Abstract
The distribution of viral genotypes in the ocean and their evolutionary relatedness remain poorly constrained. This paper presents data on the genetic diversity and evolutionary relationships of 1.2-kb DNA polymerase (pol) gene fragments from podoviruses. A newly designed set of PCR primers was used to amplify DNA directly from coastal sediment and water samples collected from inlets adjacent to the Strait of Georgia, British Columbia, Canada, and from the northeastern Gulf of Mexico. Restriction fragment length polymorphism analysis of 160 cloned PCR products revealed 29 distinct operational taxonomic units (OTUs), with OTUs within a site typically being more similar than those among sites. Phylogenetic analysis of the DNA pol gene fragments demonstrated high similarity between some environmental sequences and sequences from the marine podoviruses roseophage SIO1 and cyanophage P60, while others were not closely related to sequences from cultured phages. Interrogation of the CAMERA database for sequences from metagenomics data demonstrated that the amplified sequences were representative of the diversity of podovirus pol sequences found in marine samples. Our results indicate high genetic diversity within marine podovirus communities within a small geographic region and demonstrate that the diversity of environmental polymerase gene sequences for podoviruses is far more extensive than previously recognized.
Marine viruses are the most abundant (41) and diverse (2, 6) biological entities in the ocean. They affect community composition by causing the lysis of specific subsets of the microbial community (22, 28, 46, 47) and, by killing numerically dominant host taxa, may influence species evenness and richness (24, 28, 43, 50). Despite the abundance of bacteriophages in marine systems and their important roles in marine microbial composition, little is known about the distribution and diversity of specific groups of marine viruses. However, most marine bacteriophage isolates are tailed phages (3) belonging to the order Caudovirales (27), which comprises the families Myoviridae, Podoviridae, and Siphoviridae.
Podoviruses are classified into several groups (e.g., T7-like, P22-like, and phi-29-like) based on genome size, genome arrangement, and shared genes and can be readily isolated from seawater (11, 16, 42, 45). Genomic analysis of roseophage SIO1 (33), cyanophage P60 (7), vibriophage VpV262 (21), and cyanophage PSSP7 (40) suggests that many of the isolates are T7-like. Despite the apparently wide distribution of podoviruses in the sea, and their potential importance as agents of microbial mortality, there has been little effort to explore their diversity.
Sequence analysis of representative genes is one approach that has been used to examine the genetic diversity of specific groups of marine viruses. For example, homologues for structural genes (g20 and g23) found in T4-like phages are found in some marine myoviruses (18, 20) and have been used to examine the distribution, diversity, and evolutionary relationships among marine myoviruses (12, 14, 17, 37, 38, 49). Other studies have used DNA polymerase (pol) to examine the diversity of viruses infecting eukaryotic phytoplankton (8, 38) and have shown that phylogenies constructed with this gene are congruent with established viral taxonomy (9, 36, 37).
Although it is not universally present, family A DNA pol is a good target for examining the diversity of podoviruses (4). Our study presents a newly designed set of PCR primers that amplify a longer fragment of the DNA polymerase from a much larger suite of podoviruses and shows that the diversity within marine podoviruses as revealed by DNA pol sequences is far greater than previously realized.
MATERIALS AND METHODS
Collection and preparation of samples.
Samples were collected from the water and sediments in bays and inlets around the Strait of Georgia (labeled SOG) in British Columbia, Canada, and from water in the northeastern Gulf of Mexico (labeled GOM).
Go-Flo bottles mounted on a rosette equipped with a conductivity-temperature-depth probe were used to collect water samples (∼20 liters) from the subsurface chlorophyll maximum at 5 m in Howe Sound (49°27.30′N 123°16.88′W) on 31 July 2000, from 5 and 10 m in Malaspina Inlet (50°04.78′N 124°42.83′W) on 2 August 2000 (Malaspina 442 and 443; salinity, 26.4 and 25.0‰; 15.3 and 16.8°C, respectively), and from 25 m in the northeastern Gulf of Mexico on 21 July 2002 (29°00.037′N 87°17.836′W; salinity,. 33.3‰; 28.9°C). For each sample, the viruses were concentrated ∼100-fold (∼200-ml final volume) using ultrafiltration (42). Briefly, particulate matter was removed by pressure filtering (<17 kPa) the samples through 142-mm-diameter glass fiber (MFS GC50; nominal pore size, 1.2 μm) and polyvinylidene difluoride (Millipore GVWP; pore size, 0.22 μm) filters connected in series. The viral size fraction in the filtrate was concentrated by ultrafiltration through a 30-kDa molecular mass cutoff cartridge (Amicon S1Y30; Millipore). The concentrates were stored at 4°C in the dark for up to 3 years, until the viral DNA was extracted from 200-μl subsamples of the concentrates using a hot/cold treatment (three cycles of 2 min at 95°C and 2 min at 4°C) in a thermocycler (9). A 0.1 dilution of the extract was used as a PCR template.
Sediment cores were collected using a tribarrel gravity corer (Rigosha, Tokyo, Japan) at depths of 84 m in Sechelt Inlet (49°43.9′N 123°44.3′W) on 25 July 2001, 34 m (Malaspina sediment 1) and 50 m (Malaspina sediment 4) in Malaspina Inlet (50°04.8′N 124°42.9′W and 49°58.53′N 124°41.11′W) on 26 July 2001, and 27 m in Nanoose Bay (49°58.53′N 124°41.11′W) on 27 July 2001, all in British Columbia. Briefly, the sediments were processed as follows. Immediately after retrieval, the sediment-water interface was removed with a wide-bore serological pipette without disrupting the sediment core. Each surface sediment sample (20 cm3) was mixed with 20 ml of phosphate-buffered saline and centrifuged at 4,000 × g for 5 min at 4°C. The supernatant was filtered through 47-mm-diameter glass fiber (Whatman GF/C; nominal pore size, 1.2 μm) and polyvinylidene difluoride (Millipore HVLP; pore size, 0.45 μm) filters. Following filtration, the samples were kept in the dark at 4°C. Prior to DNA extraction, the viruses were concentrated by centrifugation at 180,000 × g for 3.5 h at 20°C. The supernatants were removed, and the pellets were stored overnight at 4°C before ∼100 μl of each pellet was resuspended in 500 μl of 50 mM Tris (pH 8.0). DNA was extracted using phenol-chloroform (10), and a 0.1 dilution of the extract was used as a PCR template.
Primer design and PCR amplification.
Degenerate primers were designed to amplify an ∼1,200-bp DNA pol fragment from a subset of viruses belonging to the Podoviridae. The primers were designed based on family A DNA pol amino acid sequences from roseophage SIO1, cyanophage P60, T7, T3, and φYeO3-12 and aligned using Clustal W. Conserved regions for primer design were manually selected using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Forward (Podo-F, 5′-GACACHCTYRTVHTGTCWMGWYTG-3′) and reverse (Podo-R2, 5′-MCKACCRTCYARDCCYTTMAK-3′) primers were inferred from their amino acid sequences and had degeneracies of 1,728 and 768, respectively.
Three microliters of either the sediment DNA extract, water column viral concentrates, or T7 lysate (control) were used as the DNA template in the first-stage PCR mixture (total volume, 25 μl). The reagents included Taq DNA polymerase assay buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl), 2.5 mM MgCl2, 160 μM of each deoxyribonucleoside triphosphate, 1.2 μM of each Podo-F and Podo-R2 primer, and 0.4 U of Platinum Taq DNA polymerase (Invitrogen Life Technologies). Negative controls contained all reagents except DNA template. The samples were denatured at 94°C for 90 s, followed by 39 cycles of denaturation at 94°C for 45 s, annealing at 56°C for 45 s, and elongation at 72°C for 60 s, with a final elongation step of 72°C for 5 min. To increase the yield for cloning, gel plugs were excised with a Pasteur pipette, added to 100 μl of buffer (10 mM Tris-Cl, pH 8.5), and heated for 5 min at 80°C. A second, 25-cycle PCR was done on 6 μl of the eluted DNA, and the product size was verified by electrophoresis. We found this approach to yield consistent and reproducible amounts of DNA for cloning across all samples.
Clone library construction and RFLP analysis.
Amplicons from the second PCR were purified with a Qiaquick kit (Qiagen), ligated into pGEM-T (Promega), and used to transform Escherichia coli DH5α. For each sample, 20 positive clones containing an insert of the correct size were verified by colony PCR. Restriction fragment length polymorphism (RFLP) analysis was done by digesting 15 μl of the colony PCR products with MboII (New England BioLabs) in a reaction mixture containing 1 U/μg of DNA and 1× NEBuffer 2 (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9) and incubating the mixture at 37°C for 1 h, followed by heat inactivation at 65°C for 20 min. One-, 2-, and 3-h incubations yielded the same restriction patterns, confirming complete digestion by 1 h. The RFLP products were separated on a 2% agarose gel in 0.5× TBE (9 mM Tris base, 9 mM boric acid, 2 mM EDTA, pH 8.0). Sequencing of three representative clones with each of four unique restriction patterns confirmed that each restriction pattern could be considered an operational taxonomic unit (OTU).
Sequencing.
Forward and reverse sequences (∼1,200 bp) were obtained for each of the 29 RFLP patterns using Big-Dye Terminator Cycle Sequencing (Applied Biosystems) and ABI 373 Stretch or ABI Prism 377 sequencers.
Phylogenetic analysis.
BLAST and the inferred amino acid sequence from the DNA polymerase gene of phage T7 were used to recover additional sequences from GenBank (Table 1) for isolates that were used for phylogenetic analysis. The sequences were trimmed to the PCR product length and aligned using ClustalW (26), and the alignment was manually edited in Se-Al Carbon (32). BLASTp was used to screen the CAMERA database for open reading frames that were homologues of family A DNA polymerase amino acid sequences from podovirus isolates, as well as the environmental PCR products. The sequences were trimmed to recover the last 240 amino acids before the reverse primer (Podo-R2); sequences that did not have the full 240 amino acids were culled. Only sequences having a podovirus as a first hit when BLAST searching in GenBank were kept for analysis (see Table S1 in the supplemental material). The maximum-likelihood analyses were performed using the RAxML Web servers (http://phylobench.vital-it.ch/raxml-bb/) with the WAG model and bootstrapping with 100 replicates (39). Bayesian phylogenetic analyses were performed with MrBayes (v3.1.2; [1], freely distributed by the authors at http://www.mrbayes.csit.fsu.edu/). The program uses a Markov chain Monte Carlo (MCMC) approach to approximate prior and posterior probabilities. Under the WAG substitution model, we ran two independent analyses of four (one cold and three heated) MCMC chains with 1,000,000 cycles and sampled every 100th cycle, and with a burn-in of 25%, distance-based calculations were run in PAUP* (PAUP* 4.0b10; Sinauer Associates, Sunderland, MA). The best tree out of 10 search replicates was estimated, and bootstrap values were calculated based on percentages of 100 replicates. The trees were viewed with Fig Tree (freely available at http://tree.bio.ed.ac.uk/software/figtree/).
TABLE 1.
Viral isolate sequences used in this study
| Virus name | Host | Accession no. | Reference |
|---|---|---|---|
| S-CBP1 | Synechococcus sp. strain CB0101 | EF535232.1 | 44 |
| S-CBP3 | Synechococcus | EF535233 | 44 |
| S-CBP42 | Synechococcus | EF535234 | 44 |
| P60 | Prochlorococcus | AF338467 | 7 |
| Syn5 | Synechococcus | YP001285436 | 31 |
| SIO1 | Roseobacter | AF189021 | 33 |
| T3 | E. coli | AJ318471 | 29 |
| T7 | E. coli | V01146 | 13 |
| φYeO3-12 | Yersinia enterocolitica (type O:3) | AJ251805 | 30 |
| Berlin | Yersinia pestis | NC_008694 | -a |
| VP4 | Vibrio | YP249581 | - |
| gh-1 | Pseudomonas putida | AF493143 | 23 |
| BA14 | E. coli | NC_011040.1 | |
| EcoDS1 | E. coli | NC_011042.1 | - |
| K11 | Klebsiella sp. strain 390a | NC_011043.1 | - |
| K1F | E. coli | NC_007456.1 | 34 |
| KvP1 | Kluyvera cryocrescens | NC_011534.1 | 25 |
| MmP1 | Morganella morganii | NC_011085.1 | - |
| Yepe2 | Y. pestis | NC_011038.1 | - |
| φA1122 | Y. pestis | NC_004777.1 | 19 |
| φSG-JL2 | Salmonella enterica subsp. enterica serovar Gallinarum | NC_010807.1 | - |
A hyphen means that the sequence was released in GenBank but is unpublished.
Nucleotide sequence accession numbers.
The sequences obtained in this study were added to GenBank and assigned the accession numbers AY258449 to AY258461 (SOG-W-A, SOG-WS-E, SOG-W-F, GOM-W-O, SOG-W-S, SOG-WS-N, SOG-W-P, SOG-S-H, SOG-WS-C, GOM-W-J, GOM-W-M, GOM-W-Q, and SOG-S-R) and AY258463 to AY258466 (SOG-S-D, SOG-S-G, SOG-S-L, and GOM-W-I).
RESULTS AND DISCUSSION
This study extends the known sequence space for viral DNA polymerase genes to reveal that podoviruses are diverse and widespread members of marine communities. Sequence analysis revealed the existence of at least three previously unrecognized broad evolutionary groups of environmental podovirus polymerase sequences that have no cultured representatives, while the genetic richness of other groups has been expanded markedly.
Our study investigated the genetic richness of marine T7-like phages by targeting family A DNA pol with the degenerate primers Podo-F and Podo-R2. These primers amplified the expected 1,430-bp product from bacteriophage T7 and generated products of ca. 1,150 to 1,250 bp (data not shown) from natural marine virus communities. Podoviruses that harbor family B DNA pol, such as the marine phage VpV262, would not be captured with this primer set.
The PCR products from each of the four water column and four sediment samples collected from the west coast of Canada and the western Gulf of Mexico were cloned, and 20 clones were analyzed from each sample. Restriction digests of 160 cloned fragments yielded 29 different RFLP patterns that were sequenced (Fig. 1); of these, 17 were at least 5% different from all others at the nucleotide level. For this reason, the sequences from the patterns B and K were removed from further analysis because they were more than 95% identical to the sequence from pattern D. Moreover, multiple sequences of clones representing the same RFLP pattern recovered the same genotype.
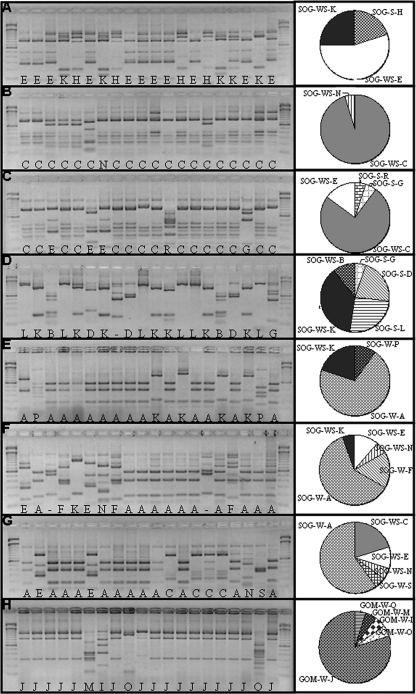
FIG. 1.
RFLP patterns generated from the MboII digestion of natural virus DNA pol clones. Shown are marine surface sediment samples from Sechelt Inlet (A), Malaspina Inlet sites 1 (B) and 4 (C), and Nanoose Bay (D) and water column samples from Howe Sound (E), Malaspina Inlet (samples 442 [F] and 443 [G]), and the Gulf of Mexico (H). Lanes 1 and 22, 100-bp ladders; lanes 2 to 21, randomly picked clones. The letters at the bottoms of the lanes represent the different patterns (OTUs) observed on the gel. The hyphens indicate OTUs for which sequence information is not available. SOG and GOM are sequences from the Strait of Georgia and Gulf of Mexico, respectively. W, S, and WS are sequences found in water, sediment, or both, respectively. The pie charts represent the proportion of each OTU in each sample.
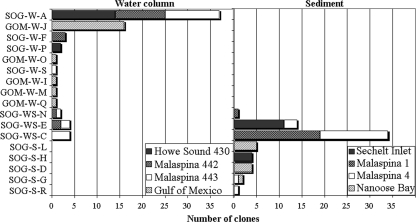
Similar to results from other studies (5, 38), the frequency with which specific genotypes occurred varied among samples, implying that viral communities are different among sites. For example, in the sediments, five genotypes were recovered from Nanoose Bay, whereas, Malaspina site 1 was mostly dominated by a single genotype (Fig. 2D and 1B, respectively). Similarly, in the clone library of 20 PCR fragments from each sample, some genotypes occurred more frequently than others. For example, sequence SOG-W-A (i.e., Strait of Georgia, water sample A) was found in three different samples, Malaspina Inlet 442 and 443, as well as Howe Sound 430 (Fig. 1E, F, and G). On the other hand, GOM-W-J was found only in water from the Gulf of Mexico (Fig. 1G). Within samples from British Columbia, six sequences occurred in more than one sample, whereas all the sequences from the Gulf of Mexico were unique to that location (Fig. 1G and 2A). Although interpretations of relative genetic richness among samples are suspect because of PCR amplification, every environment differed in terms of the sequences recovered.
FIG. 2.
Abundances of each phylotype in the clone libraries from water and sediment samples. The stacked bars identify specific sample distribution in terms of numbers of clones for each respective sequence. B and K were >95% similar at the sequence level to SOG-W-D, and are not included.
There was little genotypic overlap between OTUs in the sediment and the water (Fig. 2), implying that the virus assemblages in these environments are distinct. This is consistent with different bacterial communities being present in the water and sediments, as well (15). Nonetheless, some very similar sequences (58% at the amino acid level) were found in geographically distant locations (the Gulf of Mexico and British Columbia inlets) and different environments (water and sediment). This is similar to previous results (4), where the same Podoviridae DNA pol sequences were found in very different environments. Consequently, a larger sequencing effort, or an approach using nondegenerate primers, might reveal that sequences that were not detected are present but rare. Similarly, some Myoviridae g20 sequences (37) and cyanophage DNA pol sequences (31) are dispersed among a variety of environments, while others appear to be restricted in distribution.
Despite the high degeneracy of the primers, all of the sequences recovered were family A DNA pol genes that clustered within the Podoviridae. Sequences from T7-like phages (Fig. 2) were consistently 200 bp shorter than those of the marine phages, with nucleotides being lost throughout the gene fragment, yet, the amino acid sequences retained many regions of strong conservation. Assuming that marine phages are ancestral (21), it appears that the enterophage pol genes have become more streamlined.
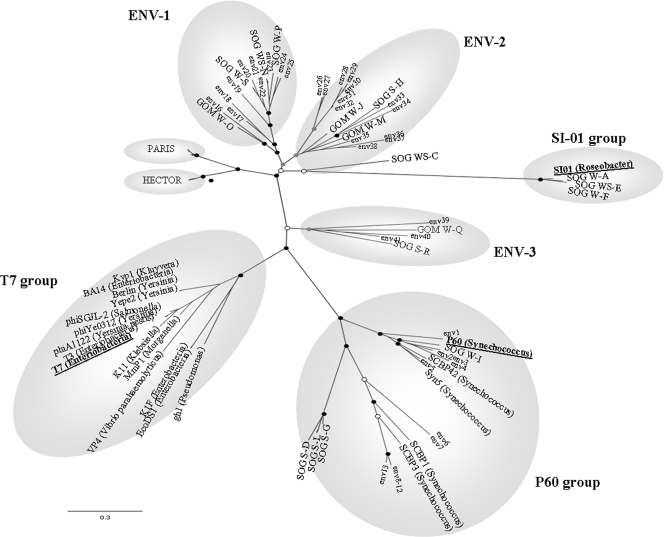
The amino acid sequence alignments showed multiple regions of conservation, as well as regions that were confined to specific groups. Phylogenetic analysis using both Bayesian (Fig. 3) and maximum-likelihood algorithms produced similar trees, with several branches clustering with known cultured representatives, including cyanophage P60, Syn5, and SCBP-1, as well as roseophage SIO1, but distinct from the T7 enterophages. Most sequences fell into the clusters ENV1 and ENV2, which are not closely related to cultured representatives. HECTOR and PARIS are PCR products obtained with nondegenerate primers that amplify a shorter part of the DNA polymerase and are representatives of podovirus sequences found in a number of environments (4); they resolve as a sister clade to the group containing roseophage SIO1 (Fig. 3). None of the HECTOR and PARIS sequences fell into other groups, and none of the sequences from this study fell into the PARIS and HECTOR groups.
FIG. 3.
Maximum-likelihood and Bayesian (1,000,000 MCMC generations; 25% burning) analyses of DNA pol inferred amino acid sequence alignments. Sequences from the major cultured representatives from each group are underlined. Sequences from the Global Ocean Survey are identified as “env” (see Table S1 in the supplemental material). The bootstrap values correspond to the frequencies of inferred trees. The black, gray, and white circles indicate internal nodes with at least 90%, 75%, and 50% bootstrap support, respectively. The envelopes (ENV) represent the different subgroups of the T7-like supergroup. The scale bar represents 0.3 amino acid substitutions per site.
A BLAST search of the CAMERA database (35) recovered many family A podovirus DNA pol sequences. The CAMERA database contains the metagenomic data from the Global Ocean Survey, a study of 23 sites along a track from the northwest Atlantic through the eastern tropical Pacific (51) that contains primarily bacterial sequences, but also many sequences from viruses (48). Only sequences that included the reverse primer Podo-R2 and had a podovirus as the first hit in GenBank were kept for phylogenetic analysis. This was important, as DNA pol sequences from some bacteria, such as Azorhizobium caulinodans ORS 57, Burkholderia oklahomensis EO147, and Pseudomonas putida KT2440 are very similar to viral DNA pol sequences. For example, there is 43% identity between the targeted fragments of the DNA polymerase of A. caulinodans and that of the cyanophage SCBP-3. However, the top hits for the bacterial sequences were not viral and, when analyzed, clustered in a separate group near the cyanophages. For this reason, the samples collected in our study were 0.2-μm-pore filtered.
Phylogenetic analysis showed that none of the Global Ocean Survey sequences fell outside of the groups circumscribed by the sequences recovered using primers. This demonstrates that these primers can be used to target the genetic diversity of Podoviridae family A pol sequences found in marine systems. The environmental sequences established three new groups, ENV1, ENV2, and ENV3, that have no cultured representatives.
This study presents a non-culture-based method using new primers that can be used to explore the diversity, evolution, and distribution of podoviruses. These results demonstrate that the genetic richness of family A DNA pol sequences associated with podoviruses is much greater than previously recognized and that some sequences are most frequently recovered from specific environments. Also, the results reveal the existence of several groups of distantly related and previously unknown podoviruses. Bringing representative host-virus systems that include representatives of these groups into culture will be essential to understanding host-virus interactions in the sea.
Supplementary Material
Acknowledgments
We thank Cindy Short for collecting samples from the Gulf of Mexico, Janice Lawrence for providing extracted DNA from sediment samples collected in British Columbia, Dany Vohl for help with the figures, and the crew of the CCGS Vector.
This research was supported in part by NSERC postgraduate scholarships to J.M.L. and K.E.R. and NSERC Discovery and Ship Time grants to C.A.S.
Footnotes
Published ahead of print on 10 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altekar, G., S. Dwarkadas, J. P. Huelsenbeck, and F. Ronquist. 2004. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407-415. [DOI] [PubMed] [Google Scholar]
- 2.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borsheim, K. Y. 1993. Native marine bacteriophages. FEMS Microbiol. Lett. 102:141-159. [Google Scholar]
- 4.Breitbart, M., J. H. Miyake, and F. Rohwer. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236:249-256. [DOI] [PubMed] [Google Scholar]
- 5.Breitbart, M., and F. Rohwer. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278-284. [DOI] [PubMed] [Google Scholar]
- 6.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, F., and C. A. Suttle. 1996. Evolutionary relationships among large double-stranded DNA viruses that infect microalgae and other organisms as inferred from DNA polymerase genes. Virology 219:170-178. [DOI] [PubMed] [Google Scholar]
- 9.Chen, F., C. A. Suttle, and S. M. Short. 1996. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl. Environ. Microbiol. 62:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cottrell, M. T., and C. A. Suttle. 1991. Wide-spread occurence and clonal variation in viruses which cause lysis of a cosmopolitain, eukaryotic marine phytoplankter, Micromonas pusilla. Mar. Ecol. Prog. Ser. 78:1-9. [Google Scholar]
- 11.DePaola, A., M. L. Motes, A. M. Chan, and C. A. Suttle. 1998. Phages infecting Vibrio vulnificus are abundant and diverse in oysters (Crassostrea virginica) collected from the Gulf of Mexico. Appl. Environ. Microbiol. 64:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorigo, U., S. Jacquet, and J. F. Humbert. 2004. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl. Environ. Microbiol. 70:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166:477-535. [DOI] [PubMed] [Google Scholar]
- 14.Filee, J., F. Tetart, C. A. Suttle, and H. M. Krisch. 2005. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc. Natl. Acad. Sci. USA 102:12471-12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippini, M., and M. Middelboe. 2007. Viral abundance and genome size distribution in the sediment and water column of marine and freshwater ecosystems. FEMS Microbiol. Ecol. 60:397-410. [DOI] [PubMed] [Google Scholar]
- 16.Frank, H., and F. Moebus. 1987. An electron microscopic study of bacteriophages from marine waters. Helgolander Meeresunters. 41:385-414. [Google Scholar]
- 17.Frederickson, C. M., S. M. Short, and C. A. Suttle. 2003. The physical environment affects cyanophage communities in British Columbia inlets. Microb. Ecol. 46:348-357. [DOI] [PubMed] [Google Scholar]
- 18.Fuller, N. J., W. H. Wilson, I. R. Joint, and N. H. Mann. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol. 64:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia, E., J. M. Elliott, E. Ramanculov, P. S. Chain, M. C. Chu, and I. J. Molineux. 2003. The genome sequence of Yersinia pestis bacteriophage φA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J. Bacteriol. 185:5248-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardies, S. C., A. M. Comeau, P. Serwer, and C. A. Suttle. 2003. The complete sequence of marine bacteriophage VpV262 infecting Vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology 310:359-371. [DOI] [PubMed] [Google Scholar]
- 22.Hennes, K. P., C. A. Suttle, and A. M. Chan. 1995. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl. Environ. Microbiol. 61:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovalyova, I. V., and A. M. Kropinski. 2003. The complete genomic sequence of lytic bacteriophage gh-1 infecting Pseudomonas putida—evidence for close relationship to the T7 group. Virology 311:305-315. [DOI] [PubMed] [Google Scholar]
- 24.Larsen, A., T. Castberg, R. A. Sandaa, C. P. D. Brussaard, J. Egge, M. Heldal, A. Paulino, R. Thyrhaug, E. J. V. Hannen, and G. Bratbak. 2001. Population dynamics and diversity of phytoplankton, bacteria and viruses in seawater enclosure. Mar. Ecol. Prog. Ser. 221:47-57. [Google Scholar]
- 25.Lingohr, E. J., A. Villegas, Y. M. She, P. J. Ceyssens, and A. M. Kropinski. 2008. The genome and proteome of the Kluyvera bacteriophage Kvp1—another member of the T7-like Autographivirinae. Virol. J. 5:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loytynoja, A., and N. Goldman. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. USA 102:10557-10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniloff, J., and H. W. Ackermann. 1998. Taxonomy of bacterial viruses: establishment of tailed virus genera and the order Caudovirales. Arch. Virol. 143:2051-2063. [DOI] [PubMed] [Google Scholar]
- 28.Middelboe, M., A. Hagstrom, N. Blackburn, B. Sinn, U. Fischer, N. H. Borch, J. Pinhassi, K. Simu, and M. G. Lorenz. 2001. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42:395-406. [DOI] [PubMed] [Google Scholar]
- 29.Pajunen, M. I., M. R. Elizondo, M. Skurnik, J. Kieleczawa, and I. J. Molineux. 2002. Complete nucleotide sequence and likely recombinatorial origin of bacteriophage T3. J. Mol. Biol. 319:1115-1132. [DOI] [PubMed] [Google Scholar]
- 30.Pajunen, M. I., S. J. Kiljunen, M. E. Soderholm, and M. Skurnik. 2001. Complete genomic sequence of the lytic bacteriophage φYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 183:1928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pope, W. H., P. R. Weigele, J. Chang, M. L. Pedulla, M. E. Ford, J. M. Houtz, W. Jiang, W. Chiu, G. F. Hatfull, R. W. Hendrix, and J. King. 2007. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned” bacteriophage of marine synechococcus. J. Mol. Biol. 368:966-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rambaut, A., N. C. Grassly, S. Nee, and P. H. Harvey. 1996. Bi-De: an application for simulating phylogenetic processes. Comput. Appl. Biosci. 12:469-471. [DOI] [PubMed] [Google Scholar]
- 33.Rohwer, F., A. M. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage Roseophage SI01 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 34.Scholl, D., and C. Merril. 2005. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J. Bacteriol. 187:8499-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshadri, R., S. A. Kravitz, L. Smarr, P. Gilna, and M. Frazier. 2007. CAMERA: a community resource for metagenomics. PLoS Biol. 5:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shackelton, L. A., and E. C. Holmes. 2004. The evolution of large DNA viruses: combining genomic information of viruses and their hosts. Trends Microbiol. 12:458-465. [DOI] [PubMed] [Google Scholar]
- 37.Short, C. M., and C. A. Suttle. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ. Microbiol. 71:480-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Short, S. M., and C. A. Suttle. 2002. Sequence analysis of marine virus communities reveals that groups of related algal viruses are widely distributed in nature. Appl. Environ. Microbiol. 68:1290-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan, M. B., M. L. Coleman, P. Weigele, F. Rohwer, and S. W. Chisholm. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 42.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1991. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl. Environ. Microbiol. 57:721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thingstad, T. F. 2000. Elements of a theory for the mechanism controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 44.Wang, K., and F. Chen. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microbiol. 34:105-116. [Google Scholar]
- 45.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 47.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 48.Williamson, S. J., D. B. Rusch, S. Yooseph, A. L. Halpern, K. B. Heidelberg, J. I. Glass, C. Andrews-Pfannkoch, D. Fadrosh, C. S. Miller, G. Sutton, M. Frazier, and J. C. Venter. 2008. The Sorcerer II Global Ocean Sampling expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS ONE 3:e1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson, W. H., N. J. Fuller, I. R. Joint, and N. H. Mann. 1999. Analysis of cyanophage diversity and population structure in a south-north transect of the Atlantic Ocean. Bull. Inst. Oceanogr. 19:209-216. [Google Scholar]
- 50.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yooseph, S., G. Sutton, D. B. Rusch, A. L. Halpern, S. J. Williamson, K. Remington, J. A. Eisen, K. B. Heidelberg, G. Manning, W. Li, L. Jaroszewski, P. Cieplak, C. S. Miller, H. Li, S. T. Mashiyama, M. P. Joachimiak, C. van Belle, J. M. Chandonia, D. A. Soergel, Y. Zhai, K. Natarajan, S. Lee, B. J. Raphael, V. Bafna, R. Friedman, S. E. Brenner, A. Godzik, D. Eisenberg, J. E. Dixon, S. S. Taylor, R. L. Strausberg, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 5:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.