Abstract
PCR techniques in combination with conventional parasite concentration procedures have potential for the sensitive and specific detection of Toxoplasma gondii oocysts in water. Three real-time PCR assays based on the B1 gene and a 529-bp repetitive element were analyzed for the detection of T. gondii tachyzoites and oocysts. Lower sensitivity and specificity were obtained with the B1 gene-based PCR than with the 529-bp repeat-based PCR. New procedures for the real-time PCR detection of T. gondii oocysts in concentrates of surface water were developed and tested in conjunction with a method for the direct extraction of inhibitor-free DNA from water. This technique detected as few as one oocyst seeded to 0.5 ml of packed pellets from water samples concentrated by Envirocheck filters. Thus, this real-time PCR may provide a detection method alternative to the traditional mouse assay and microscopy.
Toxoplasma gondii is a ubiquitous parasite found in all classes of warm-blooded vertebrates. Nearly one-third of humans have been exposed to this parasite (15). In immunocompetent adults, acute infection normally results in transient influenza-like symptoms, but in immunocompromised persons retinochoroiditis and encephalitis are more common. Infected individuals can retain the parasite as quiescent tissue cysts for long periods, but invasive infection can occur if the immune status of the infected person deteriorates (42). If women become infected during pregnancy, the parasite can cause abortion or seriously damage the fetus. The potential morbidity from the ingestion of oocysts of T. gondii and the organism's low infectious dose are a great concern for public health. There are at least four reported waterborne outbreaks of toxoplasmosis (2, 3, 14, 44), and endemic toxoplasmosis in Brazil is associated with the consumption of water or ice contaminated with T. gondii oocysts (1, 23), demonstrating the potential for the waterborne transmission of this disease (15).
There is no rapid detection method for T. gondii oocysts recovered from water or other environmental samples. Traditionally, the detection of protozoa in water required their concentration from large volumes of water by filtration or centrifugation, isolation from concentrated particulates by immunomagnetic separation (IMS) or other methods, and detection by immunofluorescence microscopy, the infection of cultured cells, biochemistry, animal infection tests, molecular techniques, or combinations of these (17, 58). For T. gondii oocysts there are no commercially available IMS techniques, no widely available immunofluorescent staining reagents, and no standardized cultivation protocols. The identification of oocysts from environmental samples has included differential floatation and mouse inoculation (27). Recently, IMS techniques have been developed for the isolation of T. gondii oocysts and sporocysts in water (16, 18). Both the oocyst and sporocyst IMS assays, however, had poor specificity, because antibodies cross-reacted with water debris and the sporocyst wall of Hammondia hammondi, Hammondia heydorni, and Neospora caninum (16).
PCR is becoming a favored technique for the detection of T. gondii oocysts in water (32, 35, 36, 46, 49, 55) over the conventional mouse bioassay (27, 55), as it reduces the detection time from weeks to 1 to 2 days. Although they have been developed for the detection of T. gondii in clinical specimens (50), no real-time PCR assays have been adapted for the detection of oocysts in water samples, possibly because of expected high concentrations of PCR inhibitors and low numbers of T. gondii oocysts in environmental samples (55).
There are several unresolved issues regarding the effectiveness of the PCR detection of T. gondii oocysts in water. The most readily available method for the isolation of T. gondii oocysts from water samples is flocculation or sucrose floatation prior to DNA extraction (35, 36, 49, 55). Because sucrose flotation and flocculation result in oocyst losses, the recovery rate of using these methods is poor. For DNA extraction, the phenol-chloroform method or QIAamp mini kit frequently is used (16, 35, 36, 46, 55). When oocysts are recovered from water either by the Environmental Protection Agency (EPA) information collection rule method (53) or EPA Method 1623 (54) without purification by IMS, neither the conventional phenol-chloroform DNA extraction nor the QIAamp mini kit is effective at removing PCR inhibitors (30, 55, 57).
Recently, a method was used effectively in the analysis of Cryptosporidium oocysts in surface water, storm water, and wastewater samples (30). This method extracted DNA directly from water concentrates without pathogen IMS, differential flotation, or enrichment cultures, and it utilized a commercial DNA extraction kit, the FastDNA spin kit for soil, and a high concentration of nonacetylated bovine serum albumin in PCR. The FastDNA soil kit has a higher capacity for PCR inhibitor removal than several other commercial extraction kits designed for environmental samples. The use of nonacetylated bovine serum in the PCR neutralizes residual PCR inhibitors that are coextracted with the DNA (30).
In the present study, the performance of two published LightCycler real-time PCR assays based on the multicopy B1 gene and 529-bp repetitive element (13, 45) and a newly developed LightCycler real-time PCR assay using a common primer set were analyzed for the detection of T. gondii, using pure DNA and DNA extracted by the aforementioned extraction method (30) from water sample concentrates seeded with known number of oocysts.
MATERIALS AND METHODS
Toxoplasma gondii strains.
Toxoplasma gondii tachyzoites of the RH strain and oocysts of the VEG strain were provided by Ynes Ortega of the University of Georgia and J. P. Dubey of the U.S. Department of Agriculture, respectively.
T. gondii oocyst stock preparation.
A hemocytometer-counted suspension of 64,000 oocysts in 2 ml of distilled water was used in an initial dilution of T. gondii oocysts. Suspensions of 1,600, 800, 400, 200, 100, 50, 25, 10, 5, and 1 oocyst then were made by serial dilution and used to evaluate the sensitivity of the B1 and 529-bp gene-based LightCycler real-time PCR assays.
Water sample concentrates and seeding.
Twenty-liter samples of stream water were collected after rain events by autosamplers as previously described (31) and were concentrated with Envirocheck HV filters (Pall Gelman Laboratory, Ann Arbor, MI) by following EPA Method 1623 to the IMS step (54). Because of the high degree of turbidity, several filters were needed to process the entire sample volume. These sample concentrates were used in seeding experiments, because they were known to have PCR inhibitors (30). Each 0.5 ml of packed pellet from the processed water samples was seeded with known numbers of T. gondii oocysts. This pellet volume was chosen because it simulates the maximum volume of water concentrates used in each IMS processing by EPA Method 1623 and the volume usually analyzed for Cryptosporidium oocysts and Giardia cysts by microscopy for each water sample.
DNA extraction.
DNA was extracted from cultured tachyzoites or water concentrates using the FastDNA spin kit for soil (Qbiogene, Irvine, CA) (30). Briefly, 6 × 106 tachyzoites or each 0.5-ml packed pellet seeded with oocysts was transferred into a 2-ml tube containing the lysing matrix E from the FastDNA spin kit for soil. After adding 978 μl of sodium phosphate buffer and 122 μl of MT buffer from the kit, the tube was vortexed in a FastPrep instrument (Qbiogene) for 30 s at a speed setting of 5.5. Each sample was processed further in accordance with the manufacture-recommended procedures, and 100 μl of DNA was eluted per sample. DNA was stored at −20°C until use in PCR. In most experiments, 2 μl of DNA was used in PCR. To determine the sensitivity of the PCR in detecting pure T. gondii DNA, 2 μl of fivefold serial dilutions of the tachyzoite DNA was used in PCR.
Target genes.
The multicopy B1 gene and the 529-bp repetitive element were the PCR targets evaluated in this study. The B1 gene consists of 35 copies of a 2,214-bp repeat, and it is highly conserved among strains of T. gondii (6). The 529-bp repetitive element consists of 200 to 300 copies in the genome of T. gondii (26).
Real-time PCR.
Three LightCycler real-time PCR assays were evaluated, each using two fluorescence resonance energy transfer (FRET) hybridization probes in PCR product detection and melting curve analysis. Two of the assays were based on the B1 gene, including one (B1-Costa PCR) by Costa et al. (13) and one developed in the present study (B1-Burg PCR). The latter used two newly designed FRET probes (Tox-B1-P1 and Tox-B1-P2) with primers Tox1 and Tox2, which were designed by Burg et al. (6). The third assay (529 PCR), based on the 529-bp repetitive element, was developed by Reischl et al. (45). The sequences of the primers and probes are shown in Table 1.
TABLE 1.
Primers and probes used in the study
| Real-time PCR | Target | Oligonucleotide | Nucleotide sequence (5′-3′) | Amplicon (bp) | Reference or source |
|---|---|---|---|---|---|
| B1-Burg | B1 gene | Primers | Tox1, GGAACTGCATCCGTTCATGAG; Tox2, TCTTTAAAGCGTTCGTGGTC | 193 | Originally by Burg et al. (6); also used by others (5, 25, 32, 37, 46, 48, 52, 55) |
| Probes | Tox-B1-P1, CCTCTGCTGGCGAAAAGTGAAATTCATGAG-fluorescein; Tox-B1-P2, Red 640-ATCTGTGCAACTTTGGTGTATTCGCAGA-phosphate | This study | |||
| B1-Costa | B1 gene | Primers | Forward, GGAGGACTGGCAACCTGGTGTCG; reverse, TTGTTTCACCCGGACCGTTTAGCAG | 126 | Costa et al. (13); also used in reference 43 |
| Probes | CGGAAATAGAAAGCCATGAGGCACTCC-fluorescein, Red 640-ACGGGCGAGTAGCACCTGAGGAGAT-phosphate | Costa et al. (13), also used in reference 43 | |||
| 529 | 529-bp repetitive element | Primers | Tox9, AGGAGAGATATCAGGACTGTAG; Tox11, GCGTCGTCTCGTCTAGATCG | 163 | Reischl et al. (45) |
| Probes | Tox-HP-1, GAGTCGGAGAGGGAGAAGATGTT-fluorescein; Tox-HP-2, Red 640-CCGGCTTGGCTGCTTTTCCTG-phosphate | Reischl et al. (45) |
After standardizations, the final PCR mixture for LightCycler real-time PCR included 2 μl of template DNA, 2 μl of GeneAmp 10× PCR buffer (Applied Biosystems, Foster City, CA), 4 mM MgCl2, 100 μM deoxynucleoside triphosphate (Applied Biosystems), 400 nM primers, 200 nM hybridization probes, 250 μg/μl of nonacetylated bovine serum albumin, and 1 U of Taq polymerase (Promega, Madison, WI) in a total volume of 20 μl. In some assay evaluations (with four replicates), the master hybridization probe kit (Roche Applied Science, Indianapolis, IN) replaced the in-house PCR mix (1× GeneAmp 10× PCR buffer, 100 μM deoxynucleoside triphosphate, and 1 U of Taq polymerase). The real-time PCR consisted of 55 cycles of denaturation at 95°C for 5 s, annealing at 52°C (B1-Burg PCR), 55°C (529 PCR), or 61°C (B1-Costa PCR) for 10 s, and extension at 72°C for 15 s, with an initial denaturation at 95°C for 3 min. The ramp rate for all transitions was 20°C/s.
To determine the sizes of the amplified products, real-time PCR products were harvested by the centrifugation of the inverted LightCycler capillaries. The PCR products were visualized by 1.5% agarose gel electrophoresis using ethidium bromide and UV transillumination.
Conventional PCR.
The B1 and 529 primers also were evaluated by conventional PCR. Each 100 μl of PCR solution contained 2.0 μl of DNA, 500 nM primers, 0.2 mM deoxynucleoside triphosphate (Applied Biosystems), 3.0 mM MgCl, 1× GeneAmp 10× PCR buffer (Applied Biosystems), and 2.5 U of Taq DNA polymerase (Promega). PCR amplification consisted of denaturation at 94°C for 4 min; 35 cycles of 94°C for 45 s, 52°C (B1-Burg primers), 55°C (529 primers), or 61°C (B1-Costa primers) for 45 s, and 72°C for 1 min; and an extension of 72°C for 10 min. PCR products were visualized by 1.5% agarose gel electrophoresis.
DNA sequencing.
After purification by Montage PCR (Millipore, Bedford, MA), selected PCR products were sequenced using an ABI BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) by following manufacturer-suggested procedures. Sequences were read on an ABI3130 genetic analyzer (Applied Biosystems). Sequence accuracy was confirmed by two-directional sequencing. Nucleotide sequences obtained were aligned with reference sequences using the ClustalX 1.81 package (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/).
Nucleotide sequence accession numbers.
Unique nucleotide sequences generated in this study were deposited in the GenBank database under accession numbers FJ656208 and FJ656209.
RESULTS
Development and testing of a B1-based real-time PCR using Burg primers.
A new LightCycler-based PCR assay was developed using published primers Tox1 and Tox2 (6) and two FRET hybridization probes (Tox-B1-P1 and Tox-B1-P2) that were designed in this study. This assay utilized the in-house PCR mix in addition to the primers and probes. In repeated analyses, the real-time PCR amplified all seven DNA preparations that each were extracted from 6 × 106 T. gondii tachyzoites, with threshold cycle (CT) values of 16 or 17 cycles. Melting curve analysis revealed that all PCR products had melting temperatures of 70.1 to 70.7°C. The agarose gel electrophoresis analysis of the real-time PCR products showed the presence of only an expected 193-bp band (data not shown).
Real-time PCR conducted using the Roche LightCycler DNA Master Hyprobe, with premixed PCR buffer, deoxynucleoside triphosphate, and DNA polymerase, resulted in PCR amplifications and CT values similar to those obtained with the in-house PCR mix. The melting temperature of PCR products in real-time PCR was slightly lower when the Roche buffer was used (68.8 to 69.1°C).
Comparison of the performance of three B1- and 529-bp-based real-time PCR assays in the detection of pure T. gondii DNA.
Dilutions of four DNA preparations from T. gondii tachyzoites were used to compare the newly established real-time PCR (B1-Burg PCR) to two previously published LightCycler PCR assays based on the B gene (B1-Costa PCR) (13) and the 529-bp repeat element (529 PCR) (45), using the in-house PCR mix. The new B1 real-time PCR detected all concentrations of T. gondii DNA, including the dilution of 1:78,125, which would represent DNA from approximately 1.5 tachyzoites. One DNA preparation produced amplification at a dilution of 1:390,625, roughly equivalent to the DNA in 0.3 tachyzoite. The CT values increased as the DNA dilutions increased (Table 2).
TABLE 2.
Sensitivity of three real-time PCR assays in the detection of the serial dilution of DNA extracted from T. gondii tachyzoitesa
| Dilution factor | B1-Burg PCRb
|
B1-Costa PCRc
|
529 PCRd
|
Pe | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive rate |
CT
|
Positive rate |
CT
|
Positive rate |
CT
|
|||||
| Mean | SD | Mean | SD | Mean | SD | |||||
| 1 | 4/4 | 16.25 | 0.50 | 4/4 | 17.00 | 0.00 | 4/4 | 13.75 | 0.96 | 1.000 |
| 5 | 4/4 | 18.25 | 0.50 | 4/4 | 20.00 | 0.00 | 4/4 | 16.50 | 1.00 | 1.000 |
| 25 | 4/4 | 20.25 | 0.50 | 4/4 | 22.00 | 0.00 | 4/4 | 18.50 | 1.00 | 1.000 |
| 125 | 4/4 | 22.25 | 0.50 | 4/4 | 24.00 | 0.00 | 4/4 | 20.50 | 1.00 | 1.000 |
| 625 | 4/4 | 24.5 | 0.58 | 4/4 | 26.25 | 0.50 | 4/4 | 22.50 | 1.00 | 1.000 |
| 3,125 | 4/4 | 27.5 | 1.00 | 4/4 | 28.75 | 0.96 | 4/4 | 24.50 | 1.00 | 1.000 |
| 15,625 | 4/4 | 29.5 | 1.00 | 3/4 | 30.33 | 0.58 | 4/4 | 27.00 | 0.82 | 0.336 |
| 78,125 | 4/4 | 32 | 0.82 | 1/4 | 32.00 | 3/4 | 29.00 | 1.00 | 0.072 | |
| 390,625 | 1/4 | 36 | 1/4 | 34.00 | 4/4 | 31.75 | 1.71 | 0.050 | ||
| 1,953,125 | 0/4 | 0/4 | 2/4 | 34.50 | 2.12 | 0.091 | ||||
Four DNA extracts of 6,000,000 tachyzoites were used in the evaluation. Each DNA was eluted with 100 μl of water. The DNA was diluted by fivefold serial dilutions, and 2 μl of the diluted DNA was used in each PCR.
Real-time PCR based on B1 primers by Burg et al. (6) and FRET probes designed in this study.
Real-time PCR based on the B1 gene by Costa et al. (12).
Real-time PCR based on the 529-bp repetitive element by Reischl et al. (45).
P values for differences in detection rates among the three PCR assays were determined by chi-square analysis.
A slightly lower detection threshold was achieved with the B1-Costa PCR. Using the same DNA dilutions, all four DNA preparations were amplified to a dilution of 1:3,125, and the equivalent of DNA extracted from approximately 37.5 tachyzoites/PCR. Lower detection rates were seen at dilutions of 1:15,525 (3/4), 1:78,125 (1/4), and 1:390,625 (1/4), roughly equivalent to the DNA extracted from approximately 7.5, 1.5, and 0.3 tachyzoites/PCR, respectively. There also was good agreement between CT values and DNA dilutions. Most PCR products had melting temperatures of 70.5 to 70.8°C. The differences between B1-Burg PCR and B1-Costa PCR were not significant (P = 0.07 at the dilution of 1:78,125).
The 529 PCR method (45) had the highest sensitivity for the detection of diluted T. gondii tachyzoite DNA. Standard CT curves were generated through a dilution of 1:390,625 for all four DNA preparations. Two of four DNA preparations produced amplification at the dilution of 1:1,953,125 (equivalent to 0.06 tachyzoite/PCR). There was very good correlation between CT values and DNA dilutions (Table 2). The melting temperatures of most PCR products were 67.0 to 67.4°C. The exponential increase in fluorescence signal occurred earlier with all four DNA preparations in the 529 PCR than in the two B1-based PCR assays (CT values of 13 to 15 cycles and 16 to 17 cycles, respectively). The differences in detection rates between the 529 PCR and two B1 PCR assays were significant at the dilution of 1:390,625 (Table 2).
Detection of T. gondii oocysts by real-time PCR assays.
The B1-Burg PCR and B1-Costa PCR were evaluated for the detection of T. gondii oocysts in seeded concentrates from storm water samples, using the in-house mix in PCR. At the 200 and 100 oocysts/pellet levels, three of six samples were amplified by the B1-Burg PCR. The B1-Burg PCR failed to amplify at lower seeding levels, with exceptions at the 25 and 1 oocyst levels, with one positive PCR for each (Table 3). The positive control DNA was amplified in all B1-Burg PCR analyses, with CT values of 16 or 17 cycles (data not shown). PCR products had melting temperatures of 70.9 to 71.9°C.
TABLE 3.
Sensitivity of three real-time PCR assays in the detection of T. gondii oocysts seeded into water sample concentrates
| Estimated no. of oocyst/ 0.5 ml water concentrate | B1-Burg PCRa
|
B1-Costa-PCRb
|
529 PCRc
|
Pd | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive rate |
CT
|
Positive rate |
CT
|
Positive rate |
CT
|
|||||
| Mean | SD | Mean | SD | Mean | SD | |||||
| 1,600 | 6/6 | 28.17 | 0.41 | 6/6 | 29.67 | 0.52 | 6/6 | 24.67 | 0.82 | 1.000 |
| 800 | 6/6 | 30.33 | 0.82 | 6/6 | 30.67 | 0.82 | 6/6 | 26.17 | 0.75 | 1.000 |
| 400 | 6/6 | 32.17 | 1.60 | 6/6 | 31.50 | 1.05 | 6/6 | 27.33 | 0.52 | 1.000 |
| 200 | 3/6 | 32.00 | 0.00 | 6/6 | 32.33 | 0.82 | 6/6 | 28.33 | 0.82 | 0.027 |
| 100 | 3/6 | 34.67 | 1.15 | 4/6 | 32.50 | 0.58 | 6/6 | 29.33 | 0.82 | 0.144 |
| 50 | 0/6 | 1/6 | 34.00 | 6/6 | 30.50 | 1.05 | 0.004 | |||
| 25 | 1/6 | 36.00 | 0/6 | 6/6 | 30.83 | 0.98 | 0.001 | |||
| 10 | 0/6 | 0/6 | 6/6 | 32.17 | 1.33 | 0.0001 | ||||
| 5 | 0/6 | 0/6 | 2/6 | 31.50 | 0.71 | 0.1054 | ||||
| 1 | 1/6 | 36.00 | 1/6 | 33.00 | 3/6 | 32.33 | 2.31 | 0.330 | ||
| 0 | 0/6 | 0/6 | 0/6 | 1.000 | ||||||
Real-time PCR based on B1 primers by Burg et al. (6) and FRET probes designed in this study.
Real-time PCR based on the B1 gene by Costa et al. (12).
Real-time PCR based on the 529-bp repetitive element by Reischl et al. (45).
P values for differences in detection rates among the three PCR assays were determined by chi-square analysis.
Slightly higher sensitivity was achieved with the B1-Costa PCR using the same DNA preparations. All samples seeded with 1,600, 800, 400, and 200 oocysts/0.5 ml pellet generated amplification, with increased CT values as the numbers of seeded oocysts decreased. At 100 and 50 oocysts/pellet, 4/6 and 1/6 samples were positive. Only 1/6 samples seeded at 1 oocyst/pellet generated PCR amplification (Table 3). Differences in sensitivity between the two B1 PCR assays were not significant. PCR products of the B1-Costa PCR had melting temperatures between 70.0 and 71.4°C.
In contrast, the 529 PCR method of Reischl et al. (45) amplified T. gondii DNA in all samples seeded with 1,600, 800, 400, 200, 100, 50, 25, and 10 oocysts/pellet. In samples with 5 and 1 oocyst/pellet, 2/6 and 3/6 samples produced PCR amplification, respectively. Correlation between the number of seeded oocysts and the CT values generated in the 529 PCR was good (Table 3). PCR products had melting temperatures between 66.7 to 67.8°C. The differences in sensitivity between the 529 PCR and two B1 PCR assays were highly significant at 50, 25, and 10 oocysts/0.5 ml pellet (P < 0.01).
Because the 529 PCR had higher sensitivity than the two B1 PCR assays, we further analyzed the performance of the in-house PCR mix with the master hybridization probe buffer. Using DNA preparations from four water concentrates seeded with known numbers of T. gondii oocysts, similar results were obtained by the 529 PCR method with both buffers (P > 0.05). Consistent amplifications of all DNA preparations were obtained when in-house mix was used in the PCR analysis of water sample concentrates seeded with 10 or more oocysts per pellet, whereas master hybridization probe buffer failed to produce amplification in one of four samples seeded with 50 or 10 oocysts/pellet (Table 4). Slightly lower melting temperatures were obtained with PCR products generated with the latter buffer (65.7 to 66.3°C versus 66.7 to 67.8°C).
TABLE 4.
Sensitivity (CT values) of 529 PCR using the Roche ready-to-use buffer (master hybridization probes) and in-house PCR mix in the detection of T. gondii oocysts seeded into storm water concentratesa
| Estimated no. of oocyst/ 0.5 ml water concentrate | Master hybridization probe kit (Roche)
|
In-house mixture
|
P | ||||
|---|---|---|---|---|---|---|---|
| Positive rate |
CT
|
Positive rate |
CT
|
||||
| Mean | SD | Mean | SD | ||||
| 1,600 | 4/4 | 22.75 | 4.27 | 4/4 | 24.75 | 0.96 | 1.000 |
| 800 | 4/4 | 25.25 | 4.57 | 4/4 | 26.25 | 0.96 | 1.000 |
| 400 | 4/4 | 26.00 | 4.00 | 4/4 | 27.25 | 0.50 | 1.000 |
| 200 | 4/4 | 27.75 | 2.87 | 4/4 | 28.25 | 0.96 | 1.000 |
| 100 | 4/4 | 33.25 | 9.84 | 4/4 | 29.25 | 0.96 | 1.000 |
| 50 | 3/4 | 30.00 | 0.00 | 4/4 | 30.25 | 0.96 | 0.500 |
| 25 | 4/4 | 35.00 | 10.03 | 4/4 | 30.75 | 0.96 | 1.000 |
| 10 | 3/4 | 31.33 | 1.15 | 4/4 | 32.25 | 1.71 | 0.500 |
| 5 | 3/4 | 34.67 | 2.31 | 2/4 | 31.50 | 0.71 | 0.500 |
| 1 | 1/4 | 33.00 | — | 2/4 | 33.00 | 2.83 | 0.500 |
| 0 | 0/4 | — | — | 0/4 | — | — | 1.000 |
Dashes indicate no amplification. P values for differences were determined by chi-square analysis.
Evaluation of B1 and 529 PCR primer specificity.
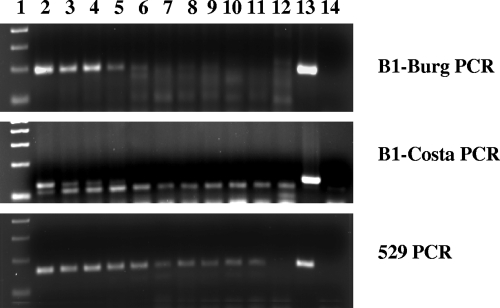
All real-time PCR products generated in the study also were analyzed by agarose gel electrophoresis to confirm the identity of the products generated. When real-time PCR was done with DNA preparations from cultured tachyzoites, all three PCR assays generated PCR products of the expected sizes (126, 163, and 193 bp for the B1-Costa PCR, 529 PCR, and B1-Burg PCR, respectively; data not shown). When PCR templates used DNA preparations from water concentrates seeded with known numbers of T. gondii oocysts, the B1-Burg PCR and 529 PCR generated only the expected band. The B1-Costa PCR, however, produced a band that was slightly smaller than expected in analyses of all DNA preparations from seeded water samples, in addition to the expected band in PCR analyses of DNA preparations from samples seeded with high numbers of T. gondii oocysts (Fig. 1). Several attempts in sequencing the nonspecific B1 PCR products failed due to the presence of underlying signals in the electropherogram.
FIG. 1.
Agarose gel electrophoresis of real-time PCR products of DNA preparations from water concentrates seeded with known numbers of T. gondii oocysts. Real-time PCR was done using the B1-Burg PCR (upper), B1-Costa PCR (middle), and 529 PCR (lower), and the PCR products were detected by conventional agarose gel electrophoresis. The B1-Burg PCR (upper) and 529 PCR (lower) generated only the expected PCR products, with noticeably higher sensitivity by the 529 PCR. In contrast, the B1-Costa PCR (middle) generated a nonspecific band that was slightly smaller than the expected PCR product, in addition to the expected band, in DNA from water concentrates seeded with high numbers of T. gondii oocysts. Lane 1, 100-bp molecular marker ladder; lanes 2 to 12, water concentrates seeded with 1,600, 800, 400, 200, 100, 50, 25, 10, 5, 1, and 0 oocyst/0.5 ml pellet, respectively; lane 13, positive control (DNA from cultured tachyzoites); and lane 14, negative control (no DNA template).
The primers used in all three real-time PCR assays were evaluated by conventional PCR. Using DNA from cultured tachyzoites, both B1 PCR assays generated only PCR products of the expected sizes. In contrast, the 529 PCR produced two bands: the expected product of 163 bp and another near 700 bp in size, even after modifications of concentrations of Mg2+, DNA dilutions, and annealing temperatures in PCR (data not shown). The DNA sequencing of the large PCR product yielded a 675-bp sequence. The alignment of the sequence obtained (FJ656209) with the reference sequence from GenBank (AF146527) showed that the product was from a tandem repeat of the 529-bp repetitive element.
When DNA preparations from seeded water concentrates were used in conventional PCR, the B1-Costa primers generated only the expected band (data not shown). The 529 PCR assay also generated the large and small PCR products of the 529-bp repetitive element for DNA preparations from water concentrates seeded with T. gondii oocysts. In contrast, the B1-Burg primers generated the expected B1 product only in samples seeded with high numbers of oocysts. In some samples, a strong, nonspecific product between 800 to 900 bp was seen (data not shown). One expected and two nonspecific large PCR products were sequenced. The sequence from the expected band was identical to the B1 reference sequence AF179171. The sequences (FJ656208) from the large PCR products were GC-rich (62.8%) and identical to each other but did not match any sequence in GenBank.
DISCUSSION
In this study, three real-time PCR assays based on the B1 gene and 529-bp repetitive element were tested for their sensitivity in detecting T. gondii oocysts in concentrates of surface water samples. These genes were selected because of their frequent use for the detection of T. gondii in clinical specimens and the high copy number of the targets (7, 20, 34, 45). The assays tested included two published LightCycler real-time PCR techniques based on these genes (13, 45) and a new LightCycler real-time PCR developed using the B1 primers designed by Burg et al. (6). The B1 primers by Burg et al. are the most widely reported primer set for the PCR detection of T. gondii (25, 32, 37, 46, 48, 52, 55). These primers amplify a 193-bp fragment, which allowed for the development of a LightCycler real-time PCR assay using two newly designed FRET probes (Table 1). This real-time PCR format was used because of its popularity in the analysis of clinical specimens (5, 7, 10-13, 20, 24, 29, 37, 38, 41, 43, 45), the time-saving nature, and the potential for quantitation (4). FRET hybridization probes were used instead of SYBR green in the detection step of the PCR assay to increase sensitivity and specificity (19, 43).
Results of the present study confirm the higher sensitivity of real-time PCR assays based on the 529-bp repetitive element compared to those based on the B1 gene (7, 8, 20, 24, 45). This likely was due to the difference in copy numbers of the two targets, as reflected by the results of analyses of pure tachyzoite DNA preparations (Table 2). The detection limit of <1.5 tachyzoites/PCR by the B1-Burg PCR is comparable to those (1 to 3 tachyzoites/PCR) of other TaqMan (33) and LightCycler (4) real-time PCR assays based on the B1 gene. The detection limit of the B1-Costa PCR was slightly higher (8 tachyzoites/PCR) but was similar to that (10 tachyzoites/PCR) of another LightCycler PCR that also is based on the Burg primers (5). The 529 PCR method was much more sensitive, with a detection limit of 0.06 to 0.3 tachyzoite/PCR, which exceeded the 0.5 tachyzoite/PCR limit of another 529-based LightCycler PCR (7).
The two B1-based real-time PCR assays were less sensitive in the detection of T. gondii oocysts seeded into water concentrates than the 529 PCR. This difference was not fully explained by the difference in copy numbers of the targets (35 versus 200 to 300 copies). One possible reason for the lower sensitivity of B1-based PCR is the nonspecificity of the primers. Both the Burg primers (6) and the Costa et al. primers (13) amplified nontarget DNA. Even though the B1 gene remains the most commonly used PCR target in the analysis of T. gondii DNA in clinical specimens (4, 5, 7-11, 20-22, 28, 33, 39, 40, 45, 47, 48, 50-52, 56) and performed well in the analysis of tachyzoites in this study, its use in the PCR detection of T. gondii oocysts of environmental samples probably is limited because of the nonspecificity of some primers and lower sensitivity.
Results of this study also indicate that the 529-bp repetitive element is a useful PCR target for the detection of T. gondii oocysts in water, with sensitivity comparable to that obtained with pure tachyzoite DNA. This finding indicates that inhibition associated with the PCR analysis of water samples was not a contributing factor for the poor sensitivity of the B1 PCR assays. One potential problem with the 529 PCR is the presence of tandem repeats in the target sequence, which could reduce the sensitivity of detection because of primer competition and the generation of multiple products. In the present study, multiple PCR products seen in the regular PCR analysis of the gene were absent in the agarose electrophoresis of real-time PCR products. The reason for the difference is not clear. Perhaps the short extension duration in the real-time PCR prevented the efficient amplification of the tandem repeats.
Results of the study suggest that the 529 PCR, originally developed for the analysis of clinical specimens (45), can be used effectively in the detection of T. gondii oocysts in water samples when the FastDNA extraction kit for soil is used in DNA extraction and high concentrations of nonacetylated bovine serum albumin are used in PCR. It is, however, important to keep in mind that the detection limit was based on the serial dilution of a hemocytometer-counted suspension and could be inaccurate. Previously, the performance of conventional or nested PCR in the detection of T. gondii oocysts was evaluated in several studies, and the detection limits varied greatly between assays and studies. The sensitivity of an 18S rRNA nested PCR was reported to be 0.1 oocyst for distilled water (35), 100 oocysts/sample for river water, and 10 oocysts/sample for well and sea water (36). Similarly, the sensitivity of a B1-based TaqMan end point assay was more than 10 oocysts/liter for drinking water and more than 1,000 oocysts/liter for raw surface water (55). Even lower sensitivity (about 50 oocysts per 50 μl of PCR mixture) was seen for river water analyzed by a B1-based PCR hybridization immunoassay (46). PCR inhibition probably was a major factor responsible for the difference in reported sensitivity (55). The DNA extraction method used in this study, the FastDNA extraction kit for soil samples, previously was shown to be better in removing PCR inhibitors than other commercial kits (30).
In conclusion, real-time PCR assays, especially the 529 PCR method, were highly effective in detecting T. gondii oocysts seeded in concentrates of stream water samples. Before the technique can be applied widely, the further standardization of procedures, including the testing of seeded water samples and field samples, and interlaboratory comparisons are needed. Nevertheless, this new approach provides an alternative to the conventional mouse inoculation assay or microscopy.
Acknowledgments
We thank Ynes Ortega of the University of Georgia and J. P. Dubey of the U.S. Department of Agriculture for providing T. gondii tachyzoites and oocysts. We also thank Kerri A. Alderisio of the New York City Department of Environmental Protection for providing packed pellets from storm water samples.
This study was supported in part by funds from the EPA and the National Oceanic and Atmospheric Administration Oceans and Human Health Initiative.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Bahia-Oliveira, L. M., J. L. Jones, J. Azevedo-Silva, C. C. Alves, F. Orefice, and D. G. Addiss. 2003. Highly endemic, waterborne toxoplasmosis in north Rio de Janeiro state, Brazil. Emerg. Infect. Dis. 9:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benenson, M. W., E. T. Takafuji, S. M. Lemon, R. L. Greenup, and A. J. Sulzer. 1982. Oocyst-transmitted toxoplasmosis associated with ingestion of contaminated water. N. Engl. J. Med. 307:666-669. [DOI] [PubMed] [Google Scholar]
- 3.Bowie, W. R., A. S. King, D. H. Werker, J. L. Isaac-Renton, A. Bell, S. B. Eng, S. A. Marion, et al. 1997. Outbreak of toxoplasmosis associated with municipal drinking water. Lancet 350:173-177. [DOI] [PubMed] [Google Scholar]
- 4.Brenier-Pinchart, M. P., V. Morand-Bui, H. Fricker-Hidalgo, V. Equy, R. Marlu, and H. Pelloux. 2007. Adapting a conventional PCR assay for Toxoplasma gondii detection to real-time quantitative PCR including a competitive internal control. Parasite 14:149-154. [DOI] [PubMed] [Google Scholar]
- 5.Buchbinder, S., R. Blatz, and A. C. Rodloff. 2003. Comparison of real-time PCR detection methods for B1 and P30 genes of Toxoplasma gondii. Diagn. Microbiol. Infect. Dis. 45:269-271. [DOI] [PubMed] [Google Scholar]
- 6.Burg, J. L., C. M. Grover, P. Pouletty, and J. C. Boothroyd. 1989. Direct and sensitive detection of a pathogenic protozoan, Toxoplasma gondii, by polymerase chain reaction. J. Clin. Microbiol. 27:1787-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderaro, A., G. Piccolo, C. Gorrini, S. Peruzzi, L. Zerbini, S. Bommezzadri, G. Dettori, and C. Chezzi. 2006. Comparison between two real-time PCR assays and a nested-PCR for the detection of Toxoplasma gondii. Acta Biomed. 77:75-80. [PubMed] [Google Scholar]
- 8.Cassaing, S., M. H. Bessieres, A. Berry, A. Berrebi, R. Fabre, and J. F. Magnaval. 2006. Comparison between two amplification sets for molecular diagnosis of toxoplasmosis by real-time PCR. J. Clin. Microbiol. 44:720-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cermakova, Z., O. Ryskova, and L. Pliskova. 2005. Polymerase chain reaction for detection of Toxoplasma gondii in human biological samples. Folia Microbiol. 50:341-344. [DOI] [PubMed] [Google Scholar]
- 10.Contini, C., M. Giuliodori, R. Cultrera, and S. Seraceni. 2006. Detection of clinical-stage specific molecular Toxoplasma gondii gene patterns in patients with toxoplasmic lymphadenitis. J. Med. Microbiol. 55:771-774. [DOI] [PubMed] [Google Scholar]
- 11.Contini, C., S. Seraceni, R. Cultrera, C. Incorvaia, A. Sebastiani, and S. Picot. 2005. Evaluation of a real-time PCR-based assay using the LightCycler system for detection of Toxoplasma gondii bradyzoite genes in blood specimens from patients with toxoplasmic retinochoroiditis. Int. J. Parasitol. 35:275-283. [DOI] [PubMed] [Google Scholar]
- 12.Costa, J. M., P. Ernault, E. Gautier, and S. Bretagne. 2001. Prenatal diagnosis of congenital toxoplasmosis by duplex real-time PCR using fluorescence resonance energy transfer hybridization probes. Prenat. Diagn. 21:85-88. [DOI] [PubMed] [Google Scholar]
- 13.Costa, J. M., C. Pautas, P. Ernault, F. Foulet, C. Cordonnier, and S. Bretagne. 2000. Real-time PCR for diagnosis and follow-up of Toxoplasma reactivation after allogeneic stem cell transplantation using fluorescence resonance energy transfer hybridization probes. J. Clin. Microbiol. 38:2929-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Moura, L., L. M. Bahia-Oliveira, M. Y. Wada, J. L. Jones, S. H. Tuboi, E. H. Carmo, W. M. Ramalho, N. J. Camargo, R. Trevisan, R. M. Graca, A. J. da Silva, I. Moura, J. P. Dubey, and D. O. Garrett. 2006. Waterborne toxoplasmosis, Brazil, from field to gene. Emerg. Infect. Dis. 12:326-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubey, J. P. 2004. Toxoplasmosis—a waterborne zoonosis. Vet. Parasitol. 126:57-72. [DOI] [PubMed] [Google Scholar]
- 16.Dumètre, A., and M. L. Darde. 2007. Detection of Toxoplasma gondii in water by an immunomagnetic separation method targeting the sporocysts. Parasitol. Res. 101:989-996. [DOI] [PubMed] [Google Scholar]
- 17.Dumètre, A., and M. L. Darde. 2003. How to detect Toxoplasma gondii oocysts in environmental samples? FEMS Microbiol. Rev. 27:651-661. [DOI] [PubMed] [Google Scholar]
- 18.Dumètre, A., and M. L. Darde. 2005. Immunomagnetic separation of Toxoplasma gondii oocysts using a monoclonal antibody directed against the oocyst wall. J. Microbiol. Methods 61:209-217. [DOI] [PubMed] [Google Scholar]
- 19.Edvinsson, B., S. Jalal, C. E. Nord, B. S. Pedersen, and B. Evengard. 2004. DNA extraction and PCR assays for detection of Toxoplasma gondii. APMIS 112:342-348. [DOI] [PubMed] [Google Scholar]
- 20.Edvinsson, B., M. Lappalainen, and B. Evengard. 2006. Real-time PCR targeting a 529-bp repeat element for diagnosis of toxoplasmosis. Clin. Microbiol. Infect. 12:131-136. [DOI] [PubMed] [Google Scholar]
- 21.Filisetti, D., M. Gorcii, E. Pernot-Marino, O. Villard, and E. Candolfi. 2003. Diagnosis of congenital toxoplasmosis: comparison of targets for detection of Toxoplasma gondii by PCR. J. Clin. Microbiol. 41:4826-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto, M., T. Takahashi, T. Kanda, and A. Iwamoto. 2004. Detection of Toxoplasma gondii by polymerase chain reaction in cerebrospinal fluid from human immunodeficiency virus-1-infected Japanese patients with focal neurological signs. J. Int. Med. Res. 32:665-670. [DOI] [PubMed] [Google Scholar]
- 23.Heukelbach, J., V. Meyer-Cirkel, R. C. Moura, M. Gomide, J. A. Queiroz, P. Saweljew, and O. Liesenfeld. 2007. Waterborne toxoplasmosis, northeastern Brazil. Emerg. Infect. Dis. 13:287-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hierl, T., U. Reischl, P. Lang, H. Hebart, M. Stark, P. Kyme, and I. B. Autenrieth. 2004. Preliminary evaluation of one conventional nested and two real-time PCR assays for the detection of Toxoplasma gondii in immunocompromised patients. J. Med. Microbiol. 53:629-632. [DOI] [PubMed] [Google Scholar]
- 25.Hill, D. E., S. Chirukandoth, J. P. Dubey, J. K. Lunney, and H. R. Gamble. 2006. Comparison of detection methods for Toxoplasma gondii in naturally and experimentally infected swine. Vet. Parasitol. 141:9-17. [DOI] [PubMed] [Google Scholar]
- 26.Homan, W. L., M. Vercammen, J. De Braekeleer, and H. Verschueren. 2000. Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int. J. Parasitol. 30:69-75. [DOI] [PubMed] [Google Scholar]
- 27.Isaac-Renton, J., W. R. Bowie, A. King, G. S. Irwin, C. S. Ong, C. P. Fung, M. O. Shokeir, and J. P. Dubey. 1998. Detection of Toxoplasma gondii oocysts in drinking water. Appl. Environ. Microbiol. 64:2278-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jalal, S., C. E. Nord, M. Lappalainen, and B. Evengard. 2004. Rapid and sensitive diagnosis of Toxoplasma gondii infections by PCR. Clin. Microbiol. Infect. 10:937-939. [DOI] [PubMed] [Google Scholar]
- 29.Jauregui, L. H., J. Higgins, D. Zarlenga, J. P. Dubey, and J. K. Lunney. 2001. Development of a real-time PCR assay for detection of Toxoplasma gondii in pig and mouse tissues. J. Clin. Microbiol. 39:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, J., K. A. Alderisio, A. Singh, and L. Xiao. 2005. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ. Microbiol. 71:1135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang, J., K. A. Alderisio, and L. Xiao. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71:4446-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, C. D., N. Okhravi, P. Adamson, S. Tasker, and S. Lightman. 2000. Comparison of PCR detection methods for B1, P30, and 18S rDNA genes of T. gondii in aqueous humor. Investig. Ophthalmol. Vis. Sci. 41:634-644. [PubMed] [Google Scholar]
- 33.Joss, A. W., R. Evans, S. Mavin, J. Chatterton, and D. Ho-Yen. 2008. The development of real time PCR to detect Toxoplasma gondii and Borrelia burgdorferi infections in postal samples. J. Clin. Pathol. 61:221-224. [DOI] [PubMed] [Google Scholar]
- 34.Kompalic-Cristo, A., C. Frotta, M. Suarez-Mutis, O. Fernandes, and C. Britto. 2007. Evaluation of a real-time PCR assay based on the repetitive B1 gene for the detection of Toxoplasma gondii in human peripheral blood. Parasitol. Res. 101:619-625. [DOI] [PubMed] [Google Scholar]
- 35.Kourenti, C., and P. Karanis. 2004. Development of a sensitive polymerase chain reaction method for the detection of Toxoplasma gondii in water. Water Sci. Technol. 50:287-291. [PubMed] [Google Scholar]
- 36.Kourenti, C., and P. Karanis. 2006. Evaluation and applicability of a purification method coupled with nested PCR for the detection of Toxoplasma oocysts in water. Lett. Appl. Microbiol. 43:475-481. [DOI] [PubMed] [Google Scholar]
- 37.Lin, M. H., T. C. Chen, T. T. Kuo, C. C. Tseng, and C. P. Tseng. 2000. Real-time PCR for quantitative detection of Toxoplasma gondii. J. Clin. Microbiol. 38:4121-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maaroufi, Y., J. M. de Bruyne, V. Duchateau, R. Scheen, and F. Crokaert. 2006. Development of a multiple internal control for clinical diagnostic real-time amplification assays. FEMS Immunol. Med. Microbiol. 48:183-191. [DOI] [PubMed] [Google Scholar]
- 39.Mahalakshmi, B., K. L. Therese, H. N. Madhavan, and J. Biswas. 2006. Diagnostic value of specific local antibody production and nucleic acid amplification technique-nested polymerase chain reaction (nPCR) in clinically suspected ocular toxoplasmosis. Ocul. Immunol. Inflamm. 14:105-112. [DOI] [PubMed] [Google Scholar]
- 40.Mahittikorn, A., H. Wickert, and Y. Sukthana. 2005. Comparison of five DNA extraction methods and optimization of a b1 gene nested PCR (nPCR) for detection of Toxoplasma gondii tissue cyst in mouse brain. Southeast Asian J. Trop. Med. Public Health 36:1377-1382. [PubMed] [Google Scholar]
- 41.Menotti, J., G. Vilela, S. Romand, Y. J. Garin, L. Ades, E. Gluckman, F. Derouin, and P. Ribaud. 2003. Comparison of PCR-enzyme-linked immunosorbent assay and real-time PCR assay for diagnosis of an unusual case of cerebral toxoplasmosis in a stem cell transplant recipient. J. Clin. Microbiol. 41:5313-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montoya, J. G., and O. Liesenfeld. 2004. Toxoplasmosis. Lancet 363:1965-1976. [DOI] [PubMed] [Google Scholar]
- 43.Nagy, B., Z. Ban, A. Beke, G. R. Nagy, L. Lazar, C. Papp, E. Toth-Pal, and Z. Papp. 2006. Detection of Toxoplasma gondii from amniotic fluid, a comparison of four different molecular biological methods. Clin. Chim. Acta 368:131-137. [DOI] [PubMed] [Google Scholar]
- 44.Palanisamy, M., B. Madhavan, M. B. Balasundaram, R. Andavar, and N. Venkatapathy. 2006. Outbreak of ocular toxoplasmosis in Coimbatore, India. Indian J. Ophthalmol. 54:129-131. [DOI] [PubMed] [Google Scholar]
- 45.Reischl, U., S. Bretagne, D. Kruger, P. Ernault, and J. M. Costa. 2003. Comparison of two DNA targets for the diagnosis of toxoplasmosis by real-time PCR using fluorescence resonance energy transfer hybridization probes. BMC Infect. Dis. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwab, K. J., and J. J. McDevitt. 2003. Development of a PCR-enzyme immunoassay oligoprobe detection method for Toxoplasma gondii oocysts, incorporating PCR controls. Appl. Environ. Microbiol. 69:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smielewska-Loœ, E. 2003. Comparison of various primer sets for detection of Toxoplasma gondii by polymerase chain reaction in fetal tissues from naturally aborted foxes. Pol. J. Vet. Sci. 6:229-234. [PubMed] [Google Scholar]
- 48.Sreekumar, C., J. R. Rao, A. K. Mishra, D. Ray, P. Joshi, and R. K. Singh. 2004. Detection of toxoplasmosis in experimentally infected goats by PCR. Vet. Rec. 154:632-635. [DOI] [PubMed] [Google Scholar]
- 49.Sroka, J., A. Wojcik-Fatla, and J. Dutkiewicz. 2006. Occurrence of Toxoplasma gondii in water from wells located on farms. Ann. Agric. Environ. Med. 13:169-175. [PubMed] [Google Scholar]
- 50.Switaj, K., A. Master, M. Skrzypczak, and P. Zaborowski. 2005. Recent trends in molecular diagnostics for Toxoplasma gondii infections. Clin. Microbiol. Infect. 11:170-176. [DOI] [PubMed] [Google Scholar]
- 51.Szénási, Z., K. Horvath, E. Sarkany, and M. Melles. 2005. Toxoplasmosis surveillance during pregnancy and quality assurance of methods in Hungary. Wien. Klin. Wochenschr. 117(Suppl. 4):29-34. [DOI] [PubMed] [Google Scholar]
- 52.Terra, M. A., A. R. Bello, O. M. Bastos, M. R. Amendoeira, J. M. Coelho, L. F. Ferreira, and A. Araujo. 2004. Detection of Toxoplasma gondii DNA by polymerase chain reaction in experimentally desiccated tissues. Mem. Inst. Oswaldo Cruz 99:185-188. [DOI] [PubMed] [Google Scholar]
- 53.U.S. Environmental Protection Agency. 1995. ICR protozoan method for detecting Giardia cysts and Cryptosporidium oocysts in water by a fluorescent antibody procedure. EPA/814-B-95-003. Office of Ground Water and Drinking Water, U.S. Environmental Protection Agency, Washington, DC.
- 54.U.S. Environmental Protection Agency. 2001. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 821-R-01-025. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 55.Villena, I., D. Aubert, P. Gomis, H. Ferte, J. C. Inglard, H. Denis-Bisiaux, J. M. Dondon, E. Pisano, N. Ortis, and J. M. Pinon. 2004. Evaluation of a strategy for Toxoplasma gondii oocyst detection in water. Appl. Environ. Microbiol. 70:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiengcharoen, J. T., R. Chiabchalard, and Y. Sukthana. 2004. PCR technique for detecting Toxoplasma gondii in animal amniotic fluid. Southeast Asian J. Trop. Med. Public Health 35:792-795. [PubMed] [Google Scholar]
- 57.Xiao, L., K. Alderisio, J. Limor, M. Royer, and A. A. Lal. 2000. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66:5492-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zarlenga, D. S., and J. M. Trout. 2004. Concentrating, purifying and detecting waterborne parasites. Vet. Parasitol. 126:195-217. [DOI] [PubMed] [Google Scholar]