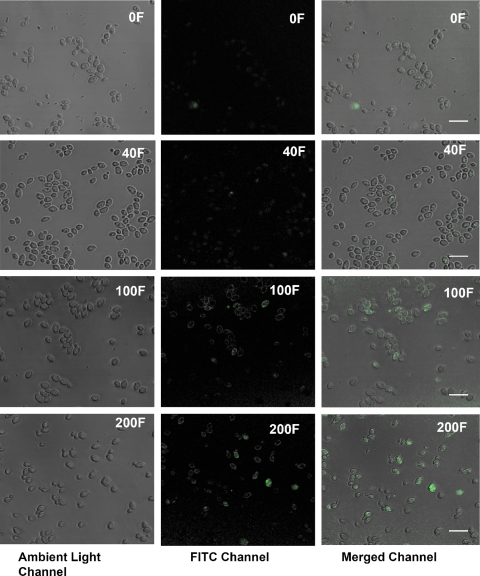
The dose-dependent increase in apoptosis in the farnesol-treated C. albicans cells was corroborated by the results from the TUNEL assay. Microscopic images revealed the presence of no or minimal green fluorescence in the cells treated with 40 μM farnesol, whereas significant fluorescence was observed in the cells treated with 100 or 200 μM farnesol (Fig. 4). In addition, flow cytometry results indicated that the percentages of positive cells as determined by TUNEL assay in the untreated samples was similar to that for the negative control (1.5 and 1.34%, respectively). Samples treated with 40, 100, or 200 μM farnesol for 24 h showed 2.0, 10.2, and 14.5% positive cells, respectively, as determined by TUNEL assay, while the positive control showed 17.3% of all cells to be positive by TUNEL assay.
FIG. 4.
TUNEL assay showing a dose-dependent increase in apoptosis, indicated by appearance of green fluorescence in farnesol-treated C. albicans cells compared to untreated cells. The amount of farnesol is indicated in each panel by a number and the letter “F.” The bar represents 20 μm.
qPCR analysis of MCA1 expression.
MCA1 encodes a homologue of a mammalian caspase in C. albicans. To determine the involvement of caspases in farnesol-induced cytotoxicity in C. albicans, the expression of MCA1 was determined by qPCR in log-phase C. albicans cells. Following a 4-h exposure to 40 μM farnesol, MCA1 expression was increased threefold relative to that for the untreated control cells. In contrast, no difference in MCA1 expression was seen in the cells exposed to 100 μM farnesol, whereas in the cells exposed to 200 μM farnesol, MCA1 expression was downregulated. Similarly, exposure to increasing concentrations of hydrogen peroxide (0, 2.5, 5.0, and 10 mM) resulted in rapid induction of MCA1 to the same levels as seen with farnesol, followed by a decrease in mRNA (data not shown).
DISCUSSION
Despite extensive studies on the involvement of farnesol in C. albicans germination and biofilm formation, the characterization of a physiological role for farnesol and its implications on the fungal cell cycle have been lacking. As eukaryotic cells, fungal and human cells share similar metabolic pathways. Hence, the inhibitory effect of farnesol on C. albicans could also involve cellular signal transduction pathways similar to the apoptotic process described in mammalian cells. Therefore, in order to elucidate the mechanisms underlying farnesol cytotoxicity and its possible involvement in an apoptotic process in C. albicans, a global two-dimensional proteomic approach was utilized to unravel altered protein expression following farnesol treatment. In addition, an assessment of standard apoptotic markers using fluorescent microscopy and gene expression analysis was also undertaken.
Proteomic analysis of the farnesol-exposed cells revealed a significant number of proteins to be differentially expressed (Table 2). Most notable among the downregulated proteins as a result of farnesol exposure were those involved in glycolysis protein synthesis and metabolism, reflecting the growth inhibition effect of farnesol on the cells (Table 2). In addition, SAM2, an enzyme suggested to increase during yeast-to-hyphal morphogenesis, was also downregulated, a finding consistent with the inhibition effect of farnesol on germination (17).
The role of the mitochondria in both apoptotic and necrotic cell death has received considerable attention, and an increase of mitochondrial membrane permeability is considered one of the key events in apoptosis (37, 40). In our study, an adverse effect of farnesol on the mitochondria was indicated by the downregulation of several proteins involved in mitochondrial electron transport and respiration (Table 2). A further indication of the presence of a stress response to farnesol for C. albicans was seen in the upregulated proteins, which included those involved in protein folding, heat shock, and protection against environmental and oxidative stress. Of interest was the upregulation of a Rho protein GDP dissociation inhibitor (RGDI1) similar to S. cerevisiae RDI1 (YDL135C) (Table 2). Rho protein GTPases are involved primarily in the reorganization of the actin cytoskeleton and are linked to morphological modifications and apoptosis in all eukaryotes (24). A study by Zhang et al. (38) demonstrated RhoGDI to be an antiapoptotic molecule that promotes the resistance of cancer cells to drug-induced toxicity where the overexpression of RhoGDI increased the resistance of cancer cells to the induction of apoptosis by chemotherapeutic agents. It is plausible, therefore, to speculate that a similar process occurs with C. albicans, where RhoGDI is overexpressed in response to the apoptotic effect of farnesol. Furthermore, in our previous work with oral squamous cell carcinoma tumor cells, RhoGDI was also found to be upregulated in these cells upon exposure to farnesol, suggesting a correlation between the cytotoxic effects of farnesol on C. albicans and human tumor cell lines (33).
The presence of a farnesol-induced drug response is also indicated by the increased expression of aryl alcohol dehydrogenase (CAD4) with 40 μM farnesol. The homologue of CAD4 in S. cerevisiae (YPL088W gene product) has been shown to contain a drug response element in its promoter that leads to induction of mRNA expression upon estradiol treatment (11). In addition, the YPL088W gene was also shown to be upregulated in microarray and proteomic studies of an azole-resistant S. cerevisiae isolate (11). Therefore, the upregulation of this protein indicates the presence of a drug response by C. albicans to farnesol.
Moreover, NADH-cytochrome b5 reductase, a homologue of S. cerevisiae MCR1 that plays a key role in the NADH-dependent reduction of the d-erythroascorbyl free radical, was also upregulated with farnesol. In S. cerevisiae, the mcr1 mutants were found to be hypersensitive to hydrogen peroxide and menadione, and overexpression of MCR1 made the cells more resistant to oxidative stress (18). Also of interest is the upregulation of acetyl coenzyme A transferase (ACH1), an enzyme representing the first step in ergosterol biosynthesis. Farnesol is a key intermediate in the ergosterol biosynthetic pathway in C. albicans; therefore, the expression of acetyl coenzyme A transferase may be regulated by the ergosterol content of the membrane, as the ACH1 gene is also responsive to ergosterol (11).
Advances in proteomic technology offer great promise in the understanding and treatment of the molecular basis of disease. Specifically, the study of dynamic protein expression has culminated in the identification of many potential new drug targets. Therefore, it is of interest to note that riboflavin synthase (RIB5), an enzyme involved in riboflavin synthesis, was among the proteins downregulated with farnesol. Bacteria and yeast are unable to incorporate riboflavin from the environment and are absolutely dependent on endogenous synthesis of this vitamin (8). Therefore, based on these findings, riboflavin synthase is a potential target for the development of antimicrobial drugs.
Mitochondrial respiration is a powerful source of superoxide radicals in yeast. ROS can damage almost every essential cellular component, resulting in enzyme inactivation, membrane disruption, and mutations, and ultimately in cell death (2, 20). Studies have revealed that perturbations in mitochondrion respiration occur early in the apoptotic process and that the mitochondrion itself serves as a control switch for apoptosis (40). We have previously shown that the farnesol-induced apoptosis in human tumor cells was mediated by mitochondria and the generation of ROS (33). Similarly, in this study mitochondrial degradation and ROS accumulation were also demonstrated by fluorescent microscopy (Fig. 3A and B).
Interestingly, the results of mitochondrial degradation, ROS accumulation, and caspase activation seem to suggest the presence of two distinct farnesol-induced processes occurring simultaneously in different cells: necrosis and apoptosis. It is important to note here that the hydrophobic nature of farnesol favors its accumulation in the cell membrane, and a disruption in membrane integrity was shown by us in both bacteria and yeast. Therefore, it is logical to speculate that in aging or damaged cells, farnesol exposure results in immediate disruption of the cell membrane, leading to necrosis. In contrast, in other cells the process of farnesol toxicity involves an apoptotic process in which increased intracellular levels of farnesol lead to ROS accumulation and ultimately to cell death.
MCA1 encodes a human homologue of caspase. In our gene expression studies, MCA1 was found to be upregulated upon exposure of cells to farnesol, with a peak in expression at 40 μM (physiological concentration) and decreasing levels of expression with increasing concentrations of farnesol. This finding could be explained by the transient expression of MCA1 over time; i.e., with increasing concentrations of farnesol, the induction of caspase expression occurs at earlier time points and is undetectable by this assay. Alternatively, we and others have observed that MCA1 expression is generally decreased in the stationary phase, and high concentrations of farnesol may possibly mimic this effect.
Furthermore, in mammalian cells, it is the posttranslational cleavage (autoactivation) of the caspase enzymes into their smaller subunits rather than their expression that is indicative of the induction of apoptosis. Hence, the overexpression of the MCA1 gene at 40 μM may be a reflection of the presence of stress to the fungal cell but not necessarily an indication of apoptosis per se. If so, activated caspases would not be expected to be detected at that concentration, as was demonstrated by the microscopic images for the presence of activated caspases (Fig. 3C).
More interestingly, fluorescent reagents designed to detect the presence of activated caspases and mitochondrial degradation in mammalian cells were applied to farnesol-exposed and non-farnesol-exposed C. albicans. Since caspases are highly conserved, the reagent successfully bound to C. albicans caspases, demonstrating the presence of activated caspases in the farnesol-exposed cells (Fig. 3C). In addition, the farnesol-treated C. albicans cells were also determined to be positive by TUNEL assay, confirming the presence of apoptosis in these cells (Fig. 3D).
Combined, our findings demonstrate that farnesol induces a caspase-mediated process of apoptosis, with the typical markers of apoptosis suggesting that a similar process mediates the cytotoxicity of this drug in both yeasts and human tumor cells. The similarities we have shown between the cytotoxic processes induced by farnesol in yeast and the cell death process triggered by this molecule in human tumor cells suggest that this mechanism is an ancestral apoptotic pathway preserved in both single-celled and metazoan organisms.
The survival of yeast depends on its ability to adapt to adverse environmental conditions in a manner that ensures survival of the clone. Based on our findings, it is conceivable to speculate that through the regulation of farnesol production, C. albicans cells may have developed a mechanism to purposely produce ROS as a regulator of apoptosis. Controlling the proliferation of cells and sparing dwindling resources, particularly in a biofilm environment where nutrients are limited and conditions unfavorable, would provide an evolutionary advantage. In addition, farnesol-mediated apoptosis might free up additional resources (derived from lysed cells) to supply the remaining cells with sufficient energy to efficiently prepare for changing, harsh conditions.
It is important to note that in our previous work, we had shown that the farnesol secreted by C. albicans in 24- to 48-h-old biofilm cultures was able to trigger apoptosis in human tumor cells through the classical apoptotic pathways, similar to the effect exerted by 30 to 60 μM synthetic farnesol. These observations support the hypothesis that the farnesol produced by C. albicans in a mature biofilm also induces apoptosis in eukaryotic cells. Further investigations exploring the role farnesol plays in orchestrating survival and continuance of a biofilm community, specifically, time course experiments monitoring the progression of farnesol secretion in biofilm and its effect on biofilm cells, are warranted and are ongoing in our laboratories.
In conclusion, the findings of this study demonstrate that farnesol inhibits the growth of C. albicans and promotes apoptosis via the induction of caspases, production of ROS, and disruption of the integrity of the mitochondria. To our knowledge, this is the first study utilizing proteomic analysis to characterize farnesol-induced changes in C. albicans protein expression, in turn identifying farnesol as a trigger of an apoptotic process in C. albicans. The characterization of the mechanistic switches that regulate active death processes in C. albicans may lead to the development of novel antifungal agents that switch on endogenous cell suicide mechanisms.
Supplementary Material
Acknowledgments
We thank H. W. de Haan-Visser for technical assistance.
This work was supported by NIH grants DE14424, DE016257, RR023250, and R01-AI69568-01A2.
Footnotes
Published ahead of print on 13 April 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa, V., and P. Moradas-Ferreira. 2001. Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol. Aspects Med. 22:217-246. [DOI] [PubMed] [Google Scholar]
- 3.Enari, M., H. Sakahira, H. Yokoyama, K. Okawa, A. Iwamatsu, and S. Nagata. 1998. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391:43-50. [DOI] [PubMed] [Google Scholar]
- 4.Enjalbert, B., A. Nantel, and M. Whiteway. 2003. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enjalbert, B., and M. Whiteway. 2005. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot. Cell 4:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fairn, G. D., K. MacDonald, and C. R. McMaster. 2007. A chemogenomic screen in Saccharomyces cerevisiae uncovers a primary role for the mitochondria in farnesol toxicity and its regulation by the Pkc1 pathway. J. Biol. Chem. 282:4868-4874. [DOI] [PubMed] [Google Scholar]
- 7.Fidel, P. L., Jr. 2006. Candida-host interactions in HIV disease: relationships in oropharyngeal candidiasis. Adv. Dent. Res. 19:80-84. [DOI] [PubMed] [Google Scholar]
- 8.Fischer, M., A.-K. Schott, K. Kemter, R. Feicht, G. Richter, B. Illarionov, W. Eisenreich, S. Gerhardt, M. Cushman, S. Steinbacher, R. Huber, and A. Bacher. 2003. Riboflavin synthase of Schizosaccharomyces pombe. Protein dynamics revealed by 19F NMR protein perturbation experiments. BMC Biochem. 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourlay, C. W., and K. R. Ayscough. 2005. Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J. Cell Sci. 118:2119-2132. [DOI] [PubMed] [Google Scholar]
- 10.Hengartner, M. O. 2001. Apoptosis: DNA destroyers. Nature 412:27-29. [DOI] [PubMed] [Google Scholar]
- 11.Hooshdaran, M. Z., K. S. Barker, G. M. Hilliard, H. Kusch, J. Morschhauser, and P. D. Rogers. 2004. Proteomic analysis of azole resistance in Candida albicans clinical isolates. Antimicrob. Agents Chemother. 48:2733-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornby, J. M., B. W. Kebaara, and K. W. Nickerson. 2003. Farnesol biosynthesis in Candida albicans: cellular response to sterol inhibition by zargozic acid B. Antimicrob. Agents Chemother. 47:2366-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jabra-Rizk, M. A., M. E. Shirtliff, C. James, and T. F. Meiller. 2006. Effect of farnesol on Candida dubliniensis biofilm formation and fluconazole resistance. FEMS Yeast Res. 6:1063-1073. [DOI] [PubMed] [Google Scholar]
- 15.Jayatilake, J. A. M. S., Y. H. Samaranayake, and L. P. Samaranayake. 2005. An ultrastructural and a cytochemical study of candidal invasion of reconstituted human oral epithelium. J. Oral Pathog. Med. 34:240-246. [DOI] [PubMed] [Google Scholar]
- 16.Lamarre, C., J.-D. LeMay, N. Deslauriers, and Y. Bourbonnais. 2001. Candida albicans expresses an unusual cytoplasmic manganese-containing superoxide dismutase (SOD3 gene product) upon the entry and during the stationary phase. J. Biol. Chem. 276:43784-43791. [DOI] [PubMed] [Google Scholar]
- 17.Lambert, R. H., and J. R. Garcia. 1990. Evidence of morphology-specific isozymes in Candida albicans. Curr. Microbiol. 20:215-221. [Google Scholar]
- 18.Lee, J.-S., W.-K. Huh, B.-H. Lee, Y.-U. Baek, C.-S. Hwang, S.-T. Kim, Y.-R. Kim, and S.-O. Kang. 2001. Mitochondrial NADH-cytochrome b5 reductase plays a crucial role in the reduction of d-erythroascorbyl free radical in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1527:31-38. [DOI] [PubMed] [Google Scholar]
- 19.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 20.Longo, V. D., L.-L. Liou, J. S. Valentine, and E. B. Gralla. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365:131-142. [DOI] [PubMed] [Google Scholar]
- 21.Madeo, F., E. Fröhlich, M. Ligr, M. Grey, S. J. Sigrist, D. H. Wolf, and K.-U. Fröhlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall, O. J. 2004. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20:2471-2472. [DOI] [PubMed] [Google Scholar]
- 23.Martchenko, M., A. M. Alarco, D. Harcus, and M. Whiteway. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell 15:456-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menotta, M., A. Amicucci, G. Basile, E. Polidori, V. Stocchi, and F. Rivero. 2008. Molecular and functional characterization of a Rho GDP dissociation inhibitor in the filamentous fungus Tuber borchii. BMC Microbiol. 8:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nailis, H., T. Coenye, F. van Nieuwerburgh, D. Deforce, and H. J. Nelis. 2006. Developmental evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh, K.-B., H. Miyazawa, T. Naito, and H. Matsuoka. 2001. Purification and characterization of an autoregulatory substance capable of regulating the morphological transition in Candida albicans. Proc. Natl. Acad. Sci. USA 98:4664-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perlroth, J., B. Choi, and B. Spellberg. 2007. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45:321-346. [DOI] [PubMed] [Google Scholar]
- 28.Phillips, A., I. Sudbery, and M. Ramsdale. 2003. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA 100:14327-14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips, A. J., J. D. Crowe, and M. Ramsdale. 2006. Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 103:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramage, G., S. P. Saville, B. L. Wickes, and J. L. Lopez-Ribot. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato, T., T. Watanabe, T. Mikami, and T. Matsumoto. 2004. Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol. Pharma. Bull. 27:751-752. [DOI] [PubMed] [Google Scholar]
- 32.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheper, M. A., M. E. Shirtliff, T. F. Meiller, B. Peters, and M. A. Jabra-Rizk. 2008. Farnesol a fungal quorum sensing molecule triggers apoptosis in human oral squamous carcinoma cells. Neoplasia 10:954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semighini, C. P., J. M. Hornby, R. Dumitru, K. W. Nickerson, and S. D. Harris. 2006. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 59:753-764. [DOI] [PubMed] [Google Scholar]
- 35.Smith, D. A., S. Nicholls, B. A. Morgan, A. J. Brown, and J. Quinn. 2004. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15:4179-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber, K., R. Sohr, B. Schulz, M. Fleischhacker, and M. Ruhnke. 2008. Secretion of E,E-farnesol and biofilm formation in eight different Candida species. Antimicrob. Agents Chemother. 52:1859-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westwater, C., E. Balish, and D. A. Schofield. 2005. Candida albicans-conditioned medium protects yeast cells from oxidative stress: a possible link between quorum sensing and oxidative stress resistance. Eukaryot. Cell 4:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, B., Y. Zhang, M. C. Dagher, and E. Shacter. 2005. Rho GDP dissociation inhibitor protects cancer cells against drug-induced apoptosis. Cancer Res. 65:6054-6062. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, J., and M. Xu. 2002. Apoptotic DNA fragmentation and tissue homeostasis. Tends Cell Biol. 12:84-89. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, S., J. H. Yang, C. K. Guo, and P. C. Cai. 2007. Gene silencing of TKTL1 by RNAi inhibits cell proliferation in human hepatoma cells. Cancer Lett. 253:108-114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.