Abstract
The first gene cassettes of integrons are involved in the last adaptation response to changing conditions and are also the most expressed. We propose a rapid method for the selection of clones carrying an integron first gene cassette that is useful for finding adaptive genes in environmental metagenomic libraries.
Integrons, genetic elements discovered in clinical environments in 1989 (26), are known to carry gene cassettes encoding adaptive proteins in different environmental contexts (17, 20); environmental pressures may thus favor the propagation of cassettes conferring a selective advantage (21, 29). Integrons contain (Fig. 1) an integrase gene, intI (6, 7, 18); a recombination site, attI (23); a set of gene cassettes formed by a coding sequence and a recombination site, attC (14, 19); and one or two promoters, allowing gene cassette expression (4, 16). Different classes of integrons were defined according to the intI gene diversity. They were found in metagenomes from various environments (9, 15, 22). New metagenomic studies always discover new integron classes, showing the importance and the diversity of such genetic elements (9, 22).
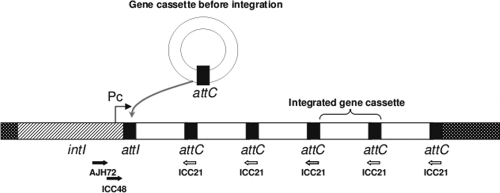
FIG. 1.
Structure of integrons and positions of the primers used. The intI gene encodes an enzyme, allowing the integration of new gene cassettes at the attI recombination site. Thus, the first gene cassette is the last one integrated. Gene cassettes are formed by a recombination site, attC, and a coding sequence. The promoter Pc allows gene cassette expression. The positions of primers AJH72, ICC21, and ICC48 used in this study are indicated.
As integrons are involved in bacterial adaptation, study of integrons would allow the finding of adaptive genes in metagenomes. But the detection of such genes among the huge abundance of gene cassettes associated with integrons is a challenge. The integration of a new gene cassette, catalyzed by the integrase, occurs by recombination between the attC site and the attI site of the integron (6, 8) (Fig. 1). The first gene cassette of an integron is, therefore, the last one integrated. As it is the closest gene to the promoter, its expression level is the highest in the integron (4). Thus, this gene cassette is a good target to find new adaptive genes in metagenomes. To amplify the first gene cassettes, a forward primer targeting the intI gene or attI site must be used. In previous studies, the determinations of gene cassette collection from environmental metagenomes did not target first gene cassettes, since they were performed by PCR methods targeting attC sites. Thus, we propose a method to construct gene cassette libraries enriched with first gene cassettes and an associated screening method for the clone selection.
The method was developed by using DNA from Xanthomonas campestris ATCC 33913T, a bacterial strain carrying an integron. DNA was extracted according to the work of Goñi-Urriza et al. (12). Coastal sediments maintained in the laboratory were used to validate this method. Total DNA (metagenomic) was extracted 1 week after addition of oil using the UltraClean soil DNA isolation kit (Mo Bio Laboratories), as previously described (24). PCR amplification, targeting the integrase gene intI in the forward direction (because attI sites are not well conserved enough to allow the good design of a primer) and the attC site in reverse, to amplify integron first gene cassettes was performed (Fig. 1). Forward primer AJH72 (10) was used for PCR of X. campestris DNA, and primer ICC48 (intB-inverted primer [25]), targeting the class 1 integron intI, was used for PCR of sediment metagenome. As the intI1 genes from environmental contexts exhibit considerable sequence diversity (11), ICC48 does not cover the entire spectrum of known intI1 genes but was chosen for its proximity to the attI site. Many primers used in previous studies targeting attC sites, such as HS286 (27), were unsuccessfully tested in the studied metagenome. Thus, ICC21, a less-degenerated primer, was designed to target the attC sites from class 1 and 2 integrons with the following sequence: 5′-GTCGGCTTGRAYGAATTGTTAGRC-3′. The PCR mixture contained DNA, 1× PCR buffer, 200 μM of each dNTP (deoxynucleoside triphosphate), 1.5 mM MgCl2, 0.2 μM of each primer, and 5 U of Taq DNA polymerase (Eurobio). The PCRs consisted of 95°C for 10 min, 40 cycles of amplification (95°C for 45 s, 52°C or 51°C for 45 s, 72°C for 1.5 min), and 72°C for 10 min. PCR products were gel purified with the GFX PCR DNA and gel band purification kit (GE Healthcare). Purified products were cloned with the TOPO TA cloning kit (Invitrogen). Clones carrying first gene cassettes were selected by colony PCR. In order to minimize time spent on and the number of PCRs, the following three primers were concomitantly used: the two TOPO TA M13 primers and the primer targeting the intI gene used in the previous PCR (AJH72 or ICC48), and this primer was fluorescently labeled by HEX (6-carboxyhexafluorescein). PCR products were separated by gel electrophoresis, and the fluorescent DNA fragments were detected with a Typhoon 9200 scanner (Amersham). The selected inserts were sequenced by using the BigDye terminator v1.1 cycle sequencing kit (Applied Biosystems). Sequences were analyzed with ORF Finder (28), BLAST (1), and ProDom (3) algorithms.
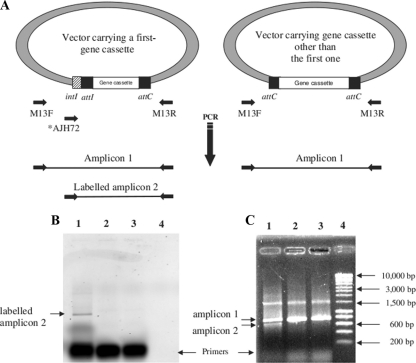
The X. campestris (ATCC 33913T) integron possesses 23 gene cassettes (10). Different concentrations of primers were tested to amplify the first gene cassette, but in all cases, several amplified fragments were obtained. Sequence analyses revealed that most of them were gene cassettes other than the first one, and in these cases, the reverse primer was also used in the forward direction. The particular structure of the attC site with inverted repeat sequences, allowing the attC primer to anneal with both strands, may explain this result. As there was no other way to amplify the first gene cassettes, their selection could not be performed by PCR only. Because sequencing all clones would represent too much work when studying metagenomes, a triplex PCR screening method was developed (Fig. 2A). Since the forward primer sequence is found only in the fragment containing the first gene cassette, the corresponding clones produce two PCR products, with one that is labeled (Fig. 2). Sequence-labeled inserts confirmed that the integron first gene cassette of X. campestris was selected. As a control, the sequences of unlabeled inserts showed that they contained integron gene cassettes but not the first gene cassettes.
FIG. 2.
Screening strategy for first gene cassette inserts. (A) Schematic of the clone library PCR screening strategy. The inserts are amplified by PCR using three primers. Only inserts carrying a first gene cassette lead to labeled amplified fragments. (B, C) Gel electrophoresis of insert PCR products from clones of X. campestris gene cassettes. (1) Insert carrying the integron first gene cassette of X. campestris; (2, 3) inserts carrying integron gene cassettes of X. campestris other than the first gene cassette; (4) molecular weight marker (SmartLadder; Eurogentec). (B) Detection of fluorescence; (C) detection of all fragments by ethidium bromide staining of the same gel.
This method was then applied to coastal mud metagenomes, in which we focused on class 1 integrons, most commonly involved in adaptive responses (13). After PCR products were cloned, among 100 clones screened, 23 fluorescent fragments were detected and sequenced. As the primer targeting intI binds at the beginning of the gene, it was nearly impossible to recognize the intI sequence, except that the intI gene is longer at the 5′ end. On the other side of the sequences, a part of the attC site must be present. Sequence analysis revealed that some fragments showed similarities to characteristic class 1 or 2 integron attC sites, but these sites could not be found in each case because of their large variability (5). A total of 29 open reading frames (ORF) were characterized as potentially transcribed by an integron promoter, but for 16 of the ORF, no similarity with any known amino acid sequences could be found. The 13 other ORF exhibited less than 40% similarity with known sequences, and no putative conserved domains were found. These observations are in accordance with previous studies showing that most of the environmental integron gene cassettes code for proteins with unknown functions (2, 17).
The first-gene cassettes of integrons appear to be good candidates to find gene cassettes, which aid bacteria in effecting a rapid adaptive response. We are now able to reveal integron last gene acquisitions of environmental bacterial communities submitted to stressful conditions. This method presents two limiting steps when working with metagenomes, as follows. (i) The primers are critical to cover the largest number of integrons. In this study, we targeted class 1 integrons because they are known to be mobile and to carry adaptive genes (29). (ii) When fragments with a large size disparity are cloned, the smallest fragments are preferentially cloned. In order to obtain a complete library with metagenomes, the cloning should be performed after fragment size separation. The PCR method combined with the screening method leads to 100% of clones carrying a first gene cassette. Thus, this new method allows the focus to be on spreading first gene cassettes in metagenomes after a specific stress.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the EMBL database with the following assigned accession numbers: FM210532 to FM210535 and FM210665 to FM210679.
Acknowledgments
This work was supported by the Aquitaine Regional Government Council (France) and the ANR/SEST (DHYVA project 06SEST09). L.H. was supported partly by a doctoral grant from the Ministère de l'Enseignement Supérieur et de la Recherche (France).
We thank all partners of the DHYVA project for their useful discussions. We particularly thank Magalie Stauffert and Laurène Fito-Boncompte for technical support. Sequence electrophoreses were performed at the Genotyping and Sequencing Facility of Bordeaux.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Boucher, Y., M. Labbate, J. E. Koenig, and H. W. Stokes. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 15:301-309. [DOI] [PubMed] [Google Scholar]
- 3.Bru, C., E. Courcelle, S. Carrere, Y. Beausse, S. Dalmar, and D. Kahn. 2005. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 33:D212-D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis, C. M., M. J. Kim, H. W. Stokes, and R. M. Hall. 1998. Binding of the purified integron DNA integrase Intl1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 6.Collis, C. M., M. J. Kim, H. W. Stokes, and R. M. Hall. 2002. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol. Microbiol. 46:1415-1427. [DOI] [PubMed] [Google Scholar]
- 7.Collis, C. M., G. D. Recchia, M. J. Kim, H. W. Stokes, and R. M. Hall. 2001. Efficiency of recombination reactions catalyzed by class 1 integron integrase intI1. J. Bacteriol. 183:2535-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demarre, G., C. Frumerie, D. N. Gopaul, and D. Mazel. 2007. Identification of key structural determinants of the IntI1 integron integrase that influence attC × attI1 recombination efficiency. Nucleic Acids Res. 35:6475-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsaied, H., H. W. Stokes, T. Nakamura, K. Kitamura, H. Fuse, and A. Maruyama. 2007. Novel and diverse integron integrase genes and integron-like gene cassettes are prevalent in deep-sea hydrothermal vents. Environ. Microbiol. 9:2298-2312. [DOI] [PubMed] [Google Scholar]
- 10.Gillings, M. R., M. P. Holley, H. W. Stokes, and A. J. Holmes. 2005. Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl. Acad. Sci. USA 102:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillings, M. R., S. Krishnan, P. J. Worden, and S. A. Hardwick. 2008. Recovery of diverse genes for class 1 integron-integrases from environmental DNA samples. FEMS Microbiol. Lett. 287:56-62. [DOI] [PubMed] [Google Scholar]
- 12.Goñi-Urriza, M., C. Arpin, M. Capdepuy, V. Dubois, P. Caumette, and C. Quentin. 2002. Type II topoisomerase quinolone resistance-determining regions of Aeromonas caviae, A. hydrophila, and A. sobria complexes and mutations associated with quinolone resistance. Antimicrob. Agents Chemother. 46:350-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall, R. M. 1997. Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found. Symp. 207:192-202; discussion, 202-205. [DOI] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, A. J., M. R. Gillings, B. S. Nield, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2003. The gene cassette metagenome is a basic resource for bacterial genome evolution. Environ. Microbiol. 5:383-394. [DOI] [PubMed] [Google Scholar]
- 16.Kim, T. E., H. J. Kwon, S. H. Cho, S. Kim, B. K. Lee, H. S. Yoo, Y. H. Park, and S. J. Kim. 2007. Molecular differentiation of common promoters in Salmonella class 1 integrons. J. Microbiol. Methods 68:453-457. [DOI] [PubMed] [Google Scholar]
- 17.Koenig, J. E., Y. Boucher, R. L. Charlebois, C. Nesbo, O. Zhaxybayeva, E. Bapteste, M. Spencer, M. J. Joss, H. W. Stokes, and W. F. Doolittle. 2008. Integron-associated gene cassettes in Halifax Harbour: assessment of a mobile gene pool in marine sediments. Environ. Microbiol. 10:1024-1038. [DOI] [PubMed] [Google Scholar]
- 18.MacDonald, D., G. Demarre, M. Bouvier, D. Mazel, and D. N. Gopaul. 2006. Structural basis for broad DNA-specificity in integron recombination. Nature 440:1157-1162. [DOI] [PubMed] [Google Scholar]
- 19.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 20.Michael, C. A., M. R. Gillings, A. J. Holmes, L. Hughes, N. R. Andrew, M. P. Holley, and H. W. Stokes. 2004. Mobile gene cassettes: a fundamental resource for bacterial evolution. Am. Nat. 164:1-12. [DOI] [PubMed] [Google Scholar]
- 21.Nemergut, D. R., A. P. Martin, and S. K. Schmidt. 2004. Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution. Appl. Environ. Microbiol. 70:1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nield, B. S., A. J. Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 23.Partridge, S. R., G. D. Recchia, C. Scaramuzzi, C. M. Collis, H. W. Stokes, and R. M. Hall. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855-2864. [DOI] [PubMed] [Google Scholar]
- 24.Precigou, S., P. Goulas, and R. Duran. 2001. Rapid and specific identification of nitrile hydratase (NHase)-encoding genes in soil samples by polymerase chain reaction. FEMS Microbiol. Lett. 204:155-161. [DOI] [PubMed] [Google Scholar]
- 25.Rosser, S. J., and H. K. Young. 1999. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J. Antimicrob. Chemother. 44:11-18. [DOI] [PubMed] [Google Scholar]
- 26.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 27.Stokes, H. W., A. J. Holmes, B. S. Nield, M. P. Holley, K. M. Nevalainen, B. C. Mabbutt, and M. R. Gillings. 2001. Gene cassette PCR: sequence-independent recovery of entire genes from environmental DNA. Appl. Environ. Microbiol. 67:5240-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wheeler, D. L., D. M. Church, R. Edgar, S. Federhen, W. Helmberg, T. L. Madden, J. U. Pontius, G. D. Schuler, L. M. Schriml, E. Sequeira, T. O. Suzek, T. A. Tatusova, and L. Wagner. 2004. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 32:D35-D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright, M. S., C. Baker-Austin, A. H. Lindell, R. Stepanauskas, H. W. Stokes, and J. V. McArthur. 2008. Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J. 2:417-428. [DOI] [PubMed] [Google Scholar]