Abstract
A hollow-fiber membrane chamber (HFMC) was developed as an in situ cultivation device for environmental microorganisms. The HFMC system consists of 48 to 96 pieces of porous hollow-fiber membrane connected with injectors. The system allows rapid exchange of chemical compounds, thereby simulating a natural environment. Comparative analysis through the cultivation of three types of environmental samples was performed using this newly designed device and a conventional agar-based petri dish. The results show that the ratios of novel phylotypes in isolates, species-level diversities, and cultivabilities in HFMC-based cultivation are higher than those in an agar-based petri dish for all three samples, suggesting that the new in situ cultivation device is effective for cultivation of various environmental microorganisms.
Although highly diverse untapped microbial consortia exist in natural environments, it is generally recognized that most microorganisms are not readily cultivable in the laboratory (1, 17). Recent advances in culture-independent molecular approaches, based on rRNA or genomic approaches that can estimate microbial composition and function, have considerably improved knowledge of microbial ecosystems (7, 11, 29, 32). However, cultivation-based approaches are still necessary for comprehensive elucidation of the physiology and ecology of these organisms and for their biotechnological applications. Recently, several attempts have been made to address these issues (19, 24). Modification of growth conditions based on conventional methods, such as controlling the substrate composition and concentration, the gelling reagent, trace additives such as signaling molecules, and the length of cultivation, has improved isolation efficiencies of rarely cultivated phyla and increased the diversity of isolates (3, 4, 6, 9, 14, 15, 26, 28, 30). Newly developed cultivation methods such as high-throughput methods have brought success with uncultivated microorganisms and improved cultivation capabilities (5, 8, 20, 22, 35). Additionally, development and use of a diffusion chamber to enable the exchange of chemical compounds during cultivation have demonstrated the importance of in situ environmental conditions for the isolation of environmental microorganisms (2, 16). Among them, a concept based on “environmental simulation” is likely to be generally effective for cultivation of environmental microorganisms because various factors that are unknown but necessary for recovery and growth can be provided to the microorganisms (10). However, very few methods have been developed that are applicable to cultivation of microorganisms under in situ environmental conditions. Consequently, it is still important to develop a new cultivation device that is particularly suitable for pure cultivation under in situ environmental conditions while maintaining simple operation. For this study, we designed a new cultivation device, called the hollow-fiber membrane chamber (HFMC), which can provide in situ environmental and liquid culture conditions while maintaining a microliter- to milliliter-scale volume of each chamber. We evaluated the effect of the new device, especially for cultivation under in situ environmental conditions, on cultivation of samples from several different environments.
HFMC.
A piece of porous hollow-fiber membrane was used to form a chamber in which microorganisms can be cultivated. The HFMC system consisted of 48- to 96-chamber units for pure cultivation of multiple samples simply and simultaneously (Fig. 1). One chamber unit consisted of a porous hollow-fiber polyvinylidene fluoride (PVDF) membrane (0.1-μm mean pore size, 67 to 70% porosity, 30-cm length, 1.2-mm outside diameter, 0.76-mm inside diameter) connected with injection and sampling devices by using syringes (Fig. 2). The hollow-fiber membrane was provided by Asahi Kasei Chemicals Co. (Tokyo, Japan). The upper part (injection and sampling part) was kept sterile during cultivation by capping it with a cover.
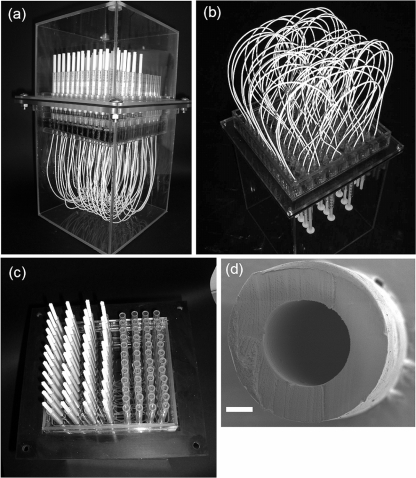
FIG. 1.
Photographs of the 48-well type of HFMC showing the overall system (a), membrane part (b), and injection part (c) and a cross-sectional scanning electron microscope image of a hollow-fiber membrane (d). The size bar represents 200 μm.
FIG. 2.
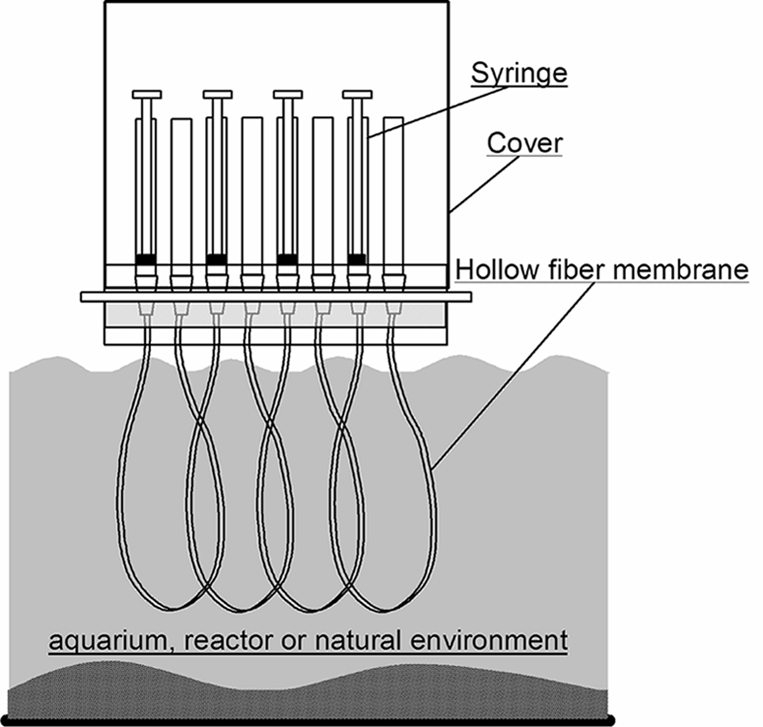
Schematic diagram of cultivation of environmental microorganisms using the HFMC under environmental conditions.
Microbial cells sampled from the environment and serially diluted were first injected into a chamber. The chamber system can be placed in a real natural or engineered environment for the desired incubation time. The membrane part of the HFMC was immersed in a liquid phase during cultivation (Fig. 2). The porous membrane allows exchange of chemical compounds, such as nutrients, metabolites, and signal molecules, but restricts movement of microbial cells. Consequently, pure cultured cells of various types can grow in each chamber under environment-simulating conditions. The chamber volume used for this study was set to 130 μl. However, it is possible to increase or decrease the volume, respectively, using a longer or shorter hollow-fiber membrane. Taking advantage of the hollow-fiber properties, each chamber possesses a high specific and membrane surface area (5.2 mm2/mm3 in the case of a 130-μl chamber volume) that is approximately 20 to 40 times higher than that of a commercial multiwell membrane plate. Consequently, rapid molecular exchange resulting from the high level of environmental imitation is expected, while maintaining the microliter-to-milliliter volume scale of the chamber. The device also allows simple handling.
Before use in this study, the HFMC was sterilized by electron beam sterilization (Nuclear Fuel Industry, Osaka, Japan), although autoclaving can also be used for sterilization. The PVDF membrane must be hydrophilized before use because PVDF originally possesses high hydrophobicity. In this study, a glycerin-dried PVDF membrane (treated with glycerin solution and dried) was used. Glycerin coated onto the membrane was removed by immersing HFMC in water or culture medium solution immediately before cultivation. Ethanol or methanol treatment of the membrane followed by replacement with water can also be used for hydrophilization.
Samples and experiments.
Several environmental samples were used for the following experiment: (i) tidal flat sediment, (ii) activated sludge from a sewage wastewater treatment plant (SWTP), and (iii) activated sludge from a laboratory-scale enhanced biological phosphorus removal (EBPR) process.
Phylogenetic distributions based on the 16S rRNA gene clone library.
Bacterial phylogenetic distributions in three samples were analyzed based on the 16S rRNA gene clone library. In all, 40 to 50 clones from the 16S rRNA gene library were analyzed to estimate the bacterial diversity in each of the three samples. Total DNA was extracted and purified from each sample—(i) tidal flat sediment, (ii) activated sludge in SWTP, and (iii) activated sludge in the EBPR reactor—using Isoplant (Nippon Gene Co., Ltd., Tokyo, Japan) according to the manufacturer's instructions. For construction of clone libraries, 16S rRNA gene PCR products obtained using the primer pair 341f and 907r (23) (tidal flat and EBPR samples) or 8f and 1492r (SWTP sample) were purified using a Wizard SV gel and a PCR cleanup system (Promega Corp., Madison, WI). Purified PCR products were cloned using a Qiagen PCR Cloning Plus kit (Valencia, CA) (tidal flat and EBPR samples) or TOPO TA cloning kit (Invitrogen, Carlsbad, CA) (SWTP sample). Then, colonies were picked up randomly and transferred to Insert Check Ready solution (Toyobo Co., Ltd., Osaka, Japan).
The cloned 16S rRNA gene fragments were sequenced using a cycle sequencing kit (Big Dye Terminator v3.1; Applied Biosystems, Foster City, CA) and a genetic analyzer (ABI PRISM 3100-Avant; Applied Biosystems) using the 341f and 907r primers (23) according to the manufacturer's instructions. Finally, the 16S rRNA gene sequences of approximately 500 bases were determined. A database search was conducted in the DNA Data Bank of Japan using BLAST. The sequences were compared with similar sequences of reference organisms using a BLAST search.
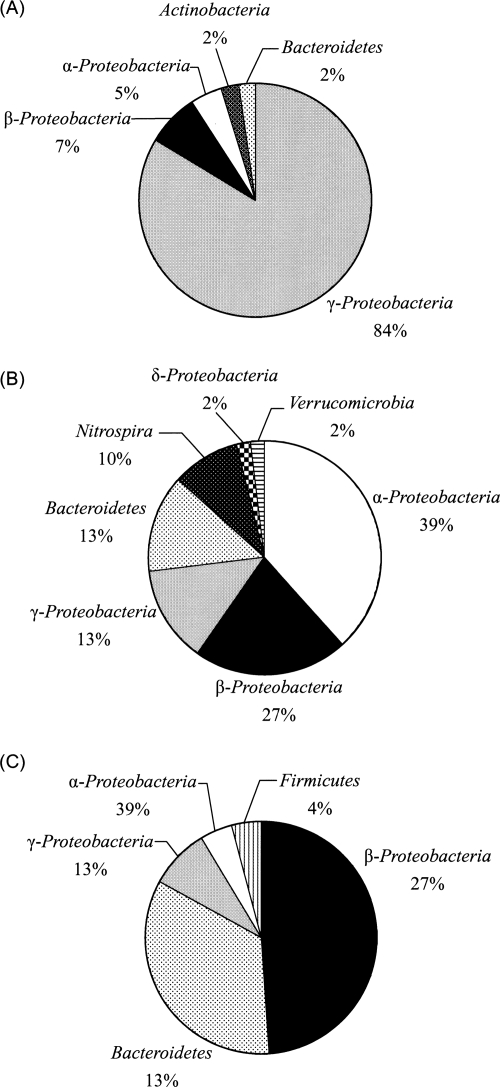
The 45-, 43-, and 50-clone libraries obtained, respectively, from tidal flat, SWTP, and EBPR samples were affiliated and constructed with 5, 7, and 5 major bacterial lineages, respectively, based on results of phylogenetic analysis (Fig. 3). Proteobacteria and the Gammaproteobacteria, Betaproteobacteria, and Alphaproteobacteria subclasses are, respectively, the most dominant groups in the tidal flat, SWTP, and EBPR samples.
FIG. 3.
Bacterial phylogenetic distributions of 16S rRNA gene clones from tidal flat (A), SWTP (B), and EBPR (C) samples.
Experimental procedures.
The diversity and components of isolates from each sample in the HFMC were compared with those of a conventional cultivation method (petri dish) using the same substrate, concentration, and incubation time.
First, the microbial cells in each sample stained with YOPRO-1 or DAPI (4′,6-diamidino-2-phenylindole) were counted using a fluorescence microscope. Microbial cell samples were dispersed using ultrasonic treatment (Sonifier II, model 150; Branson Ultrasonics Corp., Danbury, CT). They were then filtered successively with a 5- to 10-μm presized membrane filter (Omnipore; Millipore Corp., Billerica, MA) to remove large aggregates. Finally, the filtered microbial suspension was dispersed again by ultrasonic treatment to obtain a single-cell suspension followed by serial dilution to the desired concentration of 0.1 to 0.5 cell per 130 μl. After the final dilution, 130 μl of solution containing 0 or single microbial cells was injected into each membrane tube by suctioning syringes connected at the other end of the tube. Immediately after injection, the chamber was placed in a reactor or aquarium and incubated for the desired period. After the incubation, the medium containing microbial cells in each chamber was sampled, followed by staining with DAPI or YOPRO-1 to determine the growth with a fluorescence microscope.
Before the cultivation of environmental microorganisms, a model bacterial strain (Escherichia coli) was cultivated using the HFMC. Growth of microbial cells was observed inside the chamber, showing the supply of sufficient amounts of nutrient and substrate through the membrane. In fact, the efficient diffusive capability of such molecules through the membrane had also been confirmed in the prior experiment by monitoring the transfer of organic compound through the membrane.
Cultivation of environmental samples.
For the tidal flat samples, intertidal sediments were sampled from a tidal flat in Tokyo Bay, Chiba Prefecture, Japan (35°13′N, 139°79′E). Approximately 3 liters of collected sediment was placed in an aquarium in the laboratory, which was filled with 10 liters of seawater collected from the same sampling site. Immediately after the sampling, microorganisms were detached from sediment grains using ultrasonic treatment. Then, microbial cell suspensions were dispersed with combined use of ultrasonication and filtration as described above. The microbial cell suspension was diluted serially using sterilized seawater and injected into the HFMC (0.1 to 0.5 cell per chamber). The HFMC was set in the aquarium and incubated for 2 months at 23°C under aerobic conditions. The seawater was changed once a week during incubation. A part of the membrane was directly attached to surface of the sediment layer in the aquarium during the cultivation. Although setting the chambers in the correct position (inside the upper layer of sediment) was better for “in situ cultivation,” many more in situ conditions might be achieved than by the conventional method.
For the SWTP samples, activated sludge from the sewage wastewater treatment plant in Japan was sampled and dispersed, followed by serial dilution and injection into the HFMC (0.1 to 0.3 cell per chamber). The HFMC was set in a fed-batch-type bioreactor and incubated for 3 weeks at 23°C. The reactor was seeded with the activated sludge biomass and operated under aerobic conditions with an effective volume of 8 liters, while the hydraulic retention time was regulated at 24 h using continuously flowing tap water. The substrate, containing 1.8 g CH3COONa, 0.52 g peptone, 0.44 g NH4Cl, 1.28 g MgSO4·7H2O, 88 mg yeast extract, 64 mg K2HPO4, and 52 mg CaCl2·2H2O (synthetic wastewater I), was fed once a day; that is, the initial concentration of total organic carbon in the reactor was prepared to be 132 mg C per liter.
For the EBPR samples, EBPR-activated sludge was used; this sludge was enriched in a laboratory-scale sequential batch reactor operating under alternating anaerobic and feast/aerobic and famine cycling conditions (21, 27). The dispersed microbial sample was diluted serially and then injected into the HFMC (0.3 cell per chamber). The HFMC was then set in the EBPR reactor containing EBPR-activated sludge and incubated for 1 month. The reactor was operated with a 6-h cycle that consisted of 2.5-h anaerobic, 2.3-h aerobic, and 1.2-h settling/decanting periods, with an effective volume of 1 liter. Subsequently, 500 ml of the influent substrate (synthetic wastewater II, described below) was fed into the reactor every cycle (during the first 10 min of the anaerobic period), and 500 ml of treated supernatant was withdrawn during the last 5 min of the settling stage after the aerobic period. The inlet substrate solution (synthetic wastewater II) contained 0.45 g of CH3COONa, 65 mg KH2PO4, 45 mg MgSO4·7H2O, 32 mg NH4Cl, 7 mg CaCl2·2H2O, and a small amount of trace materials (18). The solids retention time was kept at 8 days by wasting biomass during the aeration period. To maintain the anaerobic and aerobic conditions, the liquid phase was bubbled respectively with N2 gas and air during the anaerobic and aerobic periods.
For petri dish cultivation of the three sample types (the tidal flat sediment, SWTP, and EBPR samples), we used sterilized seawater sampled, respectively, from the same sampling sites, synthetic wastewater I, and synthetic Wastewater II, prepared to the same concentrations as those in the reactors and containing 1.5% agarose.
Phylogenetic analyses of cultured samples.
After cultivation, the solution in each chamber was sampled, and then microbial cell growth was observed using an epifluorescence microscope. Samples that contained microbial cells were used in the next step (phylogenetic analysis). A cell concentration of at least 106 cells/ml was necessary for microscopic detection. Then 16S rRNA gene fragments from extracted total DNA of the isolated samples were amplified using bacterial primer sets of 341f and 907r (23). DNA fragments were sequenced according to the procedures used for the DNA sequencing of isolates described above. Finally, the 16S rRNA gene sequences of approximately 500 bases were determined. A database search was conducted in the DNA Data Bank of Japan using BLAST. The sequences were compared with similar sequences of reference organisms using a BLAST search. All DNA samples having 16S rRNA sequence similarity of more than 97% were grouped into an operational taxonomic unit (OTU).
Consequently, several isolates were identified from each type of sample in the HFMC (16 tidal flat, 41 SWTP, and 21 EBPR samples) presented in Tables 1, 3, and 5. Several colonies were randomly picked up from the petri dishes from each type of sample (47 tidal flat, 44 SWTP, and 41 EBPR samples), as listed in Tables 2, 4, and 6. Despite providing the same substrate composition to the microorganisms in the HFMC and in the petri dishes, the microbial compositions of the isolates with the two methods were significantly different in all three samples; that is, no isolates in the HFMC were identical to those in the petri dishes.
TABLE 1.
Phylogenetic affiliations of isolates from tidal flat with HFMC on the basis of 16S rRNA gene sequences
| Taxonomic group | Closest species | Accession no. | % Similarity | No. of isolatesa |
|---|---|---|---|---|
| Alphaproteobacteria | Loktanella atrilutea | AB246747 | 95 | 1 (1) |
| Thalassospira lucentensis | AM294944 | 98 | 1 | |
| Gammaproteobacteria | Alcanivorax borkumensis | Y12579 | 94 | 1 (1) |
| Alteromonas macleodii | AM885869 | 92 | 1 (1) | |
| Alteromonas macleodii | AM885869 | 98 | 2 | |
| Oceanospirillum kriegii | AB006767 | 92 | 1 (1) | |
| Halomonas ventosae | DQ659437 | 98-99 | 2 | |
| Marinobacter koreensis | AB274772 | 99 | 1 | |
| Marinobacter flavimaris | AY517632 | 99 | 1 | |
| Marinobacter hydrocarbonoclasticus | Y16735 | 99 | 1 | |
| Methylophaga murata | AY694421 | 93 | 1 (1) | |
| Pseudoalteromonas ganghwensis | DQ768622 | 98 | 1 | |
| Pseudoalteromonas elyakovii | AB362304 | 97 | 1 | |
| Pseudomonas pachastrellae | AY880300 | 99 | 1 |
The numbers in parentheses show the numbers of new phylotypes (a total of 5 from 16 isolates).
TABLE 3.
Phylogenetic affiliations of isolates from SWTP with HFMC on the basis of 16S rRNA gene sequences
| Taxonomic group | Closest species | Accession no. | % Similarity | No. of isolates |
|---|---|---|---|---|
| Alphaproteobacteria | Bradyrhizobium elkanii | AM179842 | 98-100 | 7 |
| Betaproteobacteria | Aquamonas fontana | AB120967 | 96 | 1 (1) |
| Hydrogenophaga intermedia | AF019037 | 98 | 1 | |
| Burkholderia fungorum | EF650018 | 100 | 2 | |
| Gammaproteobacteria | Lysobacter gummosus | DQ065753 | 99 | 2 |
| Pseudoxanthomonas japonensis | AB008507 | 99 | 3 | |
| Actinobacteria | Nocardia asteroides | DQ659898 | 96-98 | 6 (2) |
| Streptomyces roseochromogenus | AB184752 | 97-100 | 6 | |
| Cellulosimicrobium cellulans | EU287931 | 97-100 | 10 | |
| Agromyces allium | DQ673874 | 99 | 1 | |
| Spirochaetes | Leptospira illini | M88719 | 96-99 | 2 (1) |
a The numbers in parentheses show the numbers of new phylotypes (a total of 4 from a total of 41 isolates).
TABLE 5.
Phylogenetic affiliations of isolates from EBPR with HFMC on the basis of 16S rRNA gene sequences
| Taxonomic group | Closest species | Accession no. | % Similarity | No. of isolatesa |
|---|---|---|---|---|
| Alphaproteobacteria | Brevundimonas bacteroides | AJ227782 | 89 | 1 (1) |
| Betaproteobacteria | Comamonas terrigena | AM084011 | 95 | 1 (1) |
| Azospira oryzae | AF011347 | 99 | 4 | |
| Comamonas testosteroni | EF522133 | 98 | 1 | |
| Dechlorimonas agitatus | AF047462 | 92 | 1 (1) | |
| Delftia tsuruhatensis | DQ864991 | 96 | 1 (1) | |
| Delftia tsuruhatensis | DQ864991 | 97 | 1 | |
| Hydrogenophaga intermedia | AF019037 | 99 | 1 | |
| Gammaproteobacteria | Lysobacter brunescens | AB161360 | 99 | 2 |
| Pseudomonas anguilliseptica | DQ298027 | 99 | 2 | |
| Pseudoxanthomonas mexicana | AB246798 | 92 | 1 (1) | |
| Stenotrophomonas nitritireducens | DQ537219 | 92 | 1 (1) | |
| Actinobacteria | Rhodococcus erythropolis | AY281107 | 94 | 1 (1) |
| Bacteroidetes | Sphingobacterium faecium | AJ438176 | 87 | 1 (1) |
| Sphingobacterium multivorum | AB020205 | 96 | 1 (1) | |
| Sphingobacterium multivorum | EF059711 | 93 | 1 (1) |
The numbers in parentheses show the numbers of new phylotypes (a total of 10 from a total of 21 isolates).
TABLE 2.
Phylogenetic affiliations of isolates from the tidal flat with petri dish on the basis of 16S rRNA gene sequences
| Taxonomic group | Closest species | Accession no. | % Similarity | No. of isolatesa |
|---|---|---|---|---|
| Alphaproteobacteria | Antarctobacter heliothermus | Y11552 | 98-99 | 2 |
| Loktanella salsilacus | AJ582229 | 96 | 1 (1) | |
| Sphingobium yanoikuyae | AB331239 | 99 | 1 | |
| Caulobacter subvibrioides | M83797 | 99 | 1 | |
| Betaproteobacteria | Limnobacter thiooxidans | DQ922758 | 98 | 1 |
| Gammaproteobacteria | Acinetobacter johnsonii | Z93440 | 99-100 | 10 |
| Acinetobacter radioresistens | X81666 | 99-100 | 5 | |
| Stenotrophomonas maltophilia | EU430096 | 100 | 1 | |
| Actinobacteria | Micrococcus luteus | AB362253 | 99 | 22 |
The number in parentheses shows the number of new phylotypes (from a total of 44 isolates).
TABLE 4.
Phylogenetic affiliations of isolates from SWTP with petri dish on the basis of 16S rRNA gene sequences
| Taxonomic group | Closest species | Accession no. | % Similarity | No. of isolatesa |
|---|---|---|---|---|
| Alphaproteobacteria | Novosphingobium subterraneum | AY752914 | 98 | 25 |
| Sphingomonas melonis | AB334774 | 98-100 | 5 | |
| Methylobacterium jeotgali | DQ471331 | 99 | 4 | |
| Methylobacterium radiotolerans | AM910531 | 99-100 | 2 | |
| Paracoccus aminophilus | D32239 | 98 | 2 | |
| Agrobacterium tumefaciens | AF508099 | 98 | 1 | |
| Betaproteobacteria | Zoogloea resiniphila | AJ011506 | 97 | 1 |
| Duganella violaceinigra | AY376163 | 99 | 6 | |
| Acidovorax avenae | AF508114 | 98 | 1 |
Out of a total of 47 isolates, there were no new phylotypes.
TABLE 6.
Phylogenetic affiliations of isolates from EBPR with petri dish on the basis of 16S rRNA gene sequences
| Taxonomic group | Closest species | Accession no. | % Similarity | No. of isolatesa |
|---|---|---|---|---|
| Alphaproteobacteria | Bosea thiooxidans | DQ424863 | 98-99 | 7 |
| Rhizobium yanglingense | AY972370 | 99 | 3 | |
| Mesorhizobium thiogangeticum | AJ864462 | 97 | 3 | |
| Agrobacterium tumefaciens | AF508099 | 97 | 1 | |
| Sphingosinicella microcystinivorans | AB219941 | 99 | 1 | |
| Blastochloris sulfoviridis | AB245349 | 90 | 1 (1) | |
| Brevundimonas vesicularis | DQ111026 | 100 | 1 | |
| Catellibacterium nectariphilum | AB101543 | 96 | 1 (1) | |
| Betaproteobacteria | Delftia acidovorans | EU024146 | 99-98 | 2 |
| Diaphorobacter oryzae | EU342380 | 97 | 2 | |
| Gammaproteobacteria | Acinetobacter johnsonii | EF204266 | 97-99 | 19 |
| Actinobacteria | Microbacterium maritypicum | EU434520 | 98-99 | 5 |
The numbers in parentheses show the numbers of new phylotypes (a total of 2 from a total of 48 isolates).
Phylum- and subclass-level phylogenetic distributions of the isolates from the HFMC in all three samples resemble those of clone libraries compared with those of isolates from the petri dish in every sample (Fig. 3). However, species-level phylogenetic distributions in the HFMC did not match well to those of clone libraries.
Cultivation performance comparison.
We identified isolates whose 16S rRNA gene sequences were less than 97% similar to those of any known bacterial species as novel microbes in this study. The ratio of such novel phylotypes was markedly higher in the HFMC (tidal flat, 31% [5 of 16 isolates]; SWTP, 10% [4 of 41 isolates]; EBPR, 48% [10 of 21 isolates]) than those in the petri dishes (tidal flat, 2% [1 of 44 isolates]; SWTP, 0% [none of 47 isolates]; EBPR, 4% [2 of 48 isolates]) in every sample, as presented in Tables 1 to 6. For all three sample types, higher-diversity microorganisms based on the species level were cultivated in the HFMC than in the petri dishes, as shown by comparison of several diversity index scores: number of OTUs, the Shannon-Weaver diversity index, the Simpson diversity index, and evenness (Table 7). Despite the quantities of isolates in some samples in the HFMC being smaller than those in the petri dishes, the quantities of OTUs of the HFMC samples were all larger than those of the petri dish samples.
TABLE 7.
Diversity of isolates
| Sample type and method | Na | nb | n/N | Hc | 1 − Dd | Ee |
|---|---|---|---|---|---|---|
| Tidal flat | ||||||
| HFMC | 16 | 14 | 0.88 | 2.60 | 0.98 | 0.98 |
| Petri dish | 44 | 9 | 0.20 | 1.50 | 0.70 | 0.68 |
| SWTP | ||||||
| HFMC | 41 | 11 | 0.29 | 2.09 | 0.90 | 0.87 |
| Petri dish | 47 | 9 | 0.19 | 1.56 | 0.69 | 0.71 |
| EBPR | ||||||
| HFMC | 21 | 16 | 0.76 | 2.65 | 0.96 | 0.96 |
| Petri dish | 46 | 12 | 0.26 | 1.94 | 0.80 | 0.77 |
Total number of isolates.
Number of OTUs.
Shannon-Weaver diversity index, calculated as follows: 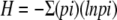 , where pi is the proportion of each phylogenetic group to n.
, where pi is the proportion of each phylogenetic group to n.
Simpson diversity index, calculated as follows: 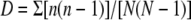 .
.
Evenness, calculated as follows from the Shannon-Weaver index: E = H/ln n.
A single dominant bacterial type, or a few types in some cases, was recovered from the petri dishes from every sample. Micrococcus luteus-related organisms (50% of isolates), Novosphingobium subterraneum-related organisms (53% of isolates), and Acinetobacter johnsonii-related organisms (approximately 40% of isolates) were dominant, respectively, in significant proportions of isolates from the tidal flat, SWTP, and EBPR samples (Tables 2, 4, and 6). The Acinetobacter-related strain is well known as the major isolate from the EBPR process, although these organisms were not major populations (21, 31). Although the colonies on the petri dish were of various sizes, the experiment was conducted carefully to avoid picking up only highly visible large colonies.
The cultivabilities of the HFMC represented 12.3%, 21.0%, and 9.2% of inoculated cells, whereas those of the petri dishes, respectively, represented 2.1%, 1.4%, and 0.2% of inoculated cells from the tidal flat, SWTP, and EBPR samples. The previous study also showed that simulated environmental conditions increase cultivability (16). Although the recoveries of the two methods should not be simply compared (one is based on dilution cultivation, and the other is in CFU), the higher cultivation capability of environmental microorganisms in the HFMC, as described above, might increase the cultivability of HFMC compared with that in the petri dishes.
Effectiveness of HFMC.
Isolation and pure culturing using an HFMC can be performed in a semiopen system and under in situ environmental conditions. These conditions provide various factors and conditions necessary for the recovery and growth of environmental microorganisms, as described below, engendering the high cultivation capability reported herein.
First, the HFMC allows the growth of microorganisms requiring syntrophic partners or interspecific or intraspecific interactions. The importance of cell-cell interactions regulated by some signaling molecules inducing the growth of previously uncultivable microorganisms has been reported recently (25).
Second, the supply of substrates of various types can be achieved in the HFMC system, such as the continuous feeding of low-concentration substrates. This kind of substrate supply might help to recover microorganisms that are recalcitrant for cultivation because a high concentration of organic or inorganic substrates is sometimes toxic to environmental microorganisms. The “K-strategist” favoring a nutrient-poor environment is thought to be a major population in a natural environment, especially among uncultivated microbes (34). Furthermore, it is possible to change the substrate media or culture conditions (such as aerobic/anaerobic) during cultivation (as with the EBPR sample in this study). The hollow-fiber membrane, possessing a large specific area, allows the immediate response of internal conditions to changes in the external conditions. In addition, specific and endemic organic and inorganic compounds in the environment functioning as substrates or growth factors (10) can be supplied to microorganisms through the membrane by this in situ environmental cultivation.
Third, it is possible to maintain stable growth conditions during cultivation. For example, metabolic byproducts and secreted materials are removed immediately and continuously by diffusion through the membrane. Reportedly, such compounds inhibit the growth and activity of microorganisms (33).
However, there are some remaining limitations that should be improved to realize more widespread use. First, the present HFMC device is restricted to fit microorganisms in the correct position under vertical sharp chemical or microbial population gradient environments, as described above. Second, an inoculation of microbial cells into the HFMC-based cultivation relied on the dilution. The microbial cells were inoculated with 0.1 to 0.5 cell per chamber in this experiment. Consequently, only small quantities of isolates could be obtained in one device. Further improvement is necessary for high-throughput cultivation. The combined use of a cell-sorting system (flow cytometry) for single-cell sorting to inoculate one cell per chamber might be effective (data not shown).
Recently developed advanced and inventive cultivation methods, such as methods using gel microdroplets (20, 35), diffusion chambers (2, 16), and microdevices (12, 13), depend on the formation of microcolonies or extremely small-scale cultivation. On the other hand, cultivation using the HFMC is based on liquid culture and enables growth of microbial cells at levels up to 109 to 1010 cells per 130-μl chamber. Thus, it can be used for a variety of purposes, such as secondary cultivation under simulated environmental conditions followed by taxonomical, physiological, and biochemical analyses. Combined use of the HFMC and the other advanced methods described above will also be a powerful approach to cultivation and to understanding the ecology and physiology of uncultivable microorganisms. Above all, the HFMC has the potential to extend the range of cultivable microorganisms in any type of environment and thus will contribute to enhanced understanding of microbial physiology and ecology, as well as providing new sources of bioproducts.
Acknowledgments
We are deeply grateful to Asahi Kasei Chemicals Co., especially Yoske Koizumi and Hiroyoshi Ohya, for various types of support, such as kindly providing us with key materials and equipment, as well as valuable advice.
This research was supported by the Industrial Technology Research Grant Program from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Footnotes
Published ahead of print on 27 March 2009.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollmann, A., K. Lewis, and S. S. Epstein. 2007. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 73:6386-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruns, A., U. Nübel, H. Cypionka, and J. Overmann. 2003. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl. Environ. Microbiol. 69:1980-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Button, D. K., F. Schut, P. Quang, R. Martin, and B. R. Robertson. 1993. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl. Environ. Microbiol. 59:881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, J.-C., and S. J. Giovannoni. 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 70:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christen, R. 2008. Global sequencing: a review of current molecular data and new methods available to assess microbial diversity. Microbes Environ. 23:253-268. [DOI] [PubMed] [Google Scholar]
- 8.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, K. E. R., S. J. Joseph, and P. H. Janssen. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari, B. C., S. J. Binnerup, and M. Gillings. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71:8714-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingham, C. J., M. van den Ende, D. Pijnenburg, P. C. Wever, and P. M. Schneeberger. 2005. Growth and multiplexed analysis of microorganisms on a subdivided, highly porous, inorganic chip manufactured from Anopore. Appl. Environ. Microbiol. 71:8978-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingham, C. J., A. Sprenkels, J. Bomer, D. Molenaar, A. van den Berg, J. E. T. van Hylckama Vlieg, and W. M. de Vos. 2007. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. USA 104:18217-18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 17.Keller, M., and K. Zengler. 2004. Tapping into microbial diversity. Nat. Rev. Microbiol. 2:141-150. [DOI] [PubMed] [Google Scholar]
- 18.Kishida, N., J. Kim, S. Tsuneda, and R. Sudo. 2006. Anaerobic/oxic/anoxic granular sludge process as an effective nutrient removal process utilizing denitrifying polyphosphate-accumulating organisms. Water Res. 40:2303-2310. [DOI] [PubMed] [Google Scholar]
- 19.Leadbetter, J. R. 2003. Cultivation of recalcitrant microbes: cells are alive, well and revealing their secrets in the 21st century laboratory. Curr. Opin. Microbiol. 6:274-281. [DOI] [PubMed] [Google Scholar]
- 20.Manome, A., H. Zhang, Y. Tani, T. Katsuragi, R. Kurane, and T. Tsuchida. 2001. Application of gel microdroplet and flow cytometry techniques to selective enrichment of non-growing bacterial cells. FEMS Microbiol. Lett. 197:29-33. [DOI] [PubMed] [Google Scholar]
- 21.Mino, T., M. C. M. van Loosdrecht, and J. J. Heinen. 1998. Microbiology and biochemistry of the enhanced biological phosphate removal process. Water Res. 32:3192-3207. [Google Scholar]
- 22.Morris, R. M., M. S. Rappé, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 23.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nichols, D. 2007. Cultivation gives context to the microbial ecologist. FEMS Microbiol. Ecol. 60:351-357. [DOI] [PubMed] [Google Scholar]
- 25.Nichols, D., K. Lewis, J. Orjala, S. Mo, R. Ortenberg, P. O'Connor, C. Zhao, P. Vouros, T. Kaeberlein, and S. S. Epstein. 2008. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 74:4889-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sangwan, P., S. Kovac, K. E. R. Davis, M. Sait, and P. H. Janssen. 2005. Detection and cultivation of soil Verrucomicrobia. Appl. Environ. Microbiol. 71:8402-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seviour, R. J., T. Mino, and M. Onuki. 2003. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 27:99-127. [DOI] [PubMed] [Google Scholar]
- 28.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Streit, W. R., and R. A. Schmitz. 2004. Metagenomics—the key to the uncultured microbes. Curr. Opin. Microbiol. 7:492-498. [DOI] [PubMed] [Google Scholar]
- 30.Tamaki, H., Y. Sekiguchi, S. Hanada, K. Nakamura, N. Nomura, M. Matsumura, and Y. Kamagata. 2005. Comparative analysis of bacterial diversity in freshwater sediment of a shallow eutrophic lake by molecular and improved cultivation-based techniques. Appl. Environ. Microbiol. 71:2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, M., R. Erhart, W. Manz, R. Amann, H. Lemmer, D. Wedi, and K.-H. Schleifer. 1994. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 60:792-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner, M., P. H. Nielsen, A. Loy, J. L. Nielsen, and H. Daims. 2006. Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr. Opin. Microbiol. 17:83-91. [DOI] [PubMed] [Google Scholar]
- 33.Watsuji, T., S. Yamada, T. Yamabe, Y. Watanabe, T. Kato, T. Saito, K. Ueda, and T. Beppu. 2007. Identification of indole derivatives as self-growth inhibitors of Symbiobacterium thermophilum, a unique bacterium whose growth depends on coculture with a Bacillus sp. Appl. Environ. Microbiol. 73:6159-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watve, M., V. Shejval, C. Sonawane, M. Rahalkar, A. Matapurkar, Y. Shouche, M. Patole, N. Phadnis, A. Champhenkar, K. Damle, S. Karandikar, V. Kshirsagar, and M. Jog. 2000. The ‘K’ selected oligotrophic bacteria: a key to uncultured diversity? Curr. Sci. 78:1535-1542. [Google Scholar]
- 35.Zengler, K., G. Toledo, M. Rappé, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]