Abstract
In the United States, total maximum daily load standards for bodies of water that do not meet bacterial water quality standards are set by each state. The presence of human polyomaviruses (HPyVs) can be used as an indicator of human-associated sewage pollution in these waters. We have developed and optimized a TaqMan quantitative PCR (QPCR) assay based on the conserved T antigen to both quantify and simultaneously detect two HPyVs; JC virus and BK virus. The QPCR assay was able to consistently quantify ≥10 gene copies per reaction and is linear over 5 orders of magnitude. HPyVs were consistently detected in human waste samples (57 of 64) and environmental waters with known human fecal contamination (5 of 5) and were not amplified in DNA extracted from 127 animal waste samples from 14 species. HPyV concentrations in sewage decreased 81.2 and 84.2% over 28 days incubation at 25 and 35°C, respectively. HPyVs results were compared to Escherichia coli, fecal coliform, and enterococci concentrations and the presence of three other human-associated microbes: Bacteroidetes, Methanobrevibacter smithii, and adenovirus. HPyVs were the most frequently detected of these in human and contaminated environmental samples and were more human specific than the Bacteroidetes (HF183) or M. smithii. HPyVs and M. smithii more closely mimicked the persistence of adenovirus in sewage than the other microbes. The use of this rapid and quantitative assay in water quality research could help regulatory agencies to identify sources of water pollution for improved remediation of contaminated waters and ultimately protect humans from exposure to pathogens.
Maintaining healthy coastal water systems is essential, since poor water quality can have detrimental effects on mangroves, seagrass beds, coral reefs, the fishing and shellfish harvesting industries, and the health of recreational water users (1, 5, 15, 17, 20, 44). Since 1972 in the United States, each state has been required to set total maximum daily loads (TMDLs) for pollutants in water bodies according to section 303(d) of the Clean Water Act (50). The probability that microbial pathogens are present is estimated by enumerating indicator bacteria, which are shed in the feces of humans and most animals. The U.S. Environmental Protection Agency recommends using Escherichia coli and enterococci to assess the quality of freshwater and saline water, respectively (47); however, Florida currently uses fecal coliforms and enterococci as indicators of fecal pollution (42).
When bacterial indicators exceed regulatory levels, a plan of action (TMDL implementation) must be developed to reduce pathogens. TMDL plans for “pathogen” reduction are particularly problematic because they rely upon surrogate indicator bacteria, which yield little or no insight as to the source of pollution. High indicator bacteria concentrations can be attributed to many sources, including agricultural runoff, storm water runoff, wildlife, pets, faulty septic systems (onsite wastewater treatment and disposal systems), and a failing central sewer infrastructure (5, 12, 28).
To address the issue of source identification, methods have been developed in which the biochemistry or genetics of certain microorganisms are used to indirectly identify probable source(s) of fecal pollution, which is termed microbial source tracking (MST) (48). MST methods based on detection of a source-associated gene (marker) by PCR have proliferated over the past 10 years due to the additional information they can provide to watershed managers on fecal contamination sources (43). Although marker detection by endpoint (binary) PCR can give important insights on the source(s) of fecal contamination, quantitative measurements can provide information about the relative magnitude of contamination from various sources. Moreover, epidemiological studies on the correlation between recreational water use, microbial contamination, and the risk of illness will greatly benefit from the ability to quantify MST markers, rather than simply assessing binary (+/−) detection.
Although many bacterial targets have been proposed for MST of human sewage (8, 39, 46a), fewer viral targets have been investigated (19, 24, 33). Polyomavirus is the sole genus in the family Polyomaviridae (22). These viruses have a 5-kbp double-stranded DNA genome surrounded by a 40- to 50-nm icosahedral capsid (38). The JCV and BKV human polyomaviruses (HPyVs) have similarly structured genomes that show ∼75% identity (21). BK virus (BKV) and JC virus (JCV) gained much attention in the late 1970s as the etiological agents of kidney nephritis (i.e., BKV reactivation in the kidneys) and progressive multifocal leukoencephalopathy (i.e., JCV reactivation in brain tissue) in the immunocompromised (16, 34). Serological studies have shown that >70% of adults harbor antibodies to BKV or JCV (27, 30, 44). These viruses are known for producing lifelong, asymptomatic viruria in immunocompetent individuals (37). In 2000 it was first suggested that JCV would be a useful indicator of human sewage in water (11). The obligate host specificity and abundance of BKV and JCV in municipal sewage has led to the successful use of these viruses to indicate human fecal pollution in environmental water samples (12, 29).
Due to the health implications of BKV and JCV, several methods have been developed to rapidly detect either BKV or JCV in clinical samples (6, 31, 35, 56). However, from an MST standpoint, it is advantageous to target both BKV and JCV. BKV has been found in feces (54), and both viruses are excreted in the urine (6, 11, 37, 55, 60) either simultaneously or individually. The focus of this research was the modification of the previously developed nested PCR protocol for HPyVs detection (29) to a TaqMan quantitative PCR (QPCR) assay to simultaneously detect and quantify both BKV and JCV. Furthermore, we compared measurements obtained with the newly developed QPCR assay to those of other water quality indicators and MST markers. These indicators included bacterial indicator concentrations (49) and PCR detection of human-associated markers currently used for MST. These included human-associated Bacteroidetes (8), Methanobrevibacter smithii (46a), and adenovirus (36). To assess the potential of HPyVs to mimic the fate of pathogens in water, the persistence of all of the water quality indicators was assessed, and relationships between bacterial indicator organisms and MST markers in both human waste samples as well as contaminated environmental samples were examined.
MATERIALS AND METHODS
Virus strains and viral DNA extraction.
BKV (ATCC VR-837), JCV (ATCC VR-1583), and adenovirus type 2 (ATCC VR-846) were obtained from the American Type Culture Collection (Manassas, VA). BKVs were propagated in HEL-299 cells (ATCC CCL-137). JCVs were propagated in COS-7 cells (ATCC CRL-1651). Adenoviruses were propagated in MRC-5 cells (ATCC CRL-171). Cell lines were grown in Eagle minimum essential medium (Sigma, St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Inc., Carlsbad, CA). Cell lines were maintained in Eagle minimum essential medium containing 2% fetal bovine serum. Viral DNA was extracted from cell culture by using a DNeasy Blood & Tissue kit (Qiagen, Inc., Valencia, CA) and stored at −20°C.
Primer and probe design.
Primers P5 and P6 have been used successfully in conventional PCRs to simultaneously detect both JCV and BKV in clinical (7) and environmental samples (29). However, based on stipulations of the TaqMan PCR assay design, P5 was modified to increase the optimum annealing temperature. The primers target a region of the T antigen in both JCV and BKC. JCV and BKV DNA sequences within the amplified partial T antigen were obtained from GenBank. Approximately 10 JCV and 10 BKV sequences were analyzed. The sequences were aligned by using CLUSTAL W (European Bioinformatics Institute, United Kingdom), and probe sequences were chosen based on identical areas of the gene sequence. Probe sequences were also chosen based on melting temperature (Tm) stipulations (5 to 10°C higher than the Tm of the primers) and base stipulations (no long runs of the same base; ≤2 G or C nucleotides in the final five bases of the 3′ end; and A, C, or T on the 5′ end). The probe was labeled with a fluorescent reporter molecule, 6-carboxyfluorescein (FAM), at the 5′ end and a minor groove binding the nonfluorescent quencher molecule (MGBNFQ) at the 3′ end. Primers were purchased from Integrated DNA Technologies (Coralville, IA), and probes were purchased from Applied Biosystems (Foster City, CA). The primers (SM2 and P6) and probe (KGJ3) sequences used in the present study are presented in Table 1.
TABLE 1.
Primers and probe sequences used in this study
| Assay | Orientation | Primer or probe | Sequence (5′-3′) | Expected product length (bp) | Source or reference |
|---|---|---|---|---|---|
| Total Bacteroidetes PCR | Forward | Bac32F | AAC GCT AGC TAC AGG CTT | 676 | 8 |
| Reverse | Bac708R | CAA TCG GAG TTC TTC GTG | 8 | ||
| Human-associated | Forward | HF183 | ATC ATG AGT TCA CAT GTC CG | 525 | 8 |
| Bacteroidetes PCR | Reverse | Bac708R | CAA TCG GAG TTC TTC GTG | 8 | |
| M. smithii PCR | Forward | Mnif-342f | AAC AGA AAA CCC AGT GAA GAG | 221 | 53 |
| Reverse | Mnif-363r | ACG TAA AGG CAC TGA AAA ACC | 53 | ||
| Adenovirus PCR | Forward | hexAA1885 | GCC GCA GTG GTC TTA CAT GCA CAT C | 301 | 36 |
| Reverse | hexAA1913 | CAG CAC GCC GCG GAT GTC AAA GT | 36 | ||
| Adenovirus nested PCR | Forward | nehexAA1893 | GCC ACC GAG ACG TAC TTC AGC CTG | 143 | 36 |
| Reverse | nehexAA1905 | TTG TAC GAG TAC GCG GTA TCC TCG CGG TC | 36 | ||
| HPyV QPCR | Forward | SM2 | AGT CTT TAG GGT CTT CTA CCT TT | 173 (JCV) | This study |
| Reverse | P6 | GGT GCC AAC CTA TGG AAC AG | 176 (BKV) | 7 | |
| Probe | KGJ3 | (FAM)-TCA TCA CTG GCA AAC AT-(MGBNFQ) | This study |
QPCR.
The QPCR mixtures were prepared using 25 μl of TaqMan Universal PCR master mix, No AmpErase UNG (Applied Biosystems), 0.5 μM primer concentrations, a 0.4 μM labeled probe concentration, and 5 μl of template DNA (5 to 15 ng/μl), and the volume was adjusted to 50 μl using reagent-grade water. Amplification was performed in a 7500 Real-Time PCR system (Applied Biosystems). Optimal temperatures and primer and probe concentrations were determined by varying conditions over both temperature and concentration gradients (data not shown). The QPCR reaction conditions were as follows: DNA polymerase activation at 95°C for 10 min was followed by 40 cycles of DNA melting at 95°C for 15 s, annealing at 55°C for 15 s, and then extension at 60°C for 60 s.
Sequencing of QPCR amplicon.
DNA was extracted from 0.1 ml of the BKV and JCV stock by using a DNeasy Blood & Tissue kit and used as a template in a real-time PCR assay. The resulting 176- and 173-bp amplicons from the BKV and JCV reactions, respectively, were purified by using a QIAquick PCR purification kit (Qiagen), cloned into pCR4-TOPO vector (Invitrogen), transferred into E. coli OneShot chemically competent cells, and plated on LB agar containing 100 μg of ampicillin/ml. Recombinant plasmids with a single copy of the partial T antigen were purified by using a GenElute Five-Minute plasmid miniprep kit (Sigma) according to the manufacturer's instructions. Plasmids containing the BKV (176 bp) or JCV (173 bp) insert sequence were sequenced at Macrogen USA (Rockville, MD). All sequences were subjected to a BLAST search (www.ncbi.nlm.nih.gov/BLAST) for comparison to published sequences. Query and matching sequences were aligned by using CLUSTAL W.
Standard curve and sensitivity of QPCR.
Purified recombinant plasmid DNA containing the BKV or JCV insert was quantified by using a Qubit fluorometer (Invitrogen). DNA quantification was performed in triplicate and averaged to determine the estimated total DNA concentration. The recombinant plasmids were serially diluted, and the limit of detection for both inserts was determined based on the lowest level of detection in the real-time PCR.
Insert copy numbers were estimated by multiplying the average DNA concentration by Avogadro's number and then dividing by the product of the insert length and average weight of a base pair (59). To produce a standard curve, the recombinant plasmid DNA was serially diluted in nuclease-free reagent-grade water to a final concentration ranging from 102 to 106 gene copies/μl. The plasmid containing the BKV insert was used to produce standard curves for both the sewage holding time experiments and the analysis of HPyV concentrations in environmental waters. Portions (5 μl) of each dilution were used as a template in the TaqMan real-time standard-curve PCRs. Each dilution was run in duplicate. The Applied Biosystems default settings for the threshold cycle (CT) were used for data analysis. The CT values were plotted against the copy number to generate the standard curve. Linear regression was used to assess the relationship between CT values and the copy number. Diluted DNA was used in all subsequent reactions to create the standard curve for quantification of HPyVs in unknown samples.
Specificity of HPyV QPCR.
Fresh animal waste samples (e.g., dog feces and cow manure) were collected from local farms, wooded areas, and households. Individual fecal samples were collected from cats (n = 5), a chicken (n = 1), cows (n = 24), cranes (n = 2), deer (n = 3), dogs (n = 55), ducks (n = 4), a fox (n = 1), horses (n = 8), a raccoon (n = 1), seagulls (n = 6), sparrows (n = 3), and feral pigs (n = 2). Samples were collected in sterile propylene tubes, placed on ice, and transported or shipped to the laboratory. All fecal samples were processed within 24 h of collection. Approximately 0.3 g of animal waste was used for DNA extraction. DNA was extracted by using a MO BIO (Carlsbad, CA) UltraClean fecal DNA kit according to the manufacturer's instructions. Composite cow and pig fecal samples were collected from flushed manure holding tanks at the University of Florida's Dairy Research Unit and Swine Unit. DNA was extracted from 1 ml of the composite sample by using a QIAamp Blood DNA Midi kit, with minor modifications. Briefly, 1 ml of the sample and 1 ml of phosphate-buffered saline were added to a 15-ml tube, and then 200 μl of Qiagen proteinase and 2.4 ml of buffer AL were added to the tube containing the sample. The tube was then incubated at 70°C for 15 min. DNA was then extracted from the lysate according to the manufacturer's instructions.
To assess the condition of samples and presence of PCR inhibitors, all animal fecal DNA was first subject to total Bacteroidetes sp. PCR (8). Briefly, the PCRs were prepared by using 12.5 μl of GoTaq Green Master Mix (Promega, Madison, WI), 0.5 μM primer concentrations (Table 1), and 2 μl of template DNA (5 to 15 ng/μl), and the volume was adjusted to 25 μl using reagent-grade water. The PCR conditions were as follows: DNA polymerase activation at 95°C for 3 min was followed by 30 cycles of DNA melting at 94°C for 45 s, annealing at 55°C for 45 s, and extension at 72°C for 60 s, with a final extension at 72°C for 5 min (Eppendorf Mastercycler thermocycler; Eppendorf International, Hamburg, Germany). PCR products were separated by agarose gel electrophoresis (2%). DNA was viewed by using ethidium bromide under UV light. Amplicons were identified visually by comparison to a BenchTop 100-bp DNA ladder (Promega) and a 676-bp positive control. DNA from fecal samples was not used for further analysis when the total Bacteroidetes target was not amplified by PCR.
Urine samples from both cats (i.e., the composite of three cats) and dogs (n = 9) were obtained from posteuthanized (within 1 h) animals at animal shelters. The bladder was expressed manually, and urine was caught directly into a sterile specimen cup. Samples were then placed on ice, shipped to the laboratory, and stored at −20°C until processed. All urine samples were processed within 7 days of collection. DNA was extracted from 1 ml of the urine sample by using a QIAamp Blood DNA Midi kit, as described above. To test for inhibition, samples were spiked with 102 BKV particles and processed in tandem with the samples. Extracted DNA was used as a template in QPCR assays as described above.
Human target samples.
A total of 90 human waste and human urine samples were collected. Of the human waste samples collected, 41 were raw sewage (wastewater treatment plant [WWTP] influents or lift stations), 9 were dechlorinated tertiary-treated WWTP effluent, 9 were from septic tank pump trucks (composite septic tank samples), and 5 were from individual septic tanks (with two to four individuals in the households). Raw sewage was collected from Brandon, FL (n = 21); Costa Mesa, CA (n = 3); Gainesville, FL (n = 10); Oldsmar, FL (n = 3); Orlando, FL (n = 2); and Tampa, FL (n = 2). Dechlorinated tertiary-treated WWTP effluent was collected from Brandon, FL (n = 9). Pump truck samples were acquired from the greater Tampa, FL, area (n = 9). Septic tank samples were collected from Cocoa, FL (n = 2), and the greater Tampa, FL, area (n = 3). All samples were collected in sterile propylene tubes or liter bottles, placed on ice, and transported or shipped to the laboratory. A total of 26 individual urine samples were analyzed. Samples were collected from healthy volunteers ranging in age from 2 to 57 years old. Volunteers collected urine in sterile specimen cups and then placed the samples at 4°C. All samples were processed within 24 h of collection.
DNA was extracted from sewage, septic tank, pump truck, and urine by using the QIAamp Blood DNA Midi kit with minor modifications, as described above. Virus particles were concentrated from 500 ml of dechlorinated tertiary-treated WWTP effluent. The pH of the water was adjusted to 3.5 by using 2.0 N HCl and was filtered through a 0.45-μm-pore-size, 47-mm-diameter nitrocellulose filter. The filter was placed into a 2-ml microcentrifuge tube and kept at −20°C. DNA was extracted from the filter within 24 h of filtration by using a MO BIO Powersoil kit according to the manufacturer's instructions. Extracted DNA was used as a template in QPCR assays as previously described. Six amplicons from human-associated samples were confirmed by sequencing and analysis by MEGA4 (as previously described). One transformed E. coli colony from each cloning reaction was analyzed.
Analysis of water quality indicators in sewage over a 28-day period (“sewage holding time experiment”).
Three 100-ml aliquots of raw sewage were placed in a dark area at room temperature (25.6 ± 0.8°C) or in an incubator (35.0 ± 0.1°C) to represent a range of ambient water temperatures in Florida during the summer months. Room temperature mimics the coastal water temperatures common to Florida coastlines, while the “hot” temperature more closely mimics the temperatures of shallow waters (e.g., ditches, small streams, etc.) that can become extremely warm during mid-summer days. One 1-ml aliquot was collected from each replicate on days 0, 7, 14, 21, and 28. DNA was extracted from the samples by using the QIAamp DNA Blood Midi kit and used as a template in QPCR assays as described above.
Environmental water samples.
Five environmental water samples with high probability of human fecal contamination were collected from various locations in Florida. DHR4a was collected from the Hillsborough River (Tampa, FL) near an overflowing lift station. Env1 and Env2 were collected from a beach (Tampa, FL) receiving contamination due to a broken sewer line. EJ3 was collected in Charlotte County, FL, near residential areas with faulty septic tanks. SJH8 was collected from a tributary of St. John's River (St. Augustine, FL) near a malfunctioning septic tank. Approximately 500 ml of water was placed into a sterile 1-liter polypropylene container and transported to the laboratory on ice. Salinity was measured by using a hand refractometer (Fisher Scientific, Pittsburgh, PA). All environmental samples were processed within 6 h of collection. To promote electrostatic interactions between the viral capsid and nitrocellulose filter, the pH of the water was adjusted to 3.5 by using 2.0 N HCl and was filtered through a 0.45-μm-pore-size nitrocellulose filter (3, 27). The filter was placed into a 2-ml microcentrifuge tube and kept at −20°C. DNA was extracted from the filter by using a MO BIO Powersoil kit in accordance with the manufacturer's instructions. HPyV copy numbers were enumerated by the QPCR assay as described above. Four amplicons from contaminated environmental water samples were confirmed by DNA sequencing and analysis by using MEGA4 (as previously described).
Comparison of HPyV QPCR to other methods.
HPyV QPCR results were compared to concentrations of indicator bacteria and PCR detection of human-associated Bacteroidetes, M. smithii, and adenovirus (see below for methods). Indicator bacteria were enumerated in all human waste, sewage holding time, and environmental water samples. PCR was used to detect human-associated Bacteroidetes, M. smithii, and adenovirus in human waste, sewage holding time, and environmental water samples. In addition, PCR was used to examine the presence or absence of human-associated Bacteroidetes and M. smithii in all animal samples.
Enumeration of indicator bacteria.
E. coli, fecal coliforms, and enterococci were enumerated in human waste samples and environmental water samples. In addition, sewage samples held at room temperature or 35°C were filtered weekly for the enumeration of all indicator bacteria. Fecal coliform concentrations were determined by membrane filtration using mFC agar (3), with incubation at 44 ± 0.5°C for 24 h. E. coli were enumerated by membrane filtration on modified mTEC agar, with incubation at 35°C for 2 h to resuscitate any injured or stressed cells, followed by incubation at 44.5°C for 22 h in a water bath (52). Enterococci were enumerated by membrane filtration on mEI agar, with incubation at 41 ± 0.5°C for 48 h (51).
Detection of human-associated Bacteroidetes.
Previously published primers specific for a partial region of 16S rRNA gene of human-associated Bacteroidetes were used in a touchdown PCR (8) (Table 1). PCRs were performed in a 25-μl mixture containing 12.5 μl of GoTaq Green Master Mix (Promega), 0.5 μM concentrations of each primer, and 2 μl of template DNA. The touchdown PCR conditions were as follows: DNA polymerase activation at 95°C for 3 min was followed by 43 cycles of DNA melting at 94°C for 45 s, annealing for 45 s, and extension at 72°C for 30 s. Annealing temperatures ranged from 65 to 55°C. Cycles were performed twice at 65 to 63°C, once at 62 to 56°C, and 30 times at 55°C; followed by a final elongation at 72°C for 5 min (using an Eppendorf Mastercycler thermocycler). PCR products were separated by 2% agarose gel electrophoresis and viewed by using ethidium bromide under UV light. Amplicons were identified visually by comparison to a BenchTop 100-bp DNA ladder (Promega) and a 525-bp positive control.
Detection of the human-associated M. smithii.
Previously published primers specific for the nifH gene of human-associated M. smithii were used in the touchdown PCR (53) (Table 1). PCRs were performed in a 25-μl mixture containing 12.5 μl of GoTaq Green Master Mix, 0.5 μM concentrations of each primer, and 2 μl of template DNA. The touchdown PCR conditions were the same as those described above. PCR products were separated by 2% agarose gel electrophoresis and viewed by using ethidium bromide under UV light. Amplicons were identified visually by comparison to a BenchTop 100-bp DNA ladder and a 221-bp positive control.
Adenovirus nested PCR.
Previously published primers specific for the hexon gene of human adenoviruses were used in the nested PCR (36) (Table 1). Amplification was carried out in a 50-μl reaction mixture containing 25 μl of GoTaq Green Master Mix, 0.08 μM concentrations of each primer, and 5 μl of template DNA. In both PCRs, the first round of denaturation was carried out for 4 min at 94°C, followed by 30 cycles of denaturing at 94°C for 90 s, annealing at 55°C for 90 s, and extension at 72°C for 120 s, followed in turn by a final elongation at 72°C for 5 min. For the nested PCR, the 50-μl reaction mixture contained 25 μl of GoTaq Green Master Mix, 0.16 μM concentrations of each primer, and 1 μl of template from the first round of PCR. The PCR products were separated by 2% agarose gel electrophoresis and viewed by using ethidium bromide under UV light. Amplicons were identified visually by comparison to a BenchTop 100-bp DNA ladder and a 143-bp positive control.
Statistical analysis.
Summary statistics were computed for variables of interest by using GraphPad InStat version 3.00 (GraphPad Software, San Diego, CA). Differences among standard deviations of HPyV and bacterial concentrations in either septic or sewage samples were determined by using the Bartlett statistic. When the Bartlett statistic P value was <0.05, then the standard deviations among the samples were considered significantly different, and means were compared by using nonparametric repeated-measures analysis of variance (ANOVA) with Dunn's multiple comparison test. When the Bartlett statistic P value was >0.05, then the standard deviations among the samples were considered not significantly different, and means were compared by using repeated-measures ANOVA with the Tukey-Kramer multiple comparison test. Means were considered significantly different when P was <0.05. Linear relationships between HPyV and indicator bacterium concentrations were determined by calculating the Pearson correlation coefficients. Differences were considered significant when P was <0.05, and two-sided tests were performed for all analyses. Comparisons of concentrations of water quality indicators over time at select temperatures were made by using repeated-measures ANOVA (InStat version 3.00). Differences of indicator concentrations among temperatures were compared by using a paired t test (InStat version 3.00). Observations of human-associated markers were converted to binary data, and binary logistic regression models (SPSS version 12.0) were used to assess the relationships between HPyV or indicator bacterium concentrations and the presence or absence of human-associated markers. Nagelkerke's R-square (R2), which can range from 0.0 to 1.0, denotes the effect size (the strength of the relationship); stronger associations have values closer to 1.0. Relationships were considered significant when the P value for the model chi-square was 0.05.
RESULTS
Sensitivity of QPCR and calculation of standard curves.
Standard curves constructed from analysis of BKV or JCV viral particles displayed a linear relationship for the QPCR assay, with an average R2 value 0.992 ± 0.008. Under these conditions, the developed QPCR protocol was able to consistently quantify ≥10 gene copies per reaction for both BKVs and JCVs (Table 2) . For all standard curves generated during the present study, the average values for the variables in the linear regression were as follows: the y intercept was 44.7 ± 2.3, and the slope was −3.61 ± 0.22.
TABLE 2.
Results of QPCR assays for known viral numbers (100, 10, 1, or 0.1 particles) and detection limits determined for BKV and JCV
| Virus | Replicate | Measured gene copy no./reaction vs an estimated gene copy no./reaction of:
|
|||
|---|---|---|---|---|---|
| 100 | 10a | 1 | 0.1 | ||
| BKV | A | 76.2 | 31.8 | 0 | 0 |
| B | 143.6 | 21.6 | 0 | 0 | |
| C | 82.1 | 13.5 | 0 | 0 | |
| Avg ± SD | 100.6 ± 37.4 | 22.3 ± 9.2 | 0 | 0 | |
| JCV | A | 76.8 | 4.3 | 0 | 0 |
| B | 77.8 | 6.7 | 0 | 0 | |
| C | 112.4 | 8.4 | 0 | 0 | |
| Avg ± SD | 89.0 ± 20.3 | 6.5 ± 2.0 | 0 | 0 | |
An estimated gene copy number of 10 was the detection limit of the assay.
Specificity of HPyV QPCR and human-associated markers.
Total Bacteroidetes, which is indicative of nonspecific fecal contamination, was determined by binary PCR in all animal fecal samples assayed (Table 3). HPyVs were not amplified in any of the individual animal fecal samples (n = 115), composite animal fecal samples (n = 2), or animal urine samples (n = 10), and the assay did not amplify adenovirus DNA (Table 3). In contrast, human-associated Bacteroidetes was detected by PCR in 13.0% (n = 15) of the animal fecal samples (cats and dogs). Moreover, human-associated M. smithii was detected in 0.9% (n = 1) of the animal fecal samples (an individual cow fecal sample). The detailed results of the marker specificity testing are presented in Table 3.
TABLE 3.
Result of human-associated markers PCR assays on nontarget and target samplesa
| Sample | Description | Source | No. of samples tested | No. of samplesb detected by:
|
||||
|---|---|---|---|---|---|---|---|---|
| Total Bacteroidetes PCR | HPyV QPCR | Human-associated Bacteroidetes PCR | M. smithii PCR | Adenovirus PCR | ||||
| Nontarget DNA virus | Adenovirus | ATCC VR-846 | 1 | ND | 0 | ND | ND | 1 |
| Target viruses | JCV | ATCC VR-1583 | 1 | ND | 1 | ND | ND | ND |
| BKV | ATCC VR-837 | 1 | ND | 1 | ND | ND | ND | |
| Animal fecal samples | Cat (Felis catus) | Individuals | 5 | 5 | 0 | 1 | 0 | ND |
| Chicken (Gallus gallus) | Individuals | 1 | 1 | 0 | 0 | 0 | ND | |
| Cow (Bos taurus) | Individuals | 24 | 24 | 0 | 0 | 1 | ND | |
| Sandhill crane (Grus canadensis) | Individuals | 2 | 2 | 0 | 0 | 0 | ND | |
| Deer (Odocoileus virginianus) | Individuals | 3 | 3 | 0 | 0 | 0 | ND | |
| Dog (Canis lupus familiaris) | Individuals | 55 | 55 | 0 | 14 | 0 | ND | |
| Duck (Anas platyrhynchos) | Individuals | 4 | 4 | 0 | 0 | 0 | ND | |
| Fox (Vulpes vulpes) | Individuals | 1 | 1 | 0 | 0 | 0 | ND | |
| Horse (Equus caballus) | Individuals | 8 | 8 | 0 | 0 | 0 | ND | |
| Racoon (Procyon lotor) | Individuals | 1 | 1 | 0 | 0 | 0 | ND | |
| Seagull (Larus atricilla) | Individuals | 6 | 6 | 0 | 0 | 0 | ND | |
| Sparrow (Ammodramus savannarum) | Individuals | 3 | 3 | 0 | 0 | 0 | ND | |
| Wild pig (Sus scrofa) | Individuals | 2 | 2 | 0 | 0 | 0 | ND | |
| Animal composite waste samples | Cow (Bos taurus) | Farm composite | 1 | 1 | 0 | 0 | 0 | ND |
| Pig (Sus scrofa domestica) | Farm composite | 1 | 1 | 0 | 0 | 0 | ND | |
| Animal urine samples | Dog (Canis lupus familiaris) | Individuals | 9 | ND | 0 | ND | ND | ND |
| Cat (Felis catus) | Composite of three cats | 1 | ND | 0 | ND | ND | ND | |
| Target human samples | Lift station | Composite | 2 | ND | 2 | 2 | 2 | 2 |
| Sewage influent | Composite | 39 | ND | 39 | 39 | 39 | 39 | |
| Septic tank pump truck | Composite of two to three septic tanks | 9 | ND | 9 | 9 | 9 | 7 | |
| Dechlorinated tertiary-treated WWTP effluent | Effluent | 9 | ND | 2 | 0 | 0 | 1 | |
| Septic tanks | Composite of two to four individuals | 5 | ND | 5 | 3 | 4 | 0 | |
| Urine samples | Individuals | 26 | ND | 6 | ND | ND | ND | |
Boldface values indicate amplification in nontarget samples.
That is, the number of samples quantifiable by QPCR or amplified by conventional PCR. ND, the sample(s) was not analyzed by the indicated assay,
HPyV QPCR analysis of human waste samples.
HPyVs were detected in 23.1% (6 of 26) of human urine samples collected from healthy individuals (Table 3). HPyVs were detected in 17% (1 of 6) of samples collected from 0- to 20-year-old age group, 25% (3 of 12) of the 21- to 40-year-old age group, and 25% (2 of 8) of the >40-year-old age group. The concentration of HPyVs in positive urine samples ranged widely, from 6.61 × 102 to 1.20 × 107 copies ml−1, with a mean concentration of 2.71 ± 4.72 × 106 copies ml−1.
HPyVs were ubiquitous in untreated sewage and septage (septic tank pump truck or individual septic tank) samples but were detected in only 22.2% of tertiary-treated WWTP effluent samples (hereafter referred to as effluent samples) (Table 3). HPyVs were found in relatively high concentrations in raw sewage samples (Table 4) . The concentrations were the same order of magnitude in raw influent samples and septic tank samples (∼104 copies ml−1); however, the mean concentration of HPyVs in raw influent was significantly higher than that in septage samples (P < 0.05). As expected, HPyVs were present at a significantly lower concentration in WWTP effluent than in the influent or septage samples.
TABLE 4.
Concentrations of HPyVs, E. coli, enterococci, and fecal coliforms in human waste samplesa
| Sourceb | Mean copies of HPyV ml −1 ± SD | Mean CFU ml −1 ± SD
|
||
|---|---|---|---|---|
| E. coli | Enterococci | Fecal coliforms | ||
| Raw sewage influent* | (3.0 ± 1.7) × 104 A | (4.6 ± 1.8) × 104 B | (2.5 ± 0.9) × 104 A | (7.4 ± 4.3) × 104 C |
| Septic tank pump truck samples* | (1.1 ± 1.0) × 104 D | (3.1 ± 1.9) × 103 D,E | (1.5 ± 1.1) × 103 E | (3.0 ± 2.0) × 104 F |
| Individual septic tanks* | (1.4 ± 1.6) × 104 G,H,I | (9.7 ± 9.1) × 103 G,J,K | (5.8 ± 7.8) × 103 H,J | (3.5 ± 2.7) × 104 I,K |
| Dechlorinated teritary-treated WWTP effluent† | (1.2 ± 2.5) × 10−1 L,M | (2.0 ± 1.4) × 10−2 L,N | (1.8 ± 1.2) × 10−1 O | (8.4 ± 4.5) × 10−2 M,N,O |
Values in the same row followed by the same capital letter (i.e., A through O) are not statistically different at P < 0.05.
*, repeated-measures ANOVA was used to determine statistical significance. †, nonparametric repeated-measures ANOVA was used to determine statistical significance.
Analysis of other water quality indicators in human waste samples.
The frequencies of human-associated Bacteroidetes, M. smithii, and adenovirus detection by PCR are summarized in Table 3. Both human-associated Bacteroidetes and M. smithii were detected in 100% of the sewage influent samples and the majority of the septage samples; however, neither marker was detected in any disinfected effluent sample. Adenoviruses were detected in 100% of the sewage influent samples. Moreover, adenoviruses were detected in 77.8 and 11.1%, respectively, of the septic tank pump truck samples and disinfected effluent samples but were not detected in any individual septic tank samples (Table 3).
The average concentrations of enterococci, E. coli, and fecal coliforms are summarized in Table 4. All statistical comparisons within a sample type were performed by repeated-measures ANOVA because several observations on one subject (i.e., sewage sample) were compared (32). In untreated sewage samples, fecal coliforms and E. coli tended to be more concentrated than enterococci; however, enterococci were more concentrated in the disinfected wastewater. The HPyV gene copy concentration was comparable to that of the indicator bacteria for all sample types (within the same order of magnitude) (Table 4).
A comparison of indicator bacteria concentrations across the sewage types revealed that E. coli and enterococcus concentrations were significantly greater in raw sewage influent than in septage samples (P < 0.05). The results for fecal coliforms were similar, except that their concentrations in raw influent were not significantly different from those in the septic tank pump truck samples. As expected, all indicator bacteria concentrations were significantly (and at least 4 orders of magnitude) lower in disinfected sewage than in influent or septage.
Correlations among bacterial indicators and HPyV concentrations in human waste samples.
Correlations among bacterial concentrations and HPyV gene copy concentrations were determined for sewage, disinfected wastewater, and septage samples (Table 5). In sewage samples, despite concentrations within the same order of magnitude, HPyV gene copy concentrations were negatively correlated with each bacterial indicator. As expected, all bacterial indicator concentrations in sewage were positively correlated with each other.
TABLE 5.
Correlation of HPyV and indicator bacteria in human sewagea
| HPyV or bacterium | Sample source | Statistical parameter | Fecal coliforms | Enterococci | E. coli |
|---|---|---|---|---|---|
| HPyV | Sewage | PCb | -0.6166 | -0.3279 | -0.5852 |
| P | 0.0001 | 0.0363 | 0.0001 | ||
| R2 | 0.3802 | 0.1075 | 0.3425 | ||
| WWTP effluent | PC | -0.7131 | 0.2812 | -0.2836 | |
| P | 0.0310 | 0.4636 | 0.4596 | ||
| R2 | 0.5085 | 0.0791 | 0.0804 | ||
| Septic tanks | PC | 0.0376 | -0.2139 | -0.3946 | |
| P | 0.9522 | 0.7297 | 0.5110 | ||
| R2 | 0.0014 | 0.0458 | 0.1557 | ||
| Pump trucks | PC | 0.4498 | -0.6062 | 0.6855 | |
| P | 0.2245 | 0.0835 | 0.0415 | ||
| R2 | 0.2023 | 0.3675 | 0.4699 | ||
| E. coli | Sewage | PC | 0.8718 | 0.3281 | |
| P | 0.0001 | 0.0362 | |||
| R2 | 0.7600 | 0.1077 | |||
| WWTP effluent | PC | 0.1035 | 0.2458 | ||
| P | 0.7910 | 0.5238 | |||
| R2 | 0.0107 | 0.0604 | |||
| Septic tanks | PC | 0.8683 | 0.8493 | ||
| P | 0.0563 | 0.0686 | |||
| R2 | 0.7539 | 0.7212 | |||
| Pump trucks | PC | 0.8145 | -0.6077 | ||
| P | 0.0075 | 0.0826 | |||
| R2 | 0.6634 | 0.3693 | |||
| Enterococci | Sewage | PC | 0.4697 | ||
| P | 0.0019 | ||||
| R2 | 0.2206 | ||||
| WWTP effluent | PC | -0.7167 | |||
| P | 0.0298 | ||||
| R2 | 0.5137 | ||||
| Septic tanks | PC | 0.9297 | |||
| P | 0.0221 | ||||
| R2 | 0.8644 | ||||
| Pump truck | PC | -0.4691 | |||
| P | 0.2027 | ||||
| R2 | 0.2201 |
Statistics were calculated for relationships between organisms in sewage, dechlorinated tertiary-treated WWTP effluent, individual septic tanks, and septic tank pump truck samples. Relationships were considered significant when P < 0.05. Values in boldface indicate significant relationships.
PC, Pearson correlation.
In disinfected wastewater samples, HPyV gene copy concentrations were negatively correlated with fecal coliforms and, while not significant, a similar tendency was observed for the relationship between HPyVs and E. coli. Enterococcus concentrations were also negatively correlated with fecal coliform concentrations in WWTP effluent, and while HPyV and enterococcus concentrations were not significantly correlated, there was a positive trend.
A small number of individual septic tank samples were collected (n = 5) which led to nonsignificant correlations among HPyV gene copy concentrations and all bacterial concentrations, as well as E. coli and both fecal coliform and enterococcus concentrations. However, enterococci were positively correlated with fecal coliform concentrations. In septic tank pump truck samples, E. coli was significantly and positively correlated with both HPyV gene copy concentrations and fecal coliforms. No other significant relationships were found within the pump truck samples.
Analysis of water quality indicators in sewage over a 28-day period.
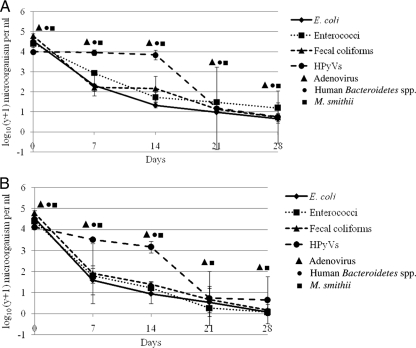
To better understand the negative correlation between indicator bacteria and HPyVs in sewage, the effects of time and temperature were assessed. There was a larger percent decrease in samples exposed to the higher temperature (35°C) compared to samples exposed to room temperature (25°C) for all water quality indicators (Fig. 1). At the higher temperature, enterococci had the largest decrease, with a change of −98.0%, followed closely by both E. coli and fecal coliforms (98.0 and 96.3% decreases, respectively). HPyVs were slightly more resilient to higher temperatures, with an 84.2% decrease. At room temperature, E. coli decreased 85.4%, fecal coliforms decreased 84.8%, HPyVs decreased 81.2%, and enterococci decreased 72.8%. Comparison of HPyV concentrations at 25°C versus 35°C showed no significant difference over the 28-day period (P = 0.4148). However, there was a significant difference between the temperatures for the concentrations of E. coli (P = 0.0068), enterococci (P < 0.0001), and fecal coliforms (P = 0.0296). Under both temperature conditions, HPyV concentrations did not significantly decrease until day 21 (P < 0.05), whereas significant decreases in indicator bacteria concentrations occurred at day 7 (P < 0.01). Human-associated Bacteroidetes, M. smithii, and adenovirus were detected by conventional PCR through day 28 at room temperature (Fig. 1A). At the high temperature both M. smithii and adenovirus were detected through day 28; however, human-associated Bacteroidetes was only detected until day 14 (Fig. 1B).
FIG. 1.
Concentration of indicator bacteria and HPyVs in sewage coupled with the PCR detection of human-associated Bacteroidetes, M. smithii, and adenovirus over a 28-day period. (A) Room temperature (25°C); (B) warm temperature (35°C).
Environmental water samples.
Salinities, bacterial and HPyV concentrations, and binary detection of human-associated markers in environmental waters with a high probability of human sewage contamination are summarized in Table 6. Environmental samples were collected over a range of salinity from fresh to marine water. All indicator bacterial concentrations exceeded regulatory standards (42, 47). Human-associated Bacteroidetes and M. smithii were detected in 80% of the environmental samples, whereas adenoviruses were detected in 60% of the samples. HPyVs were detected in all samples, at concentrations up to 1.4 × 106 copies 500 ml−1.
TABLE 6.
Physical and microbial data for all environmental samples collected in this study
| Site | Salinity (ppt) | Suspected source of contamination | CFU/100 ml ± SD
|
HPyV QPCR resulta | PCR result
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Enterococci | E. coli | Fecal coliforms | Adenovirus | Human- associated Bacteroidetes | M. smithii | ||||
| Dhr4A | 3 | Sewer cross-connection | (3.5 ± 3.2) × 104 | (3.0 ± 2.1) × 104 | (7.8 ± 3.5) × 104 | (1.4 ± 1.1) × 106 | + | + | + |
| Env1 | 33 | Faulty sewer line | (1.6 ± 0.7) × 105 | (1.1 ± 0.4) × 105 | (1.9 ± 1.1) × 105 | (3.5 ± 0.3) × 102 | + | + | + |
| Env2 | 32 | Faulty sewer line | (5.8 ± 0.3) × 104 | (4.5 ± 0.4) × 104 | (6.1 ± 2.4) × 104 | (2.2 ± 0.1) × 102 | + | + | + |
| EJ3 | 25 | Faulty septic tanks | (1.3 ± 1.0) × 104 | (1.0 ± 0.2) × 104 | (1.3 ± 0.3) × 104 | (1.2 ± 2.0) × 104 | - | + | + |
| SJH8 | 0 | Faulty septic tank | (5.8 ± 1.5) × 103 | NDb | (3.9 ± 0.1) × 103 | (9.2 ± 0.2) × 101 | - | - | - |
HPyV results are reported as gene copy number per 500-ml sample.
ND, not determined.
Binary logistic regression was used to assess the predictive relationship between HPyVs or bacterial indicators concentrations and the presence or absence of human-associated indicators (Bacteroidetes, M. smithii, and adenovirus). HPyVs, enterococci, and fecal coliform concentrations were independently strong predictors of the presence or absence of both human associated Bacteroidetes and M. smithii (Nagelkerke's R2 = 1.000, P = 0.025 for all relationships). In addition, the presence of adenovirus was strongly correlated with the concentrations of E. coli (Nagelkerke's R2 = 1.000, P = 0.034), enterococci (Nagelkerke's R2 = 1.000, P = 0.009), and fecal coliforms (Nagelkerke's R2 = 1.000, P = 0.034). The presence of adenovirus and the concentrations of HPyVs were not correlated in environmental samples (Nagelkerke's R2 = 0.090, P = 0.552). Since there must be at least one 1.0 or 0.0 when data are analyzed using binary logistic regression, the relationship between E. coli and both Bacteroidetes and M. smithii could not be determined because both human-associated markers were found in all four samples in which E. coli was enumerated.
Sequencing analysis of QPCR amplicons.
Sequences of amplicons generated from cultured JCV and BKV (n = 2), human-associated waste (n = 6), and environmental samples (n = 4) were either 173 or 176 bp in length. The 176- and 173-bp amplicon sequences showed ≥99% identity to published BKV or JCV sequences, respectively. Specifically, the sequence derived from cultured BKVs (GenBank accession no. FJ666992), sewage (GenBank accession no. FJ666993), human urine (GenBank accession no. FJ666994), and environmental water samples (GenBank accession nos. FJ666995, FJ666997, and FJ666998) showed ≥99% identity to published BKV sequences. Sequences derived from cultured JCVs (GenBank accession no. FJ666999), sewage (GenBank accession no. FJ666991), individual septic tank (GenBank accession no. FJ667002), septic tank pump truck effluent (GenBank accession no. FJ667001), tertiary-treated wastewater (GenBank accession no. FJ667000), and an environmental sample (GenBank accession no. FJ666996) showed ≥99% identity to published JCV sequences.
DISCUSSION
This study introduces a new QPCR assay for the quantification of human polyomaviruses BKV and JCV in environmental water samples and summarizes relationships among bacterial and viral water quality indicators and pathogens (adenovirus) in various human waste samples. Initial attempts in our laboratory to develop a QPCR assay for HPyVs used SYBR green PCR chemistry and previously published primers (29), but it was found that nonspecific fluorescence occurred due to primer dimer formation (data not shown). The TaqMan QPCR assay coupled with an MGBNFQ eliminates this problem and increases assay specificity. The QPCR assay can detect as few as 10 JCV and BKV particles, which is comparable to other published QPCR methods (that detect either JCV or BKV) in terms of dynamic range and precision (14, 35, 40).
Sequencing analysis of randomly chosen clones from various human and environmental samples showed that 50% of the sequences aligned with BKV sequences and 50% aligned with JCV sequences. These results indicate widespread distribution of both BKV and JCV in humans in the United States, supporting the desirability of an assay that detects both viruses. Furthermore, widespread geographic distribution provides some confidence that the assay can be useful worldwide. To date, JCV or BKV have been detected by using conventional PCR in raw sewage from Cairo, Egypt; Patras, Greece; Barcelona, Spain; Nancy, France; Pretoria, South Africa; Umea, Sweden; and Washington, DC (10, 11). The method described in the present study has quantified HPyVs in samples from California and across Florida.
The PCR results from previously published studies estimate BKV and JCV concentrations in sewage ranging from 101 to 103 BKV ml−1 and from 102 to 104 JCV ml−1 (11), which is within the range of the average concentration of HPyVs in sewage found in the present study (3.0 × 104 copies ml−1). There are no previously published data documenting the concentrations of HPyVs in onsite wastewater treatment and disposal (septic tank) systems and septic tank pump trucks. HPyVs were consistently detected in these samples, and the concentrations were consistently high (∼104 copies ml−1). These concentrations are also comparable to concentrations of culturable indicator bacteria in sewage. In contrast, the detection of human-associated Bacteroidetes and M. smithii in septic tank samples was inconsistent and detection of adenovirus in septic tank and pump truck samples was sparse. Furthermore, only HPyVs were detected in an environmental sample contaminated by faulty septic tanks. These data suggest that the HPyV assay is a useful marker for both sewage and septic system contamination.
The obligate host specificity of viruses such as HPyVs is advantageous for specific identification of human sources. It is, however, possible that animals could ingest and excrete HPyVs without infection, leading to a transient association with feces. In addition, some have speculated that kidney infection and subsequent viruria in these animals could occur despite the documented host specificity of HPyVs (41). Areas frequented by dogs (e.g., backyards or dog parks) may introduce such cosmopolitan organisms to nearby water sources during rain events, confounding attempts to identify contamination sources. In light of the recent studies reporting human-associated bacterial markers detected in dog fecal samples (2, 57), we were most concerned with cross-reactivity in dog fecal and urine samples. All of these samples, as well as all animal-derived samples tested in the present study, were negative for HPyVs, further emphasizing the potential of these viruses as indicators for human-specific fecal contamination.
The human-associated Bacteroidetes assay was less specific, since the amplicon was detected in a cat fecal sample, as well as several dog fecal samples. The cross-reactivity of the human-associated Bacteroidetes has been previously documented (2, 25). The incomplete specificity of this marker may pose problems with water quality assessment in areas frequented by these animals (e.g., dog parks and animal friendly public beaches), and it may be ideal to augment water quality analysis with a more human-specific marker. Human-associated M. smithii was more human specific than the Bacteroidetes assay, since it was detected in only one cow fecal sample. The previously published M. smithii PCR protocol developed for MST (46a) utilized a prefiltration step due to previous detection of M. smithii in ruminant ciliated protozoans (23). Prefiltration was not used in the protocol of the present study, and the M. smithii protocol may have increased the specificity with an additional prefiltration step. However, the use of prefiltration tends to increase the assay's limit of detection (unpublished data), leading to a probable tradeoff between sensitivity and specificity.
Although many MST methods focus on bacterial markers, these indicators are not accurate predictors of some human pathogens, particularly viruses and protozoa (5, 13, 18). Double-stranded DNA viruses tend to be more resilient in response to environmental stresses such as temperature, UV light, and disinfectants compared to bacteria and therefore may more accurately mimic the survival of viral and protozoan pathogens in environmental water systems (9, 30, 45, 46). Moreover, in tropical climates bacterial indicators have the potential to survive and perhaps even multiply in environmental waters (4, 26, 58), which may cause overestimation of public health risks. In the present study, we found negative correlations between HPyVs and indicator bacteria in sewage and septic tank samples (e.g., see Table 4), while all correlations among indicator bacteria were positive. The results of the sewage holding time experiment demonstrate that the kinetics of viral DNA (adenoviruses and HPyVs) decay were quite dissimilar from decay rates of culturable indicator bacteria. Furthermore, HPyVs mimicked the persistence of adenovirus at both warm and room temperatures, while the bacterial indicators did not. The fate of HPyVs in sewage may be a better predictor of the fate and persistence of pathogenic enteric viruses than indicator bacteria concentrations since, unlike the indicator bacteria, the viruses lack the potential for growth in the environment and are more similar in size and structure to each other than to bacteria. In addition, both adenovirus and HPyVs were detected in tertiary-treated wastewater, indicating the potential of these viruses to mimic the persistence of relatively disinfection-resistant microorganisms such as double-stranded DNA viruses.
The success of this QPCR assay in quantifying low numbers of HPyVs in combination with HPyV host specificity and the abundance in human-associated waste will allow for a rapid, quantitative, and cost-effective assessment of water quality. The benefits of incorporating the HPyV TaqMan QPCR assay with the expanding MST toolbox include the reduction of assay time, increased specificity for human pollution, and quantification. These benefits will in turn allow a better understanding of relationships with other MST markers and waterborne pathogens and ultimately a better perspective on the proportion of microbial contamination from human sources.
Acknowledgments
We thank Robert Ulrich for his time and insights. In addition, we thank Brittany Sears, Bina Nayak, Zach Staley, Katrina Gordon, Asja Korajkic, Phoebe Koch, Chris Staley, Brian Badgley, Miriam Brownell, Sam Farrah, Kevin Grant, John Griffith, and Kathy and Gary McQuaig, as well as the Oldsmar Wastewater Reclamation Facility, Advanced Septic Tanks, and Ted's Tanks, for help with sample collection.
Funding for this study was provided in part by a Southeastern Branch American Society of Microbiology 2007 H. Aldrich Research Award and by the Cooperative Institute for Coastal and Estuarine Environmental Technology (NA05NOS4191149, subaward 07-092).
Footnotes
Published ahead of print on 3 April 2009.
REFERENCES
- 1.Adams, S. M., A. M. Brown, and R. W. Goede. 1993. A quantitative health assessment index for rapid evaluation of fish conditions in the field. Trans. Am. Fish. Soc. 122:63-73. [Google Scholar]
- 2.Ahmed, W., J. Stewart, D. Powell, and T. Gardner. 2008. Evaluation of Bacteroides markers for the detection of human fecal pollution. Lett. Appl. Microbiol. 46:237-242. [DOI] [PubMed] [Google Scholar]
- 3.American Public Health Association. 1998. Standard methods for the examination of water and wastewater, 20th ed., p. 9.137-9.141. American Public Health Association, Washington, DC.
- 4.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnone, R. D., and J. P. Walling. 2007. Waterborne pathogens in urban watersheds. J. Water Health 5:149-162. [DOI] [PubMed] [Google Scholar]
- 6.Arthur, R. R., S. Dagostin, and K. V. Shah. 1989. Detection of BK virus and JC virus in urine and brain tissue by the polymerase chain reaction. J. Clin. Microbiol. 27:1174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Askamit, A. 1993. Diagnostic molecular microbiology principles and applications. Mayo Foundation, Rochester, MN.
- 8.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitton, G. 2005. Wastewater microbiology, 3rd ed. John Wiley & Sons, Inc., Hoboken, NJ.
- 10.Bofill-Mas, S., M. Formiga-Cruz, P. Clemente-Casares, F. Calafell, and R. Girones. 2001. Potential transmission of human polyomaviruses through the gastrointestinal tract after exposure to virions or viral DNA. J. Virol. 75:10290-10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bofill-Mas, S., S. Pina, and R. Girones. 2000. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 66:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brownell, M. J., V. J. Harwood, R. C. Kurz, S. M. McQuaig, J. Lukasik, and T. M. Scott. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res. 41:3747-3757. [DOI] [PubMed] [Google Scholar]
- 13.Dorner, S. M., W. B. Anderson, T. Gaulin, H. L. Candon, R. M. Slawson, P. Payment, and P. M. Huck. 2007. Pathogen and indicator variability in a heavily impacted watershed. J. Water Health 5:241-257. [PubMed] [Google Scholar]
- 14.Elfaitouri, A., A. Hammarin, and J. Blomberg. 2006. Quantitative real-time PCR assay for the detection of human polyomavirus infection. J. Virol. Methods 135:207-213. [DOI] [PubMed] [Google Scholar]
- 15.Frenzel, S. A. 1990. Effects of municipal wastewater discharges on aquatic communities, Boise River, Idaho. J. Am. Water Resources Assoc. 26:279. [Google Scholar]
- 16.Gardner, S. D., A. M. Field, D. V. Coleman, and B. Hulme. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253-1257. [DOI] [PubMed] [Google Scholar]
- 17.Harborne, A. R., D. C. Afzal, and M. J. Andrews. 2001. Honduras: Caribbean Coast. Mar. Pollut. Bull. 42:1221-1235. [DOI] [PubMed] [Google Scholar]
- 18.Harwood, V. J., A. D. Levine, T. M. Scott, V. Chivukula, J. Lukasik, S. R. Farrah, and J. B. Rose. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71:3163-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havelaar, A. H., W. M. Pot-Hogeboom, K. Furuse, R. Pot, and M. P. Hormann. 1990. F-specific RNA bacteriophages and sensitive host strains in feces and wastewater of human and animal origin. J. Appl. Bacteriol. 69:30-37. [DOI] [PubMed] [Google Scholar]
- 20.Henrickson, S. E., T. Wong, P. Allen, T. Ford, and P. R. Epstein. 2001. Marine swimming-related illness: implications for monitoring and environmental policy. Environ. Health Perspect. 109:645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch, H. H., and J. Steiger. 2003. Polyomavirus BK. Lancet Infect. Dis. 3:611-623. [DOI] [PubMed] [Google Scholar]
- 22.ICTVdB Management. 2006. Polyomaviridae, entry 00.047. In C. Büchen-Osmond (ed.), ICTVdB: the universal virus database, version 4. Columbia University, New York, NY.
- 23.Irbis, C., and K. Ushida. 2004. Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, S. C., W. Chu, and J. W. He. 2007. Seasonal detection of human viruses and coliphage in Newport Bay, California. Appl. Environ. Microbiol. 73:6468-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kildare, B. J., C. M. Leutenegger, B. S. McSwain, D. G. Bambic, V. B. Rajal, and S. Wuertz. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701-3715. [DOI] [PubMed] [Google Scholar]
- 26.Lasalde, C., R. Rodriguez, and G. A. Toranzos. 2005. Statistical analyses: possible reasons for unreliability of source tracking efforts. Appl. Environ. Microbiol. 71:4690-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukasik, J., T. M. Scott, D. Andryshak, and S. R. Farrah. 2000. Influence of salts on virus adsorption to microporous filters. Appl. Environ. Microbiol. 66:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsalek, J., and Q. Rochfort. 2004. Urban wet-weather flows: sources of fecal contamination impacting on recreational waters and threatening drinking-water sources. J. Toxicol. Environ. Health A 67:1765-1777. [DOI] [PubMed] [Google Scholar]
- 29.McQuaig, S. M., T. M. Scott, V. J. Harwood, S. R. Farrah, and J. O. Lukasik. 2006. Detection of human derived fecal pollution in environmental waters using a PCR-based human polyomavirus assay. Appl. Environ. Microbiol. 72:7567-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng, Q. S., and C. P. Gerba. 1996. Comparative inactivation of enteric adenoviruses, polioviruses and coliphages by ultraviolet irradiation. Water Res. 30:2665-2668. [Google Scholar]
- 31.Moret, H., V. Brodard, C. Barranger, N. Jovenin, M. Joannes, and L. Andreoletti. 2006. New commercially available PCR and microplate hybridization assay for detection and differentiation of human polyomaviruses JC and BK in cerebrospinal fluid, serum, and urine samples. J. Clin. Microbiol. 44:1305-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motulsky, H. 1995. Intuitive biostatistics, p. 260. Oxford University Press, Inc., New York, NY.
- 33.Noble, R. T., S. M. Allen, A. D. Blackwood, W. Chu, S. C. Jiang, G. L. Lovelace, M. D. Sobsey, J. R. Stewart, and D. A. Wait. 2003. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial source tracking comparison study. J. Water Health 1:195-207. [PubMed] [Google Scholar]
- 34.Padgett, B. L., D. L. Walker, G. M. Zu Rhein, R. J. Eckroade, and B. H. Dessel. 1971. Cultivation of BK virus encephalitis in an papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet 283:1363-1364. [DOI] [PubMed] [Google Scholar]
- 35.Pal, A., L. Sirota, T. Maudru, K. Peden, and A. M. Lewis, Jr. 2006. Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses. J. Virol. Methods 135:32-42. [DOI] [PubMed] [Google Scholar]
- 36.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polo, C., J. L. Perez, A. Mielnichuck, C. G. Fedele, J. Niubo, and A. Tenorio. 2004. Prevalence and patterns of polyomavirus urinary excretion in immunocompetent adults and children. Clin. Microbiol. Infect. 10:640-644. [DOI] [PubMed] [Google Scholar]
- 38.Saenz-Robles, M. T., C. S. Sullivan, and J. M. Pipas. 2001. Transforming functions of simian virus 40. Oncogene 20:7899-7907. [DOI] [PubMed] [Google Scholar]
- 39.Scott, T. M., T. M. Jenkins, J. Lukasik, and J. B. Rose. 2005. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 39:283-287. [PubMed] [Google Scholar]
- 40.Sehbani, L., B. Kabamba-Mukadi, A. T. Vandenbroucke, M. Bodeus, and P. Goubau. 2006. Specific and quantitative detection of human polyomaviruses BKV and JCV by LightCycler real-time PCR. J. Clin. Virol. 36:159-162. [DOI] [PubMed] [Google Scholar]
- 41.Shah, K. V. 1996. Polyomaviruses. Lippincott-Raven, Philadelphia, PA.
- 42.State of Florida. 2006. Surface water quality criteria. Florida Administrative Code 62-302.530. State of Florida Department of Environmental Protection, Tallahassee.
- 43.Stoeckel, D. M., and V. J. Harwood. 2007. Performance, design, and analysis in microbial source tracking studies. Appl. Environ. Microbiol. 73:2405-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szmant, A. 2002. Nutrient enrichment on coral reefs: is it a major cause of coral reef decline? Estuaries 25:743-766. [Google Scholar]
- 45.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, and C. P. Gerba. 2003. Chlorine inactivation of adenovirus type 40 and feline calicivirus. Appl. Environ. Microbiol. 69:3979-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, K. Riley, and C. P. Gerba. 2003. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 69:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Ufnar, J. A., S. Y. Wang, J. M. Christiansen, H. Yampara-Iquise, C. A. Carson, and R. D. Ellender. 2006. Detection of the nifH gene of Methanobrevibacter smithii: a potential tool to identify sewage pollution in recreational waters. J. Appl. Microbiol. 101:44-52. [DOI] [PubMed] [Google Scholar]
- 47.U.S. Environmental Protection Agency. 2003. Bacterial water quality standards for recreational waters (fresh and marine waters). EPA/823.R-03/008. U.S. Environmental Protection Agency, Office of Water, Washington, DC.
- 48.U.S. Environmental Protection Agency. 2005. Microbial source tracking guide document. EPA/600/R-05/064. U.S. Environmental Protection Agency, Office of Research and Development, Cincinnati, OH.
- 49.U.S. Environmental Protection Agency. 1986. Ambient water quality criteria for bacteria-1986. EPA440/5-84-002. U.S. Environmental Protection Agency, Washington, DC.
- 50.U.S. Environmental Protection Agency. 2005. Guidance for 2006 assessment, listing and reporting requirements pursuant to sections 303(d), 305(b), and 314 of the Clean Water Act. U.S. Environmental Protection Agency, Washington, DC.
- 51.U.S. Environmental Protection Agency. 1997. Method 1600: membrane filter test methods for enterococci in water. EPA-821/R-97/004. U.S. Environmental Protection Agency, Washington, DC.
- 52.U.S. Environmental Protection Agency. 2002. Method 1603: Escherichia coli in water by membrane filtration using modified membrane-thermotolerant E. coli agar (modified mTEC). U.S. Environmental Protection Agency, Washington, DC.
- 53.Reference deleted.
- 54.Vanchiere, J. A., R. K. Nicome, J. M. Greer, G. J. Demmler, and J. S. Butel. 2005. Frequent detection of polyomaviruses in stool samples from hospitalized children. J. Infect. Dis. 192:658-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanchiere, J. A., Z. S. White, and J. S. Butel. 2005. Detection of BK virus and simian virus 40 in the urine of healthy children. J. Med. Virol. 75:447-454. [DOI] [PubMed] [Google Scholar]
- 56.Whiley, D. M., K. E. Arden, I. M. Mackay, M. W. Syrmis, and T. P. Sloots. 2004. Simultaneous detection and differentiation of human polyomaviruses JC and BK by a rapid and sensitive PCR-ELAHA assay and a survey of the JCV subtypes within an Australian population. J. Med. Virol. 72:467-472. [DOI] [PubMed] [Google Scholar]
- 57.Whitman, R. L., K. Przybyla-Kelly, D. A. Shively, and M. N. Byappanahalli. 2007. Incidence of the enterococcal surface protein (esp) gene in human and animal fecal sources. Environ. Sci. Technol. 41:6090-6095. [DOI] [PubMed] [Google Scholar]
- 58.Wright, R. C. 1989. The survival patterns of selected faecal bacteria in tropical fresh waters. Epidemiol. Infect. 103:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yun, J. J., L. E. Heisler, I. I. Hwang, O. Wilkins, S. K. Lau, M. Hyrcza, B. Jayabalasingham, J. Jin, J. McLaurin, M. S. Tsao, and S. D. Der. 2006. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic Acids Res. 34:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, S., H. Y. Zheng, M. Suzuki, Q. Chen, H. Ikegaya, N. Aoki, S. Usuku, N. Kobayashi, S. Nukuzuma, Y. Yasuda, N. Kuniyoshi, Y. Yogo, and T. Kitamura. 2007. Age-related urinary excretion of BK polyomavirus by nonimmunocompromised individuals. J. Clin. Microbiol. 45:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]