Abstract
PCR-based methods have been developed to rapidly screen for Legionella pneumophila in water as an alternative to time-consuming culture techniques. However, these methods fail to discriminate between live and dead bacteria. Here, we report a viability assay (viability PCR [v-PCR]) for L. pneumophila that combines ethidium monoazide bromide with quantitative real-time PCR (qPCR). The ability of v-PCR to differentiate viable from nonviable L. pneumophila cells was confirmed with permeabilizing agents, toluene, or isopropanol. v-PCR suppressed more than 99.9% of the L. pneumophila PCR signal in nonviable cultures and was able to discriminate viable cells in mixed samples. A wide range of physiological states, from culturable to dead cells, was observed with 64 domestic hot-water samples after simultaneous quantification of L. pneumophila cells by v-PCR, conventional qPCR, and culture methods. v-PCR counts were equal to or higher than those obtained by culture and lower than or equal to conventional qPCR counts. v-PCR was used to successfully monitor in vitro the disinfection efficacy of heating to 70°C and glutaraldehyde and chlorine curative treatments. The v-PCR method appears to be a promising and rapid technique for enumerating L. pneumophila bacteria in water and, in comparison with conventional qPCR techniques used to monitor Legionella, has the advantage of selectively amplifying only viable cells.
Legionella organisms are ubiquitous bacteria found in many types of water sources in the environment. Their growth is especially favored in human-made warm water systems, including cooling towers, hot tubs, showerheads, and spas (3, 14, 15, 38). Legionella bacteria replicate as intracellular parasites of amoebae and persist in the environment as free-living microbes or in biofilms. In aerosol form, they enter the lungs and can cause an acute form of pneumonia known as Legionnaires' disease or a milder form of pulmonary infection called Pontiac fever. The species Legionella pneumophila is responsible for the vast majority of the most severe form of this atypical pneumonia (52, 70). Legionellosis outbreaks are associated with high mortality rates (15 to 20%) (15, 16, 38, 46), which can reach up to 50% for people with weakened immune systems (immunocompromised patients) (69). Legionella surveillance programs include regular monitoring of environmental water samples (9, 13, 66). It is generally acknowledged that Legionella represents a health risk to humans when cell densities are greater than 104 to 105 CFU per liter of water, and epidemiological data show that outbreaks of legionellosis occur at these concentrations (36, 47).
The evaluation of the risk associated with Legionella has traditionally been performed using culture-based methods (1, 24). Culture is essential for identifying and typing Legionella strains during epidemics. However, Legionella culture requires long incubation times (up to 10 days) before results can be scored. This problem makes culture unsuitable for preventive actions and rapid response in emergency situations. Moreover, under certain conditions (i.e., low-nutrient environments, oxidative or osmotic stress, etc.), Legionella cells can lose the ability to be cultured, although they are still viable (7, 17, 20, 22, 39, 45, 67). These viable but nonculturable (VBNC) Legionella cells may still represent a public health hazard because they can regain their ability to grow in new, more favorable conditions (12, 19, 23, 61).
Molecular approaches, such as quantitative real-time PCR (qPCR), are faster and can mitigate the main drawbacks of culture-based methods. qPCR is an alternative tool that offers rapid, sensitive, and specific detection of Legionella bacteria in environmental water samples (4, 5, 12, 26, 65, 68). PCR results can be obtained in hours instead of days, and VBNC Legionella cells can also be detected (12, 26). However, the major disadvantage of qPCR lies in its inability to evaluate viability due to the persistence of DNA in cells after death (27, 34). The monitoring of Legionella contamination levels by conventional qPCR may thus result in an overestimation of the risk of infection because false-positive results can be scored. However, the real risk from Legionella is limited to the live fraction of the total Legionella population. Only live or viable Legionella cells are able to replicate in pulmonary macrophages and cause severe pneumonia (14, 15). The development of more rapid, culture-independent methods capable of discriminating between live and dead cells is of major interest for measuring Legionella infection risks and preventing legionellosis. The nucleic acid-binding dye ethidium monoazide bromide (EMA), used in combination with qPCR, is an attractive alternative for selectively detecting and enumerating viable bacteria. EMA is particularly useful because it selectively penetrates cells with damaged membranes and covalently binds to DNA after photoactivation (21, 53). DNA-bound EMA molecules prevent PCR amplification and thereby lead to a strong signal reduction during qPCR. DNA from viable cells with intact cell membranes prevents EMA molecules from entering the cell and therefore can be amplified and quantified (56). Nocker et al. (41, 42) suggested that the signal reduction was due to a selective loss of genomic DNA from dead cells (rendered insoluble after cross-linkage) during the DNA extraction procedure rather than to PCR inhibition. However, Soejima et al. (59, 60) recently reported that treatment with EMA followed by visible light irradiation directly cleaves the chromosomal DNA of dead bacteria.
In this study we optimized the EMA-staining procedure in conjunction with qPCR with pure cultures of L. pneumophila. We analyzed the potential for the EMA-qPCR method to discriminate Legionella cells with compromised or intact cell membranes. We optimized this EMA-qPCR technique, viability PCR, hereafter named v-PCR, and used it to quantify viable Legionella cells in environmental water samples. We compared our results with those obtained by conventional qPCR and culture methods. In addition, we evaluated the ability of v-PCR to monitor the efficacy of different disinfection strategies.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Legionella strains used in this study were as follows: L. pneumophila strain Philadelphia-1 (ATCC 33152), L. pneumophila strain Chicago-2 (ATCC 33215), L. pneumophila strain Los Angeles-1 (ATCC 33156), L. anisa (ATCC 35292), L. gormanii (ATCC 33297), L. longbeachae (ATCC 33462), L. dumoffii (NCTC 11380), and L. bozemanii (ATCC 33217). Bacteria were grown at 37°C on buffered charcoal yeast extract (BCYE) agar supplemented with l-cysteine and ferric pyrophosphate (Oxoid Ltd., Basingstoke, United Kingdom).
Liquid cultures were prepared from single colonies of Legionella transferred to 10 ml of LB medium (Lennox L broth) (Invitrogen, Paisley, United Kingdom) containing Legionella growth supplements (Oxoid Ltd.). Cultures were grown at 37°C in an orbital shaker incubator. Cultures were then decimally diluted in 0.85% NaCl, and the number of CFU was determined by spreading 0.1 ml of appropriate dilutions onto duplicate plates of BCYE agar and incubating at 37°C for 6 days.
EMA treatment.
Solid EMA was purchased from Molecular Probes (Eugene, OR) and dissolved in dimethyl sulfoxide in the absence of light. Aliquots of 20 μl of 5-mg/ml EMA stock solution were stored at −20°C in brown, light-resistant microcentrifuge tubes until needed.
To investigate the effect of EMA on Legionella viability, a liquid culture of L. pneumophila sg1 was diluted in sterile 0.85% NaCl, as described above, and split into two fractions of identical volumes. One fraction was not heated. The other fraction was boiled for 15 min in a water bath to kill all Legionella cells. Cells were immediately placed on ice after heating. The efficacy of heat treatment was checked by plating on BCYE medium. A dilution of each fraction was subjected to EMA treatment. An aliquot from the unheated fraction was used as a control and was not treated with EMA, but the same amount of dimethyl sulfoxide that was used for the EMA-treated samples was added.
To determine the optimal concentration of EMA, dilutions of 1-ml aliquots from the treated heated and nonheated fractions were incubated in duplicate with 2.5, 5, 10, or 100 μg/ml of EMA for 10 min. After EMA treatment, tubes were exposed to a halogen light source (Osram Lum Halostar 300/500 W) to photo-cross-link EMA to DNA. During light exposure, samples were placed on ice to avoid excessive heating. To determine the optimal duration of light exposure necessary for cross-linking between EMA and DNA from dead cells, treated aliquots at each EMA concentration were exposed in duplicate to halogen light for 1 or 15 min. Control samples were also exposed to light.
EMA-treated samples and untreated controls were washed by centrifugation (16,000 × g, 5 min) with 0.85% NaCl. Pellets were resuspended in 30 to 40 μl (depending on the experiment) of the corresponding residual supernatants before DNA extraction.
DNA isolation.
After cells were washed following the EMA treatment, the lysis buffer InstaGene matrix (Bio-Rad Laboratories, Hercules, CA) was added to the sample to complete bacterial lysis and carry out subsequent DNA extraction. Tubes were heated in a boiling water bath for 15 min, then stored at −20°C for 30 min, and finally returned to 98°C for 15 min. After centrifugation, 5 μl of the resulting supernatant was used for qPCR analysis. Each sample was deposited in duplicate on each PCR plate.
Quantification by qPCR.
qPCR was performed with an iCycler iQ thermal cycler (Bio-Rad) in 96-well plates. DNA amplifications were performed using the iQ-Check Quanti L. pneumophila or iQ-Check Quanti Legionella species kits (Bio-Rad) according to the manufacturer's instructions. The validity of the quantification standard series and of the negative controls was checked. The iCycler iQ software automatically monitors the quantity of genomic units (GU) present in the sample. The cycle threshold value of the sample is compared with those of the standards by means of a standard curve. The values given for each sample in the iCycler iQ report correspond to the initial quantity of Legionella GU present in the 5-μl DNA samples transferred to each PCR well. To obtain the concentration of Legionella cells in GU/ml, the mean of the two duplicates (as calculated by the software) was multiplied by a factor representing the fraction of sample volume transferred to each well. The effect of EMA treatments was calculated by subtracting the GU/ml values (expressed in log units) of EMA-treated samples from the corresponding GU/ml values of untreated samples. The result of this subtraction represents the concentration of dead cells in the EMA-treated samples that were not amplified.
Discrimination of viable L. pneumophila cells in a mixture of viable and dead cells by v-PCR.
To check the performance of the v-PCR approach as a bacterial viability diagnostic tool, mixtures of viable and dead L. pneumophila cells were used. Heated and unheated samples were prepared from fresh diluted cultures of L. pneumophila. Tenfold dilution series were prepared for each sample. Variable numbers of viable L. pneumophila cells were mixed in defined ratios (1:1, 1:0.1, 1:0.01, and 1:0.001) with a constant number of heat-killed L. pneumophila cells and vice versa.
Membrane-permeabilizing treatments.
Two membrane-permeabilizing agents were used: isopropyl alcohol and toluene. These compounds are known to disrupt cell membranes (25, 54). By generating holes in the membrane, these agents should allow the EMA molecule to pass into the cell and covalently link to DNA, preventing amplification of DNA from dead cells. Toluene treatments at concentrations of 0.1%, 1%, and 5% were performed by adding 1, 10, and 50 μl of toluene per 1 ml of sample, respectively. Cells were exposed to toluene for 60 min at room temperature. Toluenized cells were then harvested by centrifugation and resuspended in 1 ml of 0.85% NaCl. EMA-treated samples were treated prior to DNA extraction. Cells were resuspended in 70% isopropyl alcohol and allowed to stand at room temperature for 1 h. The bacterial suspension was subsequently washed by centrifugation and resuspended in 1 ml of 0.85% NaCl before v-PCR was performed.
Application of the v-PCR method to environmental water samples.
Isolation of Legionella bacteria from natural water systems was performed as previously described (10). Briefly, 1 liter of sampled water was filtered on a 0.4-μm-pore-diameter polycarbonate membrane (Isopore; Millipore, Ireland). After filtration, bacteria collected on the membranes were resuspended in 5 ml of the sample filtrate by sonication. From this concentrated suspension, 3.1 ml was employed for culture analysis according to the 2003 NF T90-431 standardized method elaborated by the French Standardization Agency (AFNOR) (1), which conforms to the 1998 international standard ISO 11731 (24). The remaining 1.9 ml of the suspension was stored at 4°C until further analysis. Water samples in which Legionella was detected using standard culture procedures were analyzed with v-PCR and conventional qPCR (2) (EMA-untreated samples) using 0.5 ml of this stored fraction. Previous experiments had shown that defined conditions for v-PCR were also valid using 1 ml or 0.5 ml of concentrated sample (data not shown). To determine the actual correspondence between molecular and culture counts, 0.1 ml of appropriate dilutions of the stored fraction was spread on duplicate plates of glycine, vancomycin, polymyxin B, and cycloheximide (GVPC medium; Oxoid Ltd.) the same day that molecular analyses were carried out. Plates were incubated at 37°C for 10 days.
Disinfection treatments. (i) Thermal disinfection.
Thermal treatment is widely used as a disinfection method because Legionella was found to be almost instantly inactivated at >70°C (6, 29). A pure suspension spiked with 1.6 × 106 CFU/ml of L. pneumophila was treated at 70°C for 1, 3, and 5 h. Samples were simultaneously analyzed by qPCR, v-PCR, and culture before and after treatment to evaluate the effect of the disinfecting strategy on the total, viable, and culturable Legionella counts, respectively.
(ii) Chlorine treatment.
Chlorine was chosen as a representative of oxidizing biocides commonly used in disinfecting treatments of water. Although the mechanisms by which chlorine acts have not been elucidated completely, it is generally accepted that chlorine leads to perforation of the cell membrane (62, 63). Free-chlorine solution was freshly prepared by diluting commercial bleach (9.6% active chlorine) in sterile MilliQ water to obtain a 30-mg/liter working solution. The concentration of free chlorine was determined with a pocket colorimeter II analysis system (Hach test kit; Hach Co., Loveland, CO). A 0.1-mg/ml sodium thiosulfate stock solution was used for dechlorinating samples after treatment.
For chlorine experiments with L. pneumophila, a starved suspension was used in order to avoid chlorine consumption by organic matter. To prepare a mock environmental sample, 9 ml of the starved suspension was used to inoculate 1 liter of tap water. Ten bottles containing 100 ml of artificially contaminated tap water were made up. One bottle served to determine the initial L. pneumophila counts in the absence of chlorine using the v-PCR, qPCR, and culture methods. The rest of the bottles were used for the disinfection assay at different chlorine concentrations. Increasing volumes of the chlorine stock solution were added to different batches to achieve final concentrations comprising between 0.2 mg/liter and 1.6 mg/liter. The mixtures were incubated at room temperature in the dark for 60 min. After adding chlorine, 10 ml was taken from each assay bottle to measure free and total chlorine concentrations using the chlorine colorimeter kit. Following treatment, 10 ml of each sample was transferred to a tube containing sodium thiosulfate (0.02-mg/ml final concentration) to neutralize any residual chlorine. Aliquots of 1 ml were then prepared and assayed using the qPCR, v-PCR, and culture methods. A control was run simultaneously with the disinfection assay to verify that the water sample used in the experiment did not show any chlorine demand. Free and total chlorine concentrations were measured at 0, 15, 30, and 60 min after addition of different concentrations of chlorine in the tap water in the absence of L. pneumophila.
To confirm results and to analyze chlorine effects over a longer period of time, a second disinfection experiment was performed using three concentrations of chlorine (0.2, 0.5, and 1 mg/liter) for 24 h before inactivation by sodium thiosulfate.
(iii) Glutaraldehyde treatment.
Glutaraldehyde is the nonoxidizing biocide most often used in cooling water systems (18, 29, 30). A suspension containing approximately 105 CFU/ml of L. pneumophila was exposed to 500 mg/liter glutaraldehyde—a concentration reported to be effective (29)—for 30 min and 1, 6, 24, and 48 h. After treatment, Legionella cells were harvested by centrifugation and washed before being treated with EMA. Efficacy of the glutaraldehyde treatment as a biocide was assessed using plate counts on GVPC medium to confirm the absence of colonies.
Data analysis.
Standard deviations were calculated for the means of duplicate or triplicate counts. For the repeatability analysis, standard deviations were calculated from five independent replicates, and variability (the coefficient of variation) was calculated and reported as standard errors of the mean.
RESULTS
Optimization of the EMA protocol with pure suspensions of L. pneumophila.
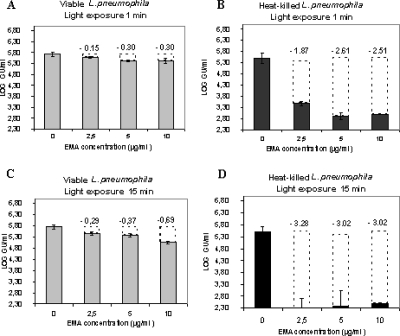
Various parameters, namely, EMA concentration and light exposure time, were optimized to selectively discriminate viable from dead L. pneumophila strain Philadelphia-1 cells. First, we used the experimental conditions established by Nogva et al. (43) and Rudi et al. (56) for discrimination between viable and dead food-borne bacteria, i.e., a treatment with 100 μg/ml EMA followed by exposure to halogen light for 1 min. In this study, this concentration was not suitable. Not only did it significantly inhibit amplification in heat-killed bacteria, but it also greatly reduced the PCR signal (by more than 3 log units) in the viable control (data not shown). We therefore then adapted the established protocol (Fig. 1). A L. pneumophila suspension containing approximately 1.8 × 105 CFU/ml was treated in duplicate with 2.5, 5, and 10 μg/ml EMA for 10 min and exposed to a halogen light for 1 min in order to obtain the EMA cross-linking. Compared to untreated controls, the EMA treatment of viable samples reduced the PCR signal from qPCRs by 0.15 log units for the 2.5-μg/ml EMA treatment and 0.30 log units for the 5- and 10-μg/ml EMA treatments. In the case of heat-killed cells, the signal reductions were 1.87 log units, 2.61 log units, and 2.51 log units for the 2.5-, 5-, and 10-μg/ml EMA concentrations, respectively (Fig. 1A and B). Increasing light exposure time to 15 min resulted in a reduction of more than 3 log units in the PCR signal of dead cell suspensions (PCR signals were close to detection limits) compared to untreated samples (Fig. 1D). The extent of inhibition in viable suspensions varied with the EMA concentration used. Increasing concentrations of EMA resulted in proportional decreases in amplification (Fig. 1C). Generally, the EMA treatment led to a slight PCR signal reduction in the EMA-treated viable controls compared with the equivalent untreated sample, suggesting that a small fraction of dead cells was present in the initial culture. Comparison of viable Legionella counts determined by solid phase cytometry (37), using the vital dye Chemchrome V6, with counts obtained using conventional qPCR confirmed the existence of dead cells in the viable cultures (data not shown). Therefore, incubation with 2.5 μg/ml EMA followed by a 15-min light exposure constituted optimal conditions in which amplification of DNA derived from dead L. pneumophila cells (more than 99.9%) was considerably suppressed, while there was no significant inhibition of amplification of DNA from viable cells. Similar results were obtained using Legionella species other than L. pneumophila. Under our established conditions, v-PCR suppressed 99.9% of the PCR signal in nonviable cultures of L. anisa, L. longbeachae, and L. gormanii (data not shown).
FIG. 1.
Optimization of EMA concentrations and light exposure times. Viable (A and C) and heat-killed (B and D) L. pneumophila cells were exposed to different EMA concentrations (2.5, 5, and 10 μg/ml) before being cross-linked with halogen light for 1 or 15 min. Black bars represent heat-killed bacteria and gray bars represent unheated bacteria. Transparent dotted bars and numerical values presented are the PCR signal reductions (in log GU/ml) observed after EMA treatment. Error bars represent standard deviations from two independent replicates.
In order to confirm our preliminary results, we evaluated the repeatability of the v-PCR technique using our defined conditions. Five independent replicates of a viable L. pneumophila strain Philadelphia-1 suspension at a cell density of approximately 1.5 × 106 CFU/ml and its corresponding heat-killed suspension were analyzed by v-PCR using 2.5 μg/ml EMA and a 15-min light exposure. Results showed that the v-PCR method has good repeatability (Table 1). The EMA treatment led to a mean signal reduction of 0.19 log units in viable cells compared to a mean signal reduction of 3.36 log units in heat-killed cells. The standard deviations of the reduction signal (expressed in log GU/ml) were 0.03 and 0.16 for viable and dead L. pneumophila cells, respectively.
TABLE 1.
Repeatability of v-PCR method
| Sample | Viable L. pneumophilaa
|
Heat-killed L. pneumophilab
|
||
|---|---|---|---|---|
| log10 GU/ml | Signal reduction (log units) | log10 GU/ml | Signal reduction (log units) | |
| Untreated control | 6.5 | 6.31 | ||
| EMA-treated replicate | ||||
| 1 | 6.32 | 0.18 | 2.97 | 3.34 |
| 2 | 6.3 | 0.2 | 2.74 | 3.58 |
| 3 | 6.26 | 0.24 | 3.17 | 3.14 |
| 4 | 6.34 | 0.16 | 3.01 | 3.31 |
| 5 | 6.34 | 0.16 | 2.89 | 3.43 |
The mean signal reduction for viable cells was 0.19 log units. The standard deviation of the reduction signal (expressed in log GU/ml) was 0.03.
The mean signal reduction for heat-killed cells was 3.36 log units. The standard deviation of the reduction signal (expressed in log GU/ml) was 0.16.
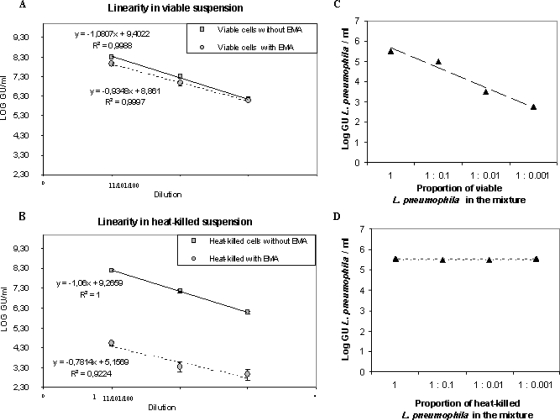
The influence of the total amount of Legionella cells present in the sample on the v-PCR results was also evaluated. For this, 10-fold serial dilutions of both unheated and heat-killed (boiled) suspensions of L. pneumophila were analyzed by v-PCR. The v-PCR method showed good linearity, i.e., PCR values were proportional to the viable or dead L. pneumophila concentrations (Fig. 2A and B). The relative detection of living and dead cells was therefore independent of cell concentration, at least in the range of tested concentrations (between 108 and 106 GU/ml). Moreover, v-PCR allowed the discrimination of viable L. pneumophila cells in mixtures containing defined ratios of viable and dead cells by v-PCR. When a variable number of viable L. pneumophila cells was mixed with a constant number of heat-killed L. pneumophila cells, the GU values determined using v-PCR increased with the proportion of viable L. pneumophila cells. The highest GU value was observed for the mixture containing equal proportions of heat-killed and viable cells (1:1). In contrast, when a constant number of viable (nonheated) L. pneumophila cells was mixed with a variable number of heat-killed L. pneumophila cells, no significant change in amplification was observed. The PCR signal appeared to be unaffected by the presence of dead L. pneumophila cells. Plots of cell concentrations (in log GU/ml) according to the proportion of viable and dead L. pneumophila cells present in mixtures are given in Fig. 2C and D, respectively.
FIG. 2.
Linearity of the v-PCR method and viable cell discrimination in mixed samples. PCR values obtained from viable (A) or heat-killed (B) serially diluted suspensions treated with EMA were compared with those obtained from untreated suspensions to analyze the influence of the total amount of Legionella cells on the v-PCR results. Error bars represent standard deviations from three independent replicates. Logs of GU values determined by v-PCR are shown as a function of the proportion of viable (C) or dead (D) L. pneumophila cells present in mixtures of defined ratios of viable and dead cells.
Permeabilizing treatments.
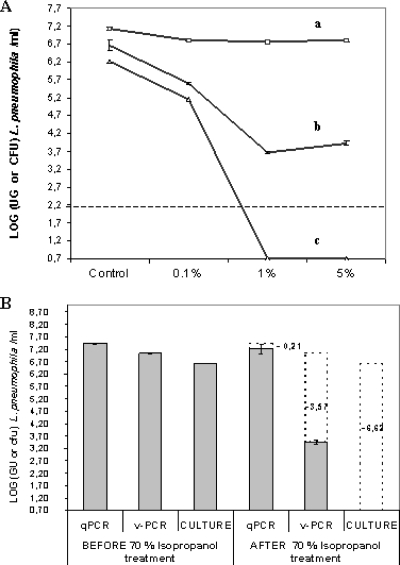
The influence of toluene on viable cell counts was evaluated using qPCR, v-PCR, and culture at three different toluene concentrations (Fig. 3A). When a suspension containing 1.61 × 106 CFU/ml L. pneumophila strain Philadelphia-1 was exposed to 0.1% toluene, the culturable cell count lowered to 5 × 105 CFU/ml. A reduction of 1 log unit was also observed using v-PCR at the same toluene concentration. The increase of the toluene concentration to 1% or 5% led to a dramatic decrease in cell viability, as measured by the ability to grow and form colonies on agar plates. No culturable L. pneumophila cells were observed after treatment at these concentrations. The EMA treatment decreased the amount of amplified DNA in toluene-treated samples up to 3 log units compared to untreated samples. As expected, the qPCR count remained constant, whatever the concentration of toluene.
FIG. 3.
Effect of permeabilizing treatments. (A) Effect of toluene treatment on L. pneumophila counts determined by qPCR (a), v-PCR (b), and culture (c) methods. L. pneumophila cells were exposed to 0.1%, 1%, or 5% toluene for 60 min. Control, without toluene treatment. The detection limit of the v-PCR assay is indicated by a dotted line. (B) Evaluation of culturable (culture) and viable (v-PCR) L. pneumophila counts compared with total count (qPCR) before and after permeabilization with 70% isopropyl alcohol. Signal reductions (in log) observed for each method after permeabilizing treatment compared with untreated control are indicated. Error bars represent standard deviations from two independent replicates.
The isopropyl alcohol treatment resulted in a reduction in viable cell numbers determined by standard plate counting and in a significant reduction in the PCR signal in cells that were stained with EMA. The results from the v-PCR method show that the EMA treatment suppressed more than 99.9% of DNA amplification from isopropyl alcohol-treated L. pneumophila strain Philadelphia-1, resulting in a reduction in copy number over 3.5 log units. In contrast, no significant differences between control and isopropyl alcohol-treated cell counts were observed using conventional qPCR (Fig. 3B). Comparable results were also obtained with two other L. pneumophila strains (strains Chicago-2 and Los Angeles-1) and four other Legionella species (L. anisa, L. dumoffii, L. bozemanii, and L. gormanii) (Table 2).
TABLE 2.
Effect of 70% isopropyl alcohol treatment on other Legionella strains and species assessed by qPCR and v-PCR
| Assay | Strain or species (strain designation)a | 70% isopropyl alcohol-killed Legionella
|
||
|---|---|---|---|---|
| Log10 GU/ml | Signal reduction (log units) | SD | ||
| qPCR | ||||
| L. pneumophila strain Los Angeles-1 (ATCC 33156) | 7.34 | 0.02 | ||
| L. pneumophila strain Chicago-2 (ATCC 33215) | 7.39 | 0.13 | ||
| L. anisa (ATCC 35292) | 7.34 | 0.03 | ||
| L. dumoffii (NCTC 11380) | 7.56 | 0.07 | ||
| L. bozemanii (ATCC 33217) | 6.87 | 0.11 | ||
| v-PCR | ||||
| L. pneumophila strain Los Angeles-1 (ATCC 33156) | 4.32 | 3.02 | 0.05 | |
| L. pneumophila strain Chicago-2 (ATCC 33215) | 4.3 | 3.09 | 0.01 | |
| L. anisa (ATCC 35292) | 4.26 | 3.08 | 0.07 | |
| L. dumoffii (NCTC 11380) | 4.08 | 3.48 | 0.28 | |
| L. bozemanii (ATCC 33217) | 3.72 | 3.12 | 0.01 | |
ATCC, American Type Culture Collection; NCTC, National Collection of Type Cultures.
Application of the v-PCR method to environmental water samples.
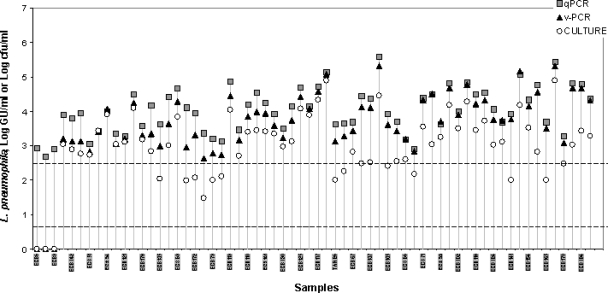
A total of 61 domestic hot-water samples yielding positive results by the standard culture method (1) were simultaneously analyzed by v-PCR, conventional qPCR, and culture methods to assess the correspondence between molecular and standard approaches. Three other samples that were deemed positive by conventional qPCR but negative by v-PCR and culture were also included in the analysis. Results of the evaluation of L. pneumophila determined for the various natural water samples are shown in Fig. 4. The three analytical methods were plotted in the same graph to facilitate comparisons, even though CFU values are not strictly comparable to GU values. As expected, qPCR values were higher than culture values for all samples analyzed. With the v-PCR technique, which removes the contribution of dead L. pneumophila cells through an EMA treatment, the viable counts varied from one sample to another one. Different patterns could be distinguished depending on results obtained using the three enumeration methods. One pattern was represented by three cases in which environmental samples were deemed positive by conventional qPCR but negative by v-PCR and culture methods. The second pattern was represented by eight samples in which the number of viable L. pneumophila cells assessed by v-PCR was very similar to the plate counts. In another six samples, counts determined by the three methods were nearly identical. However, the most common pattern was one in which v-PCR values were similar to qPCR values or intermediate between culture and qPCR counts. It should be noted that culture values used for this comparative study may be biased because they were obtained several days after sampling.
FIG. 4.
Application of the EMA-qPCR procedure to environmental water samples. The logarithms of concentrations of L. pneumophila in 64 tap water samples determined by v-PCR method (triangles) are compared to standard qPCR (squares) and culture (circles). The detection limit of the v-PCR assays is indicated by a dotted line.
The sensitivity of the v-PCR method depends on the volume of water analyzed and the number of replicates used per PCR plate. Under the experimental conditions used in this study the detection limit of the v-PCR method was 200 GU/ml for 1 ml of sample treated with EMA and two replicates per PCR plate. However, for routine applications, the detection limit is reduced to 250 GU/liter if 1 liter of sample water is used for each v-PCR assay.
Evaluation of efficacy of disinfection treatments.
To control Legionella and maintain good water quality, thermal or chemical disinfection treatments are currently employed. We checked the capacity of the v-PCR method to assess the efficacy of the most common disinfection treatments used for the inactivation of Legionella bacteria, namely, heat, chlorine, and glutaraldehyde.
(i) Thermal disinfection.
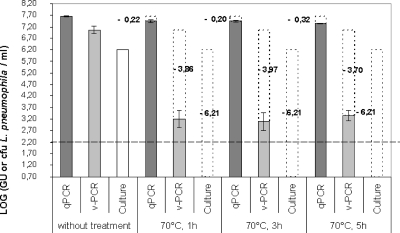
The results of the three detection methods with respect to treatment duration at 70°C are shown in Fig. 5. As expected, no Legionella cells were detected by culture after 1 h of thermal treatment. The viable Legionella count as determined by v-PCR dropped nearly 4 log units following the treatment, while the qPCR count remained practically constant before and after treatment.
FIG. 5.
Monitoring of culturable, viable, and total counts of L. pneumophila cells after exposition to 70°C for 1 h, 3 h, and 5 h. Transparent dotted bars indicate loss of culturability, as determined by plate count, or PCR signal reduction, as determined by v-PCR or conventional qPCR after thermal disinfection treatment. Log GU/ml values derived from EMA-treated suspensions were subtracted from the corresponding GU/ml values of untreated suspensions. The dotted line indicates the limit of detection of the v-PCR assay. Error bars represent standard deviations from two independent replicates.
(ii) Chlorine.
The purpose of the assay was to compare the L. pneumophila enumerations obtained using v-PCR with conventional qPCR and culture counts in artificially contaminated chlorinated tap water. Concentrations of total, viable, and culturable L. pneumophila cells in the spiked water samples were 7.6 × 106 GU/ml, 2.15 × 105 GU/ml, and 9.45 × 104 CFU/ml, respectively. A notable difference (>1.5 log units) in the number of GU determined by qPCR and v-PCR was observed, suggesting the presence of injured or dead cells in the initial inoculum.
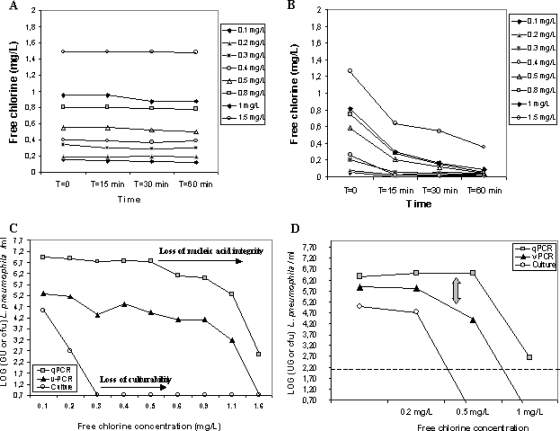
The tap water used for spiking initially contained 0.1 mg/liter free chlorine. Chlorination was carried out using concentrations of free chlorine ranging from 0.2 to 1.6 mg/liter. The kinetics of chlorine consumption over a 60-min period showed that free-chlorine concentrations in controls without bacteria remained constant, confirming that tap water used for the assay had no chlorine demand (Fig. 6A). However, the kinetics of chlorine consumption in the assay bottles showed a drop in chlorine, as a consequence of consumption by bacteria (Fig. 6B). All the plots of chlorine consumption showed a rapid decrease in chlorine concentration during the first 15 min of the assay, followed by a gradual decline.
FIG. 6.
Chlorination. Chlorine consumption over time in the tap water sample used without addition (A) of L. pneumophila and with addition (B) of L. pneumophila. (C) Effect of increasing chlorine doses on L. pneumophila counts, as assessed by qPCR (squares), v-PCR (triangles), and culture (circles) methods after 60 min of treatment. (D) Effect of 0.2 mg/liter, 0.5 mg/liter, and 1 mg/liter of chlorine after 24 h of treatment. The dotted line indicates the detection limit of the v-PCR assay.
Figure 6C summarizes the kinetics of enumerations of L. pneumophila cells using the three detection methods as a function of chlorine dose. As expected, increased chlorine doses resulted in decreased culture counts. A gradual decay in culture enumerations was observed at 0.2 mg/liter chlorine, and the application of a concentration of 0.3 mg/liter chlorine resulted in a total loss of culturability of L. pneumophila. Low concentrations of chlorine did not affect v-PCR or qPCR counts. The enumerations obtained using the two molecular methods remained constant until a threshold chlorine dose (ca. 0.5 mg/liter) was reached, after which counts dropped proportionally with chlorine concentration. It seems that from this dose upwards, chlorine diffused inside cells and induced damages that affected v-PCR as well as conventional qPCR amplification.
In a similar experiment, a tap-water sample was spiked with 1 × 105 CFU/liter of L. pneumophila, and the effect of 0.2, 0.5, and 1 mg/liter of chlorine was measured after 24 h of treatment (Fig. 6D). Culture counts began to fall at 0.2 mg/liter, and no L. pneumophila growth was observed at 0.5 mg/liter, confirming previous results. At a chlorine dose as low as 0.2 mg/liter, the kinetics of inactivation determined by v-PCR and qPCR were parallel, i.e., no reductions in GU counts were observed compared to the nonchlorinated controls, as previously observed. However, a slight difference with regard to the precedent experiment was observed at a concentration of 0.5 mg/liter. At this dose, a gap between viable and total counts was observed, and v-PCR counts dropped, whereas qPCR counts remained constant. At higher chlorine concentrations (1 mg/liter), both enumerations simultaneously dropped. These results suggest that when the threshold dose of 0.5 mg/liter chlorine is reached, EMA molecules are able to penetrate into the cells and to bind to DNA, partially preventing subsequent amplification. At higher concentrations, damages induced by chlorine seem to affect the nucleic acid integrity and their amplification with either molecular method.
(iii) Glutaraldehyde.
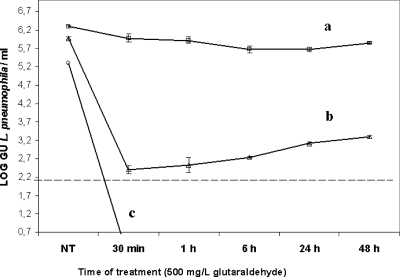
Survival curves of glutaraldehyde-treated L. pneumophila cells evaluated using culture, v-PCR, and conventional qPCR are shown in Fig. 7. No culturable L. pneumophila cells were recovered after a 30-min treatment, while qPCR counts remained invariable during the experiment. A rapid drop in viable cell concentration (over 99.9%) was observed by v-PCR during the first 30 min of the assay and was followed by a gradual increase (up to 1 log unit) in cell concentrations at 48 h of treatment. These results suggest that glutaraldehyde may cause reversible injury of L. pneumophila cells.
FIG. 7.
Survival curves of L. pneumophila cells after a 500-mg/liter glutaraldehyde disinfecting treatment evaluated by qPCR (a), v-PCR (b), and culture (c) methods. Bars represent the standard errors of the means of two independent experiments. The dotted line indicates the detection limit of the v-PCR assay.
DISCUSSION
In this study, we evaluated the potential usefulness of the v-PCR technique for discriminating viable from nonviable Legionella bacteria in water samples. One of the criteria for distinguishing viable bacteria is membrane integrity (28). Live cells with intact membranes are distinguished by their ability to exclude DNA-binding dyes, such as EMA, which can easily enter dead cells with compromised membranes. When EMA staining is combined with qPCR, selective quantification of the viable fraction of a mixed bacterial population is possible. This technique was initially employed by Nogva et al. (43) and Rudi et al. (55, 56) to differentiate between viable and dead cells of Escherichia coli, Salmonella spp., Listeria monocytogenes, and Campylobacter jejuni. Since then, the EMA-qPCR approach has been used to discriminately quantify viable cells of other bacterial types (8, 41, 51, 57, 64). The standardized protocol initially established by Nogva et al. (43) and Rudi et al. (55, 56) was not suitable to effectively discriminate live from dead Legionella cells. In this study, we developed a protocol better adapted to L. pneumophila and other Legionella spp. The optimal conditions that we defined here resulted in a PCR signal reduction of more than 99.9% in pure nonviable Legionella cultures that had been heat killed. Experiments involving mixed samples and permeabilizing compounds confirmed the selective discrimination of viable L. pneumophila by v-PCR.
Quantitative determination of viable L. pneumophila cells in environmental water samples.
The performance of the v-PCR technique was investigated with 64 domestic hot-water samples, most of them previously determined as positive by culture. v-PCR counts were compared to culture and conventional qPCR counts. The differences observed among the three techniques, depending on the water sample considered, might reflect the diverse physiological states in which Legionella cells can be found in the environment. The three analytical techniques are based on different cell properties (culturability, intact cells with amplifiable DNA, or damaged or intact cells with amplifiable DNA). Therefore, negative culture and v-PCR results observed in conjunction with a positive signal obtained by conventional qPCR suggest that in those samples, L. pneumophila cells have gone into a death phase and the qPCR signal corresponds to DNA amplification from dead or lethally damaged Legionella cells. In these kinds of samples, the potentially infectious fraction of L. pneumophila (live) bacteria was overestimated by conventional qPCR. In contrast, patterns for which v-PCR counts were between those for culture and qPCR, or in which v-PCR counts were consistently higher than those for culture, suggest that a substantial proportion of Legionella cells were stressed or in a VBNC state. The existence of Legionella doublets or chains which are counted as only 1 CFU by the culture method but quantified as individual cells by v-PCR may also explain the difference in Legionella concentrations between these techniques. Moreover, growth of Legionella on culture medium can be inhibited by the presence of other microorganisms which do not influence the v-PCR and qPCR counts. The third pattern of environmental samples showing similar culture, v-PCR, and qPCR counts may indicate that Legionella cells are in an exponential phase. Taking into account the different categories of Legionella cells, there is a priori no fundamental reason why v-PCR results should match culture results, because as noted above, each approach measures different cellular properties. It is not appropriate to try to seek correlation between v-PCR and culture nor to establish a relationship between the number of GU and the number of CFU. The main advantage of the v-PCR technique does not lie in providing the same information as culture techniques but in being more informative than the conventional qPCR technique in terms of the cell viability of Legionella.
Assessment of the efficacy of various disinfectants against Legionella using v-PCR.
Methods used to determine disinfection efficacy have traditionally been based on viability assays using conventional plate counting. A problem associated with this practice is that bacteria may become unable to form colonies on selective media because of reversible injury or because they enter into a VBNC state (17, 20, 31, 33, 35). Concerning Legionella, the monitoring of the disinfection efficacy using culture techniques requires prolonged incubation periods due to the low growth rate of these bacteria. In this context, v-PCR would identify the impact of curative treatments and provide more precise information on the physiological consequences of disinfectants more quickly than culture techniques. Disinfection by heating at 70°C is the most common strategy for controlling legionellae in hot water systems. Results obtained in this work show that the v-PCR approach successfully monitors in vitro disinfection based on thermal treatment, leading to a reduction of more than 99.9% of the L. pneumophila PCR signal in spiked samples, in contrast with conventional qPCR.
Chlorine is generally considered to be a nonselective oxidant which reacts with a variety of cellular components, such as the cytoplasmic membrane (32, 63). We evaluated the v-PCR technique for assessing the effectiveness of chlorination. The dose-effect relationship we observed (Fig. 6C) closely resembles that reported by Phe et al. (49) using nucleic acid fluorochromes and flow cytometry in chlorinated drinking water samples. These authors also showed a loss of culturability followed by nucleic acid damage, which inhibited staining with nucleic acid dyes when the chlorine level increased. Our v-PCR results were unexpected if we admit that membrane permeabilization occurs early after chlorine addition, as suggested by the absence of culturable cells with small amounts of chlorine. To explain this fact, two hypotheses can be formulated. The first hypothesis considers that the membrane is not permeabilized at low chorine levels and the loss of culturability might be a response of bacteria to environmental stress. L. pneumophila cells are stressed or partially injured rather than inactivated. Recent studies (12, 19) have reported that chlorination induced formation of VBNC L. pneumophila cells, the chlorine-treated samples being able to recover their culturability after infecting Acanthamoeba polyphaga. In the second hypothesis, we could speculate that membrane permeabilization occurs early, but holes induced by low chlorine concentrations are not large enough to allow EMA to enter into the cells and there is no reduction in the PCR signal. It has been reported that in cells with minimal membrane damage, the EMA-related compound propidium iodide has limited access to the cytoplasm (32). The concentration of 0.5 mg/liter of chlorine appeared as a threshold dose at which a difference between v-PCR and qPCR results was observed, suggesting that the degree of permeabilization of the membrane was enough to allow discrimination between live and dead cells by the v-PCR technique. At higher concentrations (>0.5 mg/liter), findings derived from v-PCR results were the same as those derived from qPCR results, although the initial difference between v-PCR and qPCR counts was maintained. At high doses, chlorine diffuses inside bacteria and exerts deleterious oxidant effects on nucleic acids (DNA damage), obviously having the same consequences for amplification in both v-PCR and qPCR techniques. Lethal DNA damage caused by chlorine has been reported previously (11, 48, 49, 50, 58). We can deduce from our results that the discrimination between live and dead cells using v-PCR after chlorination requires efficient diffusion of EMA dye through the bacterial membrane (hole sizes) and DNA integrity to allow binding.
One of the most extensively used nonoxidizing biocides is glutaraldehyde, which has been used widely to inactivate Legionella bacteria (18, 29). Among the biological mechanisms of action by which glutaraldehyde kills microorganisms, the interaction with outer membrane lipoproteins has been proposed (18). Survival curves of L. pneumophila cells treated with different concentrations of glutaraldehyde, evaluated using culture and v-PCR, demonstrated that the biocide acted rapidly. As expected, qPCR counts remained invariable after 48 h of treatment, confirming this technique's drawback of generating false-positive results. An interesting finding derived from v-PCR data is the recovery of some L. pneumophila cells after exposure to glutaraldehyde. This observation suggests that a fraction of L. pneumophila cells survived the glutaraldehyde treatment and were in a reversible injured state. If glutaradehyde-injured L. pneumophila cells are able to partially repair their damaged membranes, EMA is prevented from entering the cell, leading to a subsequent increase in the v-PCR signal. The occurrence of reversible injury caused by glutaraldehyde may explain the nonlasting effect of this biocide. The ability of injured cells to seal or to repair cellular membrane damage and recover under suitable conditions has been reported elsewhere (33, 40, 44). From a sanitary risk point of view, sublethally injured Legionella cells induced by glutaraldehyde may be capable of recovering and recolonizing water systems.
In conclusion, v-PCR represents an innovative technique that has advantages over the two other important techniques of Legionella enumeration: it can minimize false-positive results (positive signal for Legionella, although bacteria are not viable) compared to the conventional qPCR method and give rapid results compared to the current time-consuming culture method. EMA-treated samples should be more representative of the potentially pathogenic L. pneumophila population than those evaluated by qPCR. v-PCR may provide a tool for establishing an association between Legionella concentration and risk of disease because it detects culturable as well as VBNC Legionella cells. The application of v-PCR to Legionella shows promise as an excellent diagnostic assay for routine monitoring of the risk of legionellosis and a tool for providing rapid responses in emergency situations. Moreover, the v-PCR approach could allow us to better understand the way in which biocides kill cells and could help optimize disinfection strategies. Field studies would help further establish the v-PCR technique as a routine tool for regulatory monitoring of facilities and evaluation of the curative measures in human-made environments. In this context, validation of the v-PCR method is under way in combination with standardized culture and qPCR for monitoring Legionella density in a representative number of cooling towers.
Acknowledgments
This work was partially supported by the AINF (Association Interprofessionelle du Nord de la France).
We thank the Legionella Laboratory of the Institut Pasteur of Lille, France, for providing environmental samples and G. Marchand and F. Le Broc for technical assistance.
Footnotes
Published ahead of print on 10 April 2009.
REFERENCES
- 1.Association Française de Normalisation. 2003. Water quality. Detection and enumeration of Legionella spp. and Legionella pneumophila. Method by direct inoculation and after concentration by membrane filtration or centrifugation. French standard AFNOR NF T90-431. http://www.boutique.afnor.fr/boutique.asp.
- 2.Association Française de Normalisation. 2006. Water quality. Detection and quantification of Legionella and/or Legionella pneumophila by concentration and genic amplication by polymerase chain reaction (PCR). French standard AFNOR XP T90-471. http://www.boutique.afnor.fr/boutique.asp.
- 3.Atlas, R. M. 1999. Legionella: from environmental habitats to disease pathology, detection and control. Environ. Microbiol. 1:283-293. [DOI] [PubMed] [Google Scholar]
- 4.Ballard, A. L., N. K. Fry, L. Chan, S. B. Surman, J. V. Lee, T. G. Harrison, and K. J. Towner. 2000. Detection of Legionella pneumophila using a real-time PCR hybridization assay. J. Clin. Microbiol. 38:4215-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behets, J., P. Declerck, Y. Delaedt, B. Creemers, and F. Ollevier. 2007. Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J. Microbiol. Methods 68:137-144. [DOI] [PubMed] [Google Scholar]
- 6.Best, M., A. Goetz, and V. L. Yu. 1984. Heat eradication measures for control of nosocomial Legionnaires' disease. Implementation, education, and cost analysis. Am. J. Infect. Control 12:26-30. [DOI] [PubMed] [Google Scholar]
- 7.Byrd, J. J., H. S. Xu, and R. R. Colwell. 1991. Viable but nonculturable bacteria in drinking water. Appl. Environ. Microbiol. 57:875-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, B., K. Sugiyama, T. Taguri, J. Amemura-Maekawa, F. Kura, and H. Watanabe. 31 October 2008. Specific detection of viable Legionella cells by combined use of photoactivated ethidium monoazide and PCR/real-time PCR. Appl. Environ. Microbiol. doi: 10.1128/AEM.00604-08. [DOI] [PMC free article] [PubMed]
- 9.Conseil Supérieur d'Hygiène Publique de France. 2005. Le risque lié aux légionelles. Guide d'investigation et d'aide à la gestion. Rapport du Conseil Supérieur d'Hygiène Publique de France. http://www.sante.gouv.fr/htm/pointsur/legionellose/guid2005.pdf.
- 10.Delgado-Viscogliosi, P., T. Simonart, V. Parent, G. Marchand, M. Dobbelaere, E. Pierlot, V. Pierzo, F. Menard-Szczebara, E. Gaudard-Ferveur, K. Delabre, and J. M. Delattre. 2005. Rapid method for enumeration of viable Legionella pneumophila and other Legionella spp. in water. Appl. Environ. Microbiol. 71:4086-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dukan, S., and D. Touati. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 178:6145-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dusserre, E., C. Ginevra, S. Hallier-Soulier, F. Vandenesch, G. Festoc, J. Etienne, S. Jarraud, and M. Molmeret. 2008. A PCR-based method for monitoring Legionella pneumophila in water samples detects viable but noncultivable legionellae that can recover their cultivability. Appl. Environ. Microbiol. 74:4817-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Surveillance Scheme for Travel Associated Legionnaires' Disease and the European Working Group for Legionella Infections. 2005. European guidelines for control and prevention of travel associated Legionnaires' disease. Treatment methods. http://www.ewgli.org/data/european_guidelines/european_guidelines_jan05.pdf.
- 14.Fields, B. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 15.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, D. W., T. R. Tsai, W. Orenstein, W. E. Parkin, H. J. Beecham, R. G. Sharrar, J. Harris, G. F. Mallison, S. M. Martin, J. E. McDade, C. C. Shepar, and P. S. Brachman. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189-1197. [DOI] [PubMed] [Google Scholar]
- 17.Fry, J. C., and T. Zia. 1982. A method for estimating viability of aquatic bacteria by slide culture. J. Appl. Bacteriol. 53:189-198. [Google Scholar]
- 18.Ganzer, G. A. 2001. Glutaraldehyde: a versatile microbiocide for use in water treatment applications. Analyst VIII 2:23-28. [Google Scholar]
- 19.García, M. T., S. Jones, C. Pelaz, R. D. Millar, and Y. Abu Kwaik. 2007. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. 9:1267-1277. [DOI] [PubMed] [Google Scholar]
- 20.Hay, J., D. V. Seal, B. Billcliffe, and J. H. Freer. 1995. Non-culturable Legionella pneumophila associated with Acanthamoeba castellanii: detection of the bacterium using DNA amplification and hybridization. J. Appl. Bacteriol. 78:61-65. [DOI] [PubMed] [Google Scholar]
- 21.Hixon, S. C., W. E. White, and K. L. Yielding. 1975. Selective covalent binding of an ethidium analog to mitochondrial DNA with production of petite mutants in yeast by photoaffinity labeling. J. Mol. Biol. 92:319-329. [DOI] [PubMed] [Google Scholar]
- 22.Hussong, D., R. R. Colwell, M. O. O'Brien, E. Weiss, A. D. Pearson, R. M. Weiner, and W. D. Burge. 1987. Viable Legionella pneumophila not detectable by culture on agar media. BioTechnology 5:947-950. [Google Scholar]
- 23.Hwang, M. G., H. Katayama, and S. Ohgaki. 2006. Effect of intracellular resuscitation of Legionella pneumophila in Acanthamoeba polyphage cells on the antimicrobial properties of silver and copper. Environ. Sci. Technol. 10:7434-7439. [DOI] [PubMed] [Google Scholar]
- 24.International Standards Organisation. 1998. Water quality-detection and enumeration of Legionella. International standard ISO 11731. International Standards Organisation (International Organization for Standardization), Geneva, Switzerland.
- 25.Jackson, R. W., and J. A. DeMoss. 1965. Effects of toluene on Escherichia coli. J. Bacteriol. 90:1420-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joly, P., P. A. Falconnet, J. André, N. Weill, M. Reyrolle, F. Vandenesch, M. Maurin, J. Etienne, and S. Jarraud. 2006. Quantitative real-time Legionella PCR for environmental water samples: data interpretation. Appl. Environ. Microbiol. 72:2801-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joux, F., and P. Lebaron. 2000. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect. 2:1523-1535. [DOI] [PubMed] [Google Scholar]
- 29.Kim, B. R., J. E. Anderson, S. A. Mueller, W. A. Gaines, and A. M. Kendall. 2002. Literature review—efficacy of various disinfectants against Legionella in water systems. Water Res. 36:4433-4444. [DOI] [PubMed] [Google Scholar]
- 30.Leung, H. W. 2001. Aerobic and anaerobic metabolism of glutaraldehyde in river water-sediment systems. Arch. Environ. Contam. Toxicol. 41:267-273. [DOI] [PubMed] [Google Scholar]
- 31.Lisle, J. T., S. C. Broadaway, A. M. Prescott, B. H. Pyle, C. Fricker, and G. A. McFeters. 1998. Effects of starvation on physiological activity and chlorine disinfection resistance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lisle, J. T., B. H. Pyle, and G. A. McFeters. 1999. The use of multiple indices of physiological activity to access viability in chlorine disinfected Escherichia coli O157:H7. Lett. Appl. Microbiol. 29:42-47. [DOI] [PubMed] [Google Scholar]
- 33.Mackey, B. M. 2000. Injured bacteria, p. 315-341. In B. M. Lund, T. C. Baird-Parker, and G. W Gould (ed.), The microbiological safety and quality of foods. Aspen Publishers, Gaithersburg, MD.
- 34.Masters, C. I., J. A. Shallcross, and B. M. Mackey. 1994. Effect of stress treatments on the detection of Listeria monocytogenes and enterotoxigenic Escherichia coli by the polymerase chain reaction. J. Appl. Bacteriol. 77:73-79. [DOI] [PubMed] [Google Scholar]
- 35.McFeters, G. A. 1990. Enumeration, occurrence, and significance of injured indicator bacteria in drinking water, p. 478-492. In G. A. McFeters (ed.), Drinking water microbiology. Springer-Verlag, New York, NY.
- 36.Meenhorst, P. L., A. L. Reingold, D. G. Groothuis, G. W. Gorman, H. W. Wilkinson, R. M. McKinney, J. C. Feeley, D. J. Brenner, and R. van Furth. 1985. Water-related nosocomial pneumonia caused by Legionella pneumophila serogroups 1 and 10. J. Infect. Dis. 152:356-364. [DOI] [PubMed] [Google Scholar]
- 37.Mignon-Godefroy, K., J. C. Guillet, and C. Butor. 1997. Solid-phase cytometry for detection of rare events. Cytometry 27:336-344. [PubMed] [Google Scholar]
- 38.Miquel, P. H., S. Haeghebaert, D. Che, C. Campese, C. Guitard, T. Brigaud, M. Thérouanne, G. Panié, S. Jarraud, and D. Ilef. 2004. Epidémie communautaire de légionellose, Pas-de-Calais, France, Novembre 2003-Janvier 2004. Bull. Epidemiol. Hebd. 37:179-181. [Google Scholar]
- 39.Murga, R., T. S. Forster, E. Brown, J. M. Pruckler, B. S. Fields, and R. M. Donlan. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121-3126. [DOI] [PubMed] [Google Scholar]
- 40.Nebe-von Caron, G., P. Stephens, and R. A. Badley. 1998. Assessment of bacterial viability status by flow cytometry and single cell sorting. J. Appl. Microbiol. 84:988-998. [DOI] [PubMed] [Google Scholar]
- 41.Nocker, A., C. Y. Cheung, and A. K. Camper. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310-320. [DOI] [PubMed] [Google Scholar]
- 42.Nocker, A., and A. K. Camper. 2006. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72:1997-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogva, H. K., S. M. Dromtorp, H. Nissen, and K. Rudi. 2003. Ethidium monoazide for DNA-based differentiation of viable and dead bacteria by 5′-nuclease PCR. BioTechniques 34:804-813. [DOI] [PubMed] [Google Scholar]
- 44.Novo, D. J., N. G. Perlmutter, R. H. Hunt, and H. M. Shapiro. 2000. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 44:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliver, J. D. 2000. The public health significance of viable but nonculturable bacteria, p. 277-300. In R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, DC.
- 46.O'Mahony, M. C., R. E. Stanwell-Smith, H. E. Tillett, D. Harper, J. G. Hutchison, I. D. Farrell, D. N. Hutchinson, J. V. Lee, P. J. Dennis, H. V. Duggal, et al. 1990. The Stafford outbreak of Legionnaires' disease. Epidemiol. Infect. 104:361-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson, W. J., D. V. Seal, E. Curran, T. M. Sinclair, and J. C. McLuckie. 1994. Fatal nosocomial Legionnaires' disease: relevance of contamination of hospital water supply by temperature-dependent buoyancy-driven flow from spur pipes. Epidemiol. Infect. 112:513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phe, M. H., M. Dossot, and J. C. Block. 2004. Chlorination effect on the fluorescence of nucleic acid staining dyes. Water Res. 38:3729-3737. [DOI] [PubMed] [Google Scholar]
- 49.Phe, M. H., M. Dossot, H. Guilloteau, and J. C. Block. 2005. Nucleic acid fluorochromes and flow cytometry prove useful in assessing the effect of chlorination on drinking water bacteria. Water Res. 39:3618-3628. [DOI] [PubMed] [Google Scholar]
- 50.Phe, M. H., M. Dossot, H. Guilloteau, and J. C. Block. 2007. Highly chlorinated Escherichia coli cannot be stained by propidium iodide. Can. J. Microbiol. 53:664-670. [DOI] [PubMed] [Google Scholar]
- 51.Pisz, J. M., J. R. Lawrence, A. N. Schafer, and S. D. Siciliano. 2007. Differentiation of genes extracted from non-viable versus viable micro-organisms in environmental samples using ethidium monoazide bromide. J. Microbiol. Methods 71:312-318. [DOI] [PubMed] [Google Scholar]
- 52.Reingold, A. L., B. M. Thomason, B. J. Brake, L. Thacker, H. W. Wilkinson, and J. N. Kuritsky. 1984. Legionella pneumonia in the United States: the distribution of serogroups and species causing human illness. J. Infect. Dis. 149:819. [DOI] [PubMed] [Google Scholar]
- 53.Riedy, M. C., K. A. Muirhead, C. P. Jensen, and C. C. Stewart. 1991. Use of a photolabeling technique to identify nonviable cells in fixed homologous or heterologous cell populations. Cytometry 12:133-139. [DOI] [PubMed] [Google Scholar]
- 54.Roth, B. L., M. Poot, S. T. Yue, and P. J. Millard. 1997. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63:2421 2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rudi, K., K. Naterstad, S. M. Dromtorp, and H. Holo. 2005. Detection of viable and dead Listeria monocytogenes on gouda-like cheeses by real-time PCR. Lett. Appl. Microbiol. 40:301-306. [DOI] [PubMed] [Google Scholar]
- 56.Rudi, K., B. Moen, S. M. Dromtorp, and A. L. Holck. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl. Environ. Microbiol. 71:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rueckert, A., R. S. Ronimus, and H. W. Morgan. 2005. Rapid differentiation and enumeration of the total, viable vegetative cell and spore content of thermophilic bacilli in milk powders with reference to Anoxybacillus flavithermus. J. Appl. Microbiol. 99:1246-1255. [DOI] [PubMed] [Google Scholar]
- 58.Saby, S., I. Sibille, L. Mathieu, J. L. Paquin, and J. C. Block. 1997. Influence of water chlorination on the counting of bacteria with DAPI (4′,6-diamidino-2-phenylindole). Appl. Environ. Microbiol. 63:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soejima, T., K. Iida, T. Qin, H. Taniai, M. Seki, A. Takade, and S. Yoshida. 2007. Photoactivated ethidium monoazide directly cleaves bacterial DNA and is applied to PCR for discrimination of live and dead bacteria. Microbiol. Immunol. 51:763-775. [DOI] [PubMed] [Google Scholar]
- 60.Soejima, T., K. Iida, T. Qin, H. Taniai, M. Seki, and S.-I. Yoshida. 2008. Method to detect only live bacteria during PCR amplification. J. Clin. Microbiol. 46:2305-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steinert, M., L. Emödy, R. Amann, and J. Hacker. 1997. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl. Environ. Microbiol. 63:2047-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venkobachar, C., L. Iyengar, and A. V. S. P. Rao. 1977. Mechanism of disinfection: effect of chlorine on cell membrane functions. Water Res. 11:727-729. [Google Scholar]
- 63.Virto, R., P. Mañas, I. Alvarez, S. Condon, and J. Raso. 2005. Membrane damage and microbial inactivation by chlorine in the absence and presence of a chlorine-demanding substrate. Appl. Environ. Microbiol. 71:5022-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, S., and R. E. Levin. 2006. Discrimination of viable Vibrio vulnificus cells from dead cells in real-time PCR. J. Microbiol. Methods 64:1-8. [DOI] [PubMed] [Google Scholar]
- 65.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.World Health Organization. 2006. Guidelines for safe recreational water environments. http://www.who.int/water_sanitation_health/bathing/srwe1/en/.
- 67.Yamamoto, H., Y. Hashimoto, and T. Ezaki. 1996. Study of nonculturable Legionella pneumophila cells during multiple-nutrient starvation. FEMS Microbiol. Ecol. 20:149-154. [Google Scholar]
- 68.Yaradou, D. F., S. Hallier-Soulier, S. Moreau, F. Poty, Y. Hillion, M. Reyrolle, J. André, G. Festoc, K. Delabre, F. Vandenesch, J. Etienne, and S. Jarraud. 2007. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Appl. Environ. Microbiol. 73:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, V. L. 2000. Legionella pneumophila, p. 2424-2435. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed., vol. 2. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 70.Yu, V. L., J. F. Plouffe, M. C. Pastoris, J. E. Stout, M. Schousboe, A. Widmer, J. Summersgill, T. File, C. M. Heath, D. L. Paterson, and A. Chereshsky. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127-128. [DOI] [PubMed] [Google Scholar]