Abstract
Versaphilic Anaeromyxobacter dehalogenans strains implicated in hexavalent uranium reduction and immobilization are present in the fractured saprolite subsurface environment at the U.S. Department of Energy Integrated Field-Scale Subsurface Research Challenge (IFC) site near Oak Ridge, TN. To provide insight into the in situ distribution of Anaeromyxobacter strains in this system with a nonuniform groundwater flow, 16S rRNA gene-targeted primers and linear hybridization (TaqMan) probes were designed for Oak Ridge IFC Anaeromyxobacter isolates FRC-D1 and FRC-W, along with an Anaeromyxobacter genus-targeted probe and primer set. Multiplex quantitative real-time PCR (mqPCR) was applied to samples collected from Oak Ridge IFC site areas 1 and 3, which are not connected by the primary groundwater flow paths; however, transport between them through cross-plane fractures is hypothesized. Strain FRC-W accounted for more than 10% of the total quantifiable Anaeromyxobacter community in area 1 soils, while strain FRC-D1 was not detected. In FeOOH-amended enrichment cultures derived from area 1 site materials, strain FRC-D1 accounted for 30 to 90% of the total Anaeromyxobacter community, demonstrating that this strain was present in situ in area 1. The area 3 total Anaeromyxobacter abundance exceeded that of area 1 by 3 to 5 orders of magnitude, but neither strain FRC-W- nor FRC-D1-like sequences were quantifiable in any of the 33 area 3 groundwater or sediment samples tested. The Anaeromyxobacter community in area 3 increased from <105 cells/g sediment outside the ethanol biostimulation treatment zone to 108 cells/g sediment near the injection well, and 16S rRNA gene clone library analysis revealed that representatives of a novel phylogenetic cluster dominated the area 3 Anaeromyxobacter community inside the treatment loop. The combined applications of genus- and strain-level mqPCR approaches along with clone libraries provided novel information on patterns of microbial variability within a bacterial group relevant to uranium bioremediation.
Molecular analyses enable specific detection of target organisms, providing insight into microbial biogeography and the factors controlling microbial community structure and function over temporal and spatial scales (reviewed in references 16, 31, and 37). Spatial isolation in disconnected environments has been demonstrated for plant rhizospheres (36), saturated soils versus unsaturated soils (47, 55), and undisturbed (pristine) top soils (8, 10). One unresolved issue of biogeography involves the spatial distribution of distinct populations (of a given species) in physically connected environments, such as heterogeneous subsurface media with nonuniform flow characteristics (e.g., fractured saprolite). A complex matrix of aged bedrock makes up the uranium-contaminated subsurface environment at the U.S. Department of Energy Integrated Field-Scale Subsurface Research Challenge (IFC) site, near Oak Ridge, TN (formerly known as the Field Research Center [FRC]). Connectivity and transport between two distinct Oak Ridge IFC study areas (a near-source contaminant plume in area 3 and a farther-source plume in area 1) have been hypothesized based on evidence of flow through fractured bedding planes that extend from the vicinity of area 3 (near the S-3 waste disposal ponds) to area 1 (35). Whether microbes are transported between these two subsurface areas is unclear, but the implications are important for understanding spatially variable biogeochemical processes that control contaminant fate and migration at the site (35).
Anaeromyxobacter dehalogenans populations are relevant to bioremediation at the Oak Ridge IFC site due to their capacity to metabolically reduce soluble U(VI) to sparingly soluble, immobile U(IV) (30, 41, 50). More than a dozen different Anaeromyxobacter 16S rRNA gene sequences have been identified in contaminated site materials derived from the Oak Ridge IFC site (7, 32, 33). Distinct A. dehalogenans strains were isolated from IFC site materials from area 1 (GenBank accession numbers FJ190048 to FJ190062 [16S rRNA gene sequences]). Laboratory characterization of A. dehalogenans strains that are very closely related (>99.9% 16S rRNA gene similarity) demonstrated metabolic variability in terms of growth rates as well as substrates (18, 40, 46). This is consistent with observations from other studies that have demonstrated that exploring diversity at the subspecies level is crucial for understanding microbial interactions and processes (9, 10, 14, 23, 24, 38). Tools that capture the distribution and abundance of A. dehalogenans strains in spatially and temporally heterogeneous subsurface environments are desirable to comprehensively describe biogeochemical processes controlling contaminant migration. Quantitative real-time PCR (qPCR) approaches using TaqMan probe detection chemistry offer high specificity (i.e., distinguish sequences that differ by only 1 or 2 bp) and allow the quantification of multiple targets in a single reaction mix (2, 29). The multiplex qPCR (mqPCR) technique reduces material consumption, labor, and the probability for experimental errors and has been applied successfully for discrimination of pathogenic bacteria, including Listeria monocytogenes strains (25) and Brucella isolates (42), as well as for simultaneous identification of four bioterrorism agents (48). Despite successful applications in the medical and biodefense fields, mqPCR approaches have had limited application to monitoring bioremediation processes. To demonstrate that strain-specific resolution of Anaeromyxobacter strains is feasible and provides relevant information about microbial distribution, we designed and applied an mqPCR approach to characterize and monitor the Anaeromyxobacter community at the Oak Ridge IFC site across areas 1 and 3. The results from this study provide new information about microbial, and hence functional, heterogeneity in a uranium-contaminated, saturated subsurface environment with nonuniform flow.
MATERIALS AND METHODS
Selection of strains.
Oak Ridge IFC site isolates FRC-D1 and FRC-W were derived from area 1 soil core samples collected from boreholes FW032 and FW034, respectively. Strain FRC-D1 was isolated via FeOOH enrichment (33), whereas strain FRC-W was obtained via enrichment with 2-chlorophenol, as were the first Anaeromyxobacter isolates (40). Strains FRC-D1 and FRC-W share 99.99% 16S rRNA gene sequence similarity over a stretch of 1,550 bp and share >99% 16S rRNA gene sequence similarity with the previously characterized Anaeromyxobacter strains 2CP-1 and 2CP-C (11, 40).
Culture conditions.
A. dehalogenans strains were routinely grown at 25°C without shaking in 60-ml (nominal capacity) glass serum bottles (Wheaton, Millville, NJ) with 40 ml of reduced, 30 mM bicarbonate-buffered mineral salts medium and a N2-CO2 headspace (80:20 [vol:vol]) (26). The bottles were sealed with butyl rubber stoppers (Geo-Microbial Technologies, Inc., Ochelata, OK) and aluminum crimp caps (Wheaton). Acetate (5 mM) (Sigma-Aldrich, St. Louis, MO) was provided as an electron donor, and fumarate (10 mM) (Sigma-Aldrich) served as the electron acceptor.
Coculture experiments were initiated with 300 μl of FRC-W culture and 400 μl of FRC-D1 culture, which were both in late exponential phase (optical densities at 600 nm of 0.20 and 0.15, respectively). Each inoculum contained 1.0 × 107 to 1.5 × 107 cells. Coculture samples were removed periodically for 16S rRNA gene quantification and organic acid (i.e., acetate, fumarate, and succinate) analysis.
Oak Ridge IFC site description.
Subsurface soil cores and sediment slurries were acquired from the Oak Ridge IFC site, which is heavily contaminated with a mixture of metals, radionuclides, and ligands (49). The site resides near the former S-3 waste disposal ponds, which were unlined surface impoundments used in the disposal of over 300 million liters of concentrated uranium and nitric acid waste during the period of 1951 to 1983 (6). Infiltration was the primary release mechanism to soils and groundwater, and vast subsurface domains have been contaminated, creating a massive legacy waste problem (49).
Contaminant fate and transport at the Oak Ridge IFC site are controlled by complex, nonlinear, and transient hydrological, geochemical, and microbial processes (49). The dynamics of these three interacting processes are controlled by the highly structured nature of the subsurface media that underlie the site (20, 21). Oak Ridge IFC site subsurface soils and sediments are acidic inceptisols consisting of highly fractured saprolites that have weathered from interbedded shale-limestone sequences. The limestone has been weathered to massive clay lenses that may contain residual carbonate, and the more resistant shale has weathered to an extensively fractured saprolite. Fractures are highly interconnected, with densities in the range of 200 fractures per meter (13). The fracture network consists of (i) fractures along bedding planes, (ii) two sets of orthogonal extensional fractures that are perpendicular to bedding planes, and (iii) shear fractures. Extensional fractures are either parallel or perpendicular to the strike of bedding planes and form an orthogonal fracture network with the bedding plane fractures. Under saturated conditions, strike parallel fractures control the direction of groundwater and contaminant migration, but bedding plane fractures dominate the fracture network of the media and may also contribute, to a significantly lesser extent, to the migration tendency of contaminants.
Soil, sediment, and groundwater samples were obtained from two spatially distinct Oak Ridge IFC site locations, areas 1 and 3 (35). Area 3 is near the S-3 waste disposal pond waste source. Materials in the vicinity of area 3 are up-dip from subsurface material residing in area 1, which is tens of meters away. The two subsurface environments are not connected by the main groundwater flow path, which is strike parallel from the waste source, but they may be connected by less conductive bedding plane parallel and orthogonal extension fractures. Single-well push-pull tests and downstream samplers have been utilized during biostimulation experiments using ethanol, acetate, and/or glucose in area 1 (19, 32, 33). Area 3 is the location of a pilot-scale in situ uranium bioremediation study that involved groundwater conditioning and periodic ethanol (i.e., 88.12% ethanol, 4.65% methanol, and 7.23% water) biostimulation (7, 51-53).
Field samples, microcosms, and enrichment cultures.
Soil core samples were collected from area 1 boreholes as described previously (3, 33; http://public.ornl.gov/orifc/map_area1_inset.cfm). Microcosms were established with homogenized soil core samples collected from six area 1 wells and boreholes (FWB030, FWB302, FWB032, FB061, FWB027, and FWB034) by Petrie et al. (33), and FeOOH enrichment cultures were provided by Denise Akob and Joel Kostka, Florida State University. In addition, the Kostka group provided area 1 soil core samples, including multiple homogenized-depth-interval samples from boreholes FB074 (96, 115, 130, 132, 148, 168, 186, 188, 204, 228, 230, and 240 in. below ground surface [bgs]), FB089 (21, 27, and 33 in. bgs), and FB090 (17, 23, and 29 in. bgs), for DNA extraction and qPCR analysis. Soil cores included in molecular analysis were collected adjacent to those used for enrichment and isolation efforts (33, 49).
Area 3 sediment samples were collected from wells FW024, FW026, FW100, FW101, FW102, FW103, and FW104, inside the pilot-scale U(VI) bioreduction demonstration plot (51-53; http://public.ornl.gov/orifc/map_area3_inset_a.cfm). Sediment samples were collected on 5 October 2005, during the ethanol biostimulation phase, using a well surging procedure as described previously (7). Groundwater samples were collected by Sue Carroll, Weimin Wu, Jack Carley, and Terry Gentry from wells FW104, FW101, and FW102, inside the area 3 pilot-scale demonstration plot (5, 51-53). Area 3 microcosms were established by Youlboong Sung from site samples obtained on 4 August 2005 from well FW104, as described previously (5). In an anoxic chamber filled with N2-H2 (97:3 [vol:vol]), 160-ml glass serum bottles received 15 ml of mineral salts medium and were amended with 2 g of sediment, 10 mM acetate, 5 mM fumarate, and 5 mM nitrate.
DNA extraction.
Aliquots (0.5 ml) from microcosms, enrichment cultures, and cocultures were removed with a syringe and transferred to 1.5-ml Eppendorf plastic tubes. Cells were collected by centrifugation at 16,000 × g for 3 min at room temperature, the supernatant was decanted, and the pellets were frozen at −20°C for at least 24 h. To extract DNA, 0.2 ml InstaGene Matrix solution (Bio-Rad Laboratories, Hercules, CA) was added to each frozen pellet, and DNA was prepared according to the manufacturer's specifications. The DNA was used immediately for qPCR analysis or stored at −20°C.
DNAs from 14 area 1 samples collected from boreholes FB074, FB089, and FB090 were extracted using a MoBio Power Soil DNA kit (MoBio, Carlsbad, CA) following the manufacturer's protocol. Additional DNA samples from 44 area 1 enrichment cultures and 18 area 3 sediment samples were provided by Denise Akob (Florida State University) and by Mary Beth Leigh and Erick Cardenas (Michigan State University), respectively. Dry DNA pellets from area 3 sediments were suspended in 100 μl of sterile, nuclease-free, deionized water to yield a DNA concentration of 1 ng μl−1. Fifteen DNA samples from area 3 groundwater collected from wells FW102 and FW101 were provided by Joy Van Nostrand and Jizhong Zhou (University of Oklahoma). Quantification of laboratory cultures, soil and sediment samples, and groundwater samples was normalized to ml of culture, g of soil or sediment, and liters of groundwater, respectively.
Design of A. dehalogenans strain-specific primers and probes.
Thirty-one Anaeromyxobacter-like 16S rRNA gene sequences from the NCBI nonredundant database (GenBank accession numbers AJ504428 to AJ504437, AF382396 to AF382400, AF482687, AKYG1825, DQ451451, DQ145119, DQ145125, DQ110017, DQ110097, AY360608, AY527735, AY527764, AY527784, AY527785, AY527791, and AY527798) were aligned using the ClustalW method (MegAlign; DNA STAR Inc., Madison, WI). Included in the alignment were sequences of related deltaproteobacteria, including several myxobacteria (GenBank accession numbers AJ233935, AJ233908, AJ233897, CEX23393, AJ233913, M34114, and AF503460) and two Geobacter species (GenBank accession numbers L07834 and U13928).
Linear hybridization (TaqMan) probes and primers were designed using Primer Express software (Applied Biosystems [ABI], Foster City, CA). All primer/probe sets were designed to meet the criterion for multiplex application (i.e., probes and primers share similar melting temperatures) (Table 1) (1). qPCR mixtures contained a forward primer, a reverse primer (Integrated DNA Technologies, Coralville, IA), and a probe that carried 6-carboxyfluorescein (FAM), NED, or VIC as a reporter dye on the 5′ end and a nonfluorescent quencher with a minor-groove binder on the 3′ end (ABI). Two strain-specific TaqMan mqPCR probes targeting the variable V3 region of the 16S rRNA gene were designed for the same primer pair (Table 1). The specificity of all probes and primers was evaluated using BLAST for “short, nearly exact nucleotide” sequence matches (4). To verify probe specificity, each strain-specific probe was used in reaction mixtures with target and/or nontarget (i.e., the cloned 16S rRNA gene fragment of the other strain) plasmid DNA. The FAM-labeled FRC-W probe targets A. dehalogenans strains FRC-W, 2CP-C, 2CP-3, and 2CP-5. The NED-labeled FRC-D1 probe targets strains FRC-D1, 2CP-1, and 2CP-2. A VIC-labeled genus-specific total Anaeromyxobacter (TAna) probe targets a separate region of the 16S rRNA gene with a different primer pair and was used to quantify the total Anaeromyxobacter community (Table 1). The optimal probe and primer concentrations were determined experimentally following the protocol provided in the ABI Chemistry Guide (1). For enumeration of total bacterial 16S rRNA gene copies, primers Bac1055YF and Bac1392R combined with Bac1115Probe were used as described previously (38).
TABLE 1.
Probes and primers used for TaqMan mqPCR
| Primer or probe | Target group | Sequence (5′-3′)b |
|---|---|---|
| Ade399 Fwd | Anaeromyxobacter genus | GCA ACG CCG CGT GTG T |
| Ade466 Rev | Anaeromyxobacter genus | TCC CTC GCG ACA GTG CTT |
| TAna VIC probe | Anaeromyxobacter genus | VIC-ATG AAG GTC TTC GGA TCG T-NFQ |
| 2CP444 Fwd | 2CP-like strainsa | TCG CGA GGG ACG AAT AAG G |
| 2CP513 Rev | 2CP-like strainsa | CGG TGC TTC CTC TCG AGG TA |
| FRC-D1 NED probe | 2CP-1, 2CP-2, FRC-D1 | NED-ACA GTC CGT TTC GAT GAC-NFQ |
| FRC-W FAM probe | 2CP-C, 2CP-5, FRC-W | FAM-ACA GTC CGT CAC GAT GA-NFQ |
Includes all Anaeromyxobacter strains described by Sanford et al. (40).
Strain-specific nucleotides are underlined. NFQ, nonfluorescent quencher.
qPCR analysis.
qPCR master mix (ABI), 2.5 μM of each probe (ABI), and 5 μM of each primer were combined in sterile, nuclease-free water prior to the addition of any DNA template. Aliquots (18 μl) of the reaction mix were dispensed into an ABI MicroAmp fast optical 96-well reaction plate held on ice. Template DNA (2 μl) was added to each well, the contents were mixed, and the wells were sealed with an ABI optical adhesive cover. An ABI 7500 real-time PCR system was used to detect FAM, VIC, and/or NED fluorescence, either individually or simultaneously (multiplex). Anaeromyxobacter-targeted mqPCRs were carried out with the following thermal program: 50°C for 2 min (1 cycle); 95°C for 10 min (1 cycle); and 95°C for 15 s and 60°C for 1 min (40 cycles). Fluorescence data were collected at 60°C during each cycle. qPCR to enumerate total bacteria was carried out as described previously (38).
qPCR calibration curves.
In order to obtain calibration standard curves (i.e., gene copy number versus the cycle number at which the fluorescence intensity reaches a set cycle threshold value), the 16S rRNA genes of strains 2CP-1 and 2CP-C, which share identical primer and probe binding sites with strains FRC-D1 and FRC-W, respectively (Table 1), were cloned into plasmids, using an Invitrogen Topo TA cloning kit (Invitrogen, Carlsbad, CA). A 10-fold dilution series of purified plasmid was used for each qPCR plate, and copy numbers of unknown samples were calculated as described previously (38). Samples and standards were run in triplicate qPCRs. Amplification efficiencies were calculated based on standard curve slope values as described previously (34). If necessary, samples were diluted so that the copy number values fell within the quantification range of the calibration curve for each individual reaction plate. The lowest value for the linear calibration curve for each plate was reported as the quantification limit, which ranged from 1.0 × 101 to 3.0 × 101 gene copies per reaction (see Table S1 in the supplemental material). Samples falling above the quantification limit produced fluorescence that crossed the threshold value within 38 PCR cycles in every replicate. A result of below the detection limit was reported when probe fluorescence did not exceed the threshold fluorescence (i.e., ΔRn = 0) for any reaction cycle. “Detected but not quantified” was reported when fluorescence values crossed the threshold value after more than 38 PCR cycles (1). The quantification of 16S rRNA gene copies was utilized as an approximation of cell numbers. The four sequenced Anaeromyxobacter genomes (GenBank accession numbers CP001131, CP000769, CP000251, and ABKC00000000) harbor duplicate rRNA operons (45), suggesting that members of the Anaeromyxobacter group contain two 16S rRNA gene copies.
Clone libraries.
Three 16S rRNA gene clone libraries were established using the Anaeromyxobacter 16S rRNA gene-specific forward primer 60F, described previously (33), and the reverse primer 1450-1470 REV (5′-TTG GCG CGG CCA CTT CT-3′). The primer pair yielded 1,410-bp amplicons. One library was established with DNAs extracted from biomass collected on a 0.2-μm membrane filter from 2 liters of groundwater obtained from the Oak Ridge IFC site area 3 injection well FW104 on 4 August 2005 (gro8 clones). Another library was generated with DNAs extracted from microcosms established with materials collected from well FW104 (micA clones). A third library was established with DNAs extracted from sediment collected from well FW104 at the end of the ethanol biostimulation phase before a U(IV) reoxidation experiment was initiated (52) (sedO clones). The total volume of each PCR mixture was 25 μl and contained (final concentrations) 1× reaction buffer, 2.5 mM MgCl2 (GeneAmp PCR kit; ABI), 50 nM of each primer, 250 μM of each deoxynucleoside triphosphate (Invitrogen), 13 mg ml−1 of bovine serum albumin (Invitrogen), 2.5 U of AmpliTaq polymerase (ABI), and 2 μl of 1:10 diluted template DNA (10 to 20 ng ml−1). PCRs were carried out in a 9600 GeneAmp PCR system (Perkin-Elmer, Waltham, MA) with the following temperature program: 94°C for 2 min 10 s (1 cycle); 94°C for 30 s, 57.5°C for 45 s, and 72°C for 2 min 10 s (30 cycles); and 72°C for 6 min. Fresh PCR products were cloned into the TOPO vector pCR 2.1 (TA cloning kit; Invitrogen) according to the manufacturer's instructions. Transformants from each of the three Anaeromyxobacter 16S rRNA gene clone libraries were selected and screened for the presence of the expected 16S rRNA gene insert by using the vector-targeted primers M13F and M13R (54) and the Anaeromyxobacter 16S rRNA gene-targeted primers 60F and 461R (33). In order to detect distinct sequences, amplified fragments from 10 clones of each library were digested with the restriction endonucleases MspI, HhaI, and RsaI (Gibco) at 37°C for 3 h according to the manufacturer's recommendations. Restriction fragments were resolved in 3% (wt/vol) Metaphor agarose (FMC Bioproducts, Rockland, ME) gels, using fresh TAE buffer (40 mM Tris base in 20 mM acetic acid, 1 mM EDTA, pH 8.5) at 4°C, and were stained with ethidium bromide (1 μg ml−1). Fragment sizes were estimated by comparison with Invitrogen 1kb Plus DNA molecular size markers.
Sequence and phylogenetic analysis.
To obtain sequence information on cloned Anaeromyxobacter 16S rRNA gene fragments, plasmids were extracted with a Qiagen plasmid mini kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. Plasmid templates (∼270 ng of DNA) from representative clones were sequenced by Nevada Genomics Center (University of Nevada, Reno). Sequences were aligned and analyzed with Clustal W and tested for possible chimera artifacts with the RDP Chimera Check tool (12). Phylogenetic relationships of Anaeromyxobacter 16S rRNA gene sequences were inferred using the neighbor-joining method in MEGA4 (39, 43). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown at each branching point (15). The evolutionary distances were calculated using the maximum composite likelihood method (44) and are given as the number of base substitutions per site. All missing data and positions containing gaps were eliminated from the data set (MEGA4 complete deletion option).
Modeling spatial distribution of Anaeromyxobacter strains in area 3.
The area 3 Anaeromyxobacter population distribution was visualized using U.S. Department of Defense Groundwater Modeling System 3.1 (http://chl.erdc.usace.army.mil/Media/3/7/2/GMSFactSheet.pdf). Input parameters were Anaeromyxobacter abundance data measured for samples obtained from the hydraulically connected depths of each well sampled in area 3 (well screen depth of 12 to 14 m bgs for multilevel sampling wells [i.e., depth interval 2] [51]). Interpolation of scatter point data was performed using the natural neighbor method and the software's default parameters.
Statistical analysis.
The average slope and y intercept of each qPCR calibration curve were determined by regression analysis and used to calculate the number of gene copies per ml of culture or sample. All significance tests, regression analyses, and 95% confidence interval calculations were performed using GraphPad Prism 5 with default settings. An analysis of variance repeated-measures test with the Tukey posttest (95% confidence) was used to determine whether the standard curves generated for the three primer-probe combinations were consistent with one another and between multiplex and single experiments. Samples for qPCR analysis were run with three replicates per reaction. Coculture experiments were performed in triplicate. The symbols and error bars for coculture analyses thus represent the averages for nine qPCR data points and standard deviations for three replicate culture data points per time point.
Chemical analyses.
Organic acids for coculture experiments were quantified using a Waters 1525 high-performance liquid chromatography system equipped with an Aminex HPX-87H ion-exclusion column and a Waters 2487 dual-wavelength absorbance detector as previously described (17).
RESULTS
Sensitivity and specificity of mqPCR approach.
TaqMan detection chemistry enabled quantification of Anaeromyxobacter target 16S rRNA gene copies over a large dynamic range spanning 8 orders of magnitude (i.e., 101 to 109 copies per reaction). The quantification limit was fewer than 10 copies of target gene per reaction in the strain- and Anaeromyxobacter genus-specific assays. No cross-reactivity was observed between the strain FRC-W-specific probe and DNA from the closely related, nontarget strain FRC-D1, or vice versa, even though they share 99.99% 16S rRNA gene sequence identity. mqPCR analysis with FRC-W-FAM, FRC-D1-NED, and TAna-VIC TaqMan probes (all with a nonfluorescent quencher) produced results similar to those of singleplex analyses (see Fig. S1 in the supplemental material), thus validating the multiplex analysis. Analysis of variance comparisons of linear regression slopes and y intercepts for multiple standard curves indicated that singleplex (individual) and multiplex qPCR results were not significantly different, with P values ranging from 0.14 to 0.67 (see Table S1 in the supplemental material). Regression analysis of multiplex data indicated a slope of −3.27 ± 0.10 for amplification with the NED-labeled FRC-D1 probe, which is close to the slope value of −3.32 corresponding to 100% amplification efficiency (1). Amplification efficiencies were calculated based on the slope of the line describing cycle number versus log gene copy number for 20 multiplex standard curves and 3 individual standard curves per probe (see Table S1 in the supplemental material). Amplification with the VIC-labeled TAna probe was less efficient, with a slope of −3.76 ± 0.12 and an efficiency value of 1.84, which is comparable to previously published qPCR efficiency values (34, 38). Standard curves obtained with the VIC-labeled probe had higher y intercepts, indicating that more gene copies were needed to achieve equivalent fluorescence intensities (i.e., lower sensitivity was achieved). Hence, the TAna primer/probe set provided slightly reduced sensitivity and efficiency compared to the strain-specific assays.
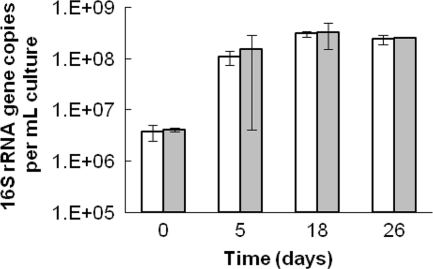
The analysis of strain FRC-W and strain FRC-D1 cocultures verified that strain-specific and genus-level TAna TaqMan probes quantified both strains along with the total Anaeromyxobacter community simultaneously in multiplex format (Fig. 1). Since coculture experiments contained only strains FRC-D1 and FRC-W, the sum of cells enumerated using the strain-specific probes was expected to equal the enumeration by the TAna probe. Over the course of the experiment, the total Anaeromyxobacter cell numbers and the sum of copies enumerated with each strain-specific probe remained within 1 standard deviation for triplicate culture data (representative data points are shown in Fig. 1). The agreement between the TAna probe data and the numbers generated with the strain-specific probes corroborated that all three assays were suitable for application in the mqPCR format.
FIG. 1.
Comparison of strain-specific probe quantification and genus-targeted TAna probe quantification of coculture 16S rRNA gene copies. White bars represent the sum of 16S rRNA gene copies enumerated with both strain-specific probes. Gray bars represent the copies enumerated with the TAna probe.
Application of the mqPCR approach to Oak Ridge IFC site-derived samples.
Upon validation of the mqPCR method with defined cocultures, the approach was used to quantify Anaeromyxobacter 16S rRNA gene sequences in DNAs derived from Oak Ridge IFC site materials. The multiplex tool successfully amplified DNA from enrichment cultures, microcosms, and field samples, including sediment, groundwater, and soil. The mqPCR tool demonstrated that Anaeromyxobacter strains were distributed heterogeneously at the Oak Ridge IFC site. In FeOOH enrichment cultures derived from area 1 well FWB032 supplied with acetate, strain FRC-D1-like sequences accounted for approximately one-third of the Anaeromyxobacter community and became dominant when lactate was supplied as an electron donor. These findings suggest that FRC-D1-like organisms are present in situ in area 1; however, analysis of three soil samples from area 1 failed to yield any FRC-D1-like sequences, indicating that these populations are present in numbers below the detection limit (i.e., no fluorescence signal in qPCRs) (Table 2). In contrast, FRC-W-like sequences were not detected in FeOOH enrichment cultures but ranged from 6.40 × 102 ± 3.5 × 101 to 3.5 × 104 ± 1.2 × 104 16S rRNA gene copies per g of soil in area 1 boreholes FB074 and FB089, respectively. TAna probe results for area 1 samples indicated that total Anaeromyxobacter 16S rRNA gene copies ranged from 5.2 × 102 ± 2.2 × 102 to 1.13 × 105 ± 5.4 × 104 per g of soil for boreholes FB074 and FB089, respectively. The cell number for each target strain ranged from 13% ± 1% to 122% ± 36% of the total Anaeromyxobacter community in all area 1-derived materials (soil cores or enrichment cultures) (Table 2).
TABLE 2.
Geochemical characteristics and presence of A. dehalogenans strains FRC-D1 and FRC-W in Oak Ridge IFC site materials (area 1)
| Area 1 location | Site geochemistrya
|
Sample type (collection date) | % of total Anaeromyxobacter communityc
|
||||
|---|---|---|---|---|---|---|---|
| Uranium (mg/liter) | Nitrate (mM) | Fe(II) (mM) | pH | FRC-D1 | FRC-W | ||
| FB074 | — | 1.99-21.85b | — | 2.91-6.25b | Soil core (18 October 2004) | BDL | 77.4 ± 6.6-122 ± 36d |
| FB089 | <0.015e | 62.42e | <0.587e | 5.48e | Soil core (17 May 2005) | BDL | 13 ± 1-31 ± 10d |
| FB090 | 0.05e | 16.39e | 3.21e | 6.02e | Soil core (17 May 2005) | BDL | 31.2 ± 19.5f |
| FWB032 | 1b | 11.8b | — | 5.4b | FeOOH-acetate microcosm, pH 7g | 34 ± 14 | BDL |
| 1b | 11.8b | — | 5.4b | FeOOH-lactate microcosm, pH 7g | 91 ± 21 | BDL | |
—, no data available.
Values represent site data at the time of sample collection (34; David Watson, personal communication).
BDL, below detection limit (indicates that no fluorescence signal was detected above the background).
Ranges of values represent samples from multiple homogenized soil core depth intervals for a single borehole.
Value measured in groundwater collected from an adjacent monitoring well in 2004 (http://public.ornl.gov).
Only a single depth interval contained a quantifiable number of Anaeromyxobacter 16S rRNA gene sequences.
Microcosms were established by Petrie et al. (33) from site materials collected in 2001.
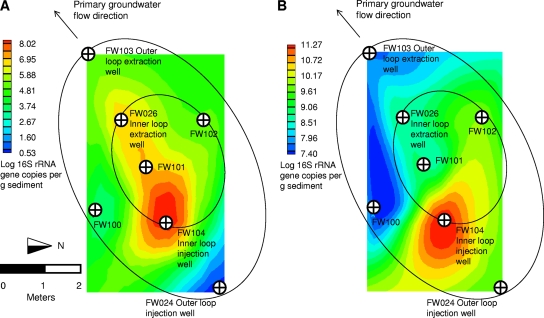
mqPCR results for sediment and groundwater samples from the area 3 bioremediation pilot-scale treatment zone indicated that total Anaeromyxobacter cell numbers were elevated throughout the subsurface region influenced by ethanol biostimulation, as were total bacterial cell numbers (Fig. 2A and B). The Anaeromyxobacter community increased from below the detection limit to 7.8 × 104 ± 2.8 × 104 16S rRNA gene copies per g sediment outside the treatment zone and to values as high as 3.5 × 108 ± 1.0 × 108 16S rRNA gene copies per g of sediment near the injection well (FW104) (Fig. 2A and B and Table 3). Relative to total bacterial 16S rRNA gene copies, Anaeromyxobacter 16S rRNA gene copies increased from 0.0002% ± 0.0001% outside the treatment zone to 2.3% ± 0.8% in the middle multiport well located downstream of the injection well (Table 3). Although mqPCR assessment of 33 area 3 samples (18 sediment and 15 groundwater samples) demonstrated the presence of Anaeromyxobacter in this subsurface region, neither strain FRC-D1 nor strain FRC-W 16S rRNA gene sequences could be quantified in any tested sample from area 3; however, FRC-W-like sequences were detected but not quantified (i.e., present below the quantification limit of 1.2 × 101 16S rRNA gene copies per reaction) in samples collected from wells FW100-3 and FW103, located outside the ethanol stimulation loop.
FIG. 2.
Spatial representation of Anaeromyxobacter (TAna probe) (A) and total bacterial (B) 16S rRNA gene abundances at the Oak Ridge IFC site area 3, based on qPCR analysis of samples from seven wells. Circles schematically represent the inner and outer recirculation loops, based on tracer studies and hydrologic models (28). FW100, FW101, and FW102 are multiport sampling wells.
TABLE 3.
Comparison of Anaeromyxobacter 16S rRNA gene copy numbers to total bacterial 16S rRNA gene copy numbers
| Location (well no. [description]) | No. of total Anaeromyxobacter 16S rRNA genes per g sediment | Proportion of total Anaeromyxobacter strains in total bacteria (%) |
|---|---|---|
| FW105 (outside the treatment loops) | 7 × 104 ± 3 × 104 | 0.0002 ± 0.0001 |
| FW104 (inner loop injection well) | 1.8 × 108 ± 5 × 107 | 0.06 ± 0.04 |
| FW103 (outer loop extraction well) | 1.2 × 105 ± 2 × 104 | 0.13 ± 0.09 |
| FW102-3 (multiport well in inner loop) | 1.3 × 105 ± 1 × 104 | 0.0020 ± 0.0019 |
| FW101-2 (multiport well in inner loop) | 1.4 × 107 ± 2 × 106 | 2.3 ± 0.8 |
| FW100-2 (multiport well in outer loop) | 4.2 × 103 ± 8 × 102 | 0.016 ± 0.003 |
| FW026 (inner loop extraction well) | 7.0 × 106 ± 7 × 105 | 1.2 ± 0.1 |
| FW024 (outer loop injection well) | BDLa | BDLa |
BDL, below the detection limit.
Clone libraries from area 3 site materials.
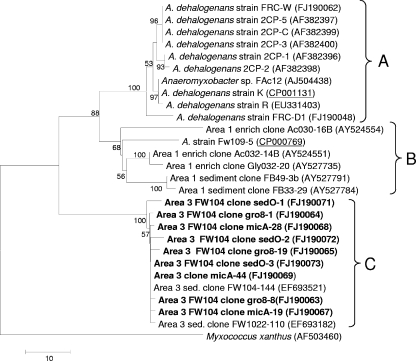
To explore the Anaeromyxobacter 16S rRNA gene sequences dominant in area 3, three Anaeromyxobacter 16S rRNA gene clone libraries were established with DNAs derived from samples of the ethanol injection well (FW104). Restriction analysis of nearly complete 16S rRNA genes (1,410 bp) yielded identical patterns, and 11 fragments were sequenced. Pairwise comparisons of evolutionary divergence among the 16S rRNA gene sequences recovered from area 3 with the A. dehalogenans type strain 2CP-1 indicated sequence similarities of 93.3 to 94%. The 16S rRNA gene sequences from uncharacterized Anaeromyxobacter spp. in area 3 were detected and quantified with the TAna probe but not with either of the strain-specific probes. Consistent with the qPCR results, the 16S rRNA gene target region (Escherichia coli positions 474 to 490) of strains detected in area 3 (GenBank accession numbers CP000769 and FJ190063 to FJ190073) was not complementary to the FRC-W- and FRC-D1-specific minor-groove-binding probes. Evolutionary distance analysis revealed that the Anaeromyxobacter strains detected in area 1 and the available Anaeromyxobacter isolates are phylogenetically distinct from the dominant area 3 Anaeromyxobacter population(s) (Fig. 3). Bootstrap values reinforce that representatives of at least three distinct Anaeromyxobacter clusters, designated A, B, and C, exist at the Oak Ridge IFC site (Fig. 3). Cluster A contains characterized Anaeromyxobacter isolates (11, 40, 45, 46), including type strain 2CP-1 (11), as well as the Oak Ridge IFC site isolates targeted in this study (strains FRC-D1 and FRC-W). Cluster A sequences were detected in area 1 but not in area 3, whereas cluster C sequences, which were >99.4% similar to each other, were unique to area 3.
FIG. 3.
16S rRNA gene-based phylogeny of characterized Anaeromyxobacter strains, Oak Ridge IFC site isolates, and environmental clone sequences. Area 3 sequences from cloned 16S rRNA gene fragments determined in this study are indicated in bold. The Myxococcus xanthus 16S rRNA gene sequence (GenBank accession number AF503460) was used to root the tree. The scale bar represents the number of differences in the nucleotide sequence. Bootstrap values are based on 500 replications and are not shown at nodes with <50% bootstrap support. GenBank accession numbers of genomes (underlined) and 16S rRNA gene sequences are indicated in parentheses. Distinct phylogenetic clusters are indicated with the letters A, B, and C.
DISCUSSION
Consistent with previous community analyses (7, 32, 33), Anaeromyxobacter 16S rRNA gene sequences are present in uranium-contaminated areas 1 and 3 at the Oak Ridge IFC site. The spatial analysis of samples across seven wells of area 3 demonstrated that Anaeromyxobacter abundance correlated with the zone influenced by ethanol biostimulation and the zone of decreased dissolved U(VI) concentrations (Fig. 2) (52, 53). Anaeromyxobacter spp. grow with a variety of electron acceptors, including U(VI), nitrate, ferric iron, and manganese oxide (18, 40, 50). Hence, respiratory substrates other than U(VI) may have supported Anaeromyxobacter growth, although nitrate concentrations in the treatment zone were generally low during active bioremediation (52) and nitrate was not detected in area 3 groundwater samples collected concurrently with the area 3 sediment samples used for this study (collected 5 October 2005) (7). Iron (0.01 to 0.03 mM) and manganese (0.06 to 0.08 mM) were present in area 3 groundwater at the time of sampling (7), and respiration of these oxidized metals may have contributed to growth in situ. The spatial correlation between Anaeromyxobacter and decreased soluble uranium concentrations is consistent with laboratory studies demonstrating that Anaeromyxobacter spp. respire U(VI) (41, 50) and suggests that Anaeromyxobacter spp. grow with U(VI) as an electron acceptor in situ. Interestingly, the mqPCR and clone library analyses demonstrated an uneven distribution of three distinct Anaeromyxobacter clades across areas 1 and 3. Within the Anaeromyxobacter genus, only cluster C 16S rRNA gene sequences were detected in the area 3 biostimulation treatment zone. The samples examined in this study provided no evidence for the presence of cluster A or cluster B sequences in the area 3 treatment zone, although cluster A and B representatives occur in area 3. For example, strain FW109-5 (cluster B) was isolated from area 3 site materials outside the treatment zone (Matthew Fields, personal communication), and 16S rRNA gene sequences of strain FRC-W (cluster A) were detected but not quantified (<1.2 × 101 gene copies per reaction) just outside the treatment zone. Conversely, cluster C sequences have not been detected in area 1 and strain FRC-W (cluster A) sequences were quantifiable in all area 1 soil cores tested. Strain FRC-W uses amorphous ferric iron as an electron acceptor (S. H. Thomas and F. E. Löffler, unpublished data), but area 1 FeOOH enrichment cultures were dominated by FRC-D1 16S rRNA gene sequences. This was unexpected because FRC-D1-like 16S rRNA gene sequences were not detected in any area 1 samples examined, suggesting that this strain is not abundant under in situ conditions but responds to laboratory enrichment with FeOOH. These findings demonstrate that strain differences affect field-scale processes, thus emphasizing the need for strain-specific monitoring tools. TaqMan probes with a conjugated minor-groove-binding tripeptide (2) discriminated strain FRC-W and strain FRC-D1 16S rRNA gene sequences, with only two mismatches in the probe target sequence. TaqMan probes allow multiplex detection, which requires that primers and probes have similar melting characteristics, and the data obtained with different quencher and fluorophore combinations must be compared to individual quantification results (1). Although mqPCR requires careful design and evaluation, the multiplex format reduces the cost of the analyses and eliminates one element of experimental uncertainty by utilizing a single aliquot of template for quantification of the gene targets.
Based on previous principles of microbial biogeography (31, 37), the heterogeneous distribution of Anaeromyxobacter strains at the Oak Ridge IFC site may be a result of several factors. The level of metabolic diversity within the Anaeromyxobacter community is unclear due to a limited number of isolated representatives. Characterization of cluster A Anaeromyxobacter pure cultures indicates that physiological differences exist between strains that share nearly identical 16S rRNA gene sequences (S. H. Thomas, unpublished data). Traits relevant for field-scale processes include metabolic versatility (i.e., metal and nitrate respiration), growth rates, and tolerance to elevated concentrations of electron acceptors. The characterized cluster A representatives share the ability to reduce U(VI), but the rates can be quite variable (unpublished data). Another shared trait among characterized isolates is the ability to reduce nitrate, but the tolerance to nitrate (and nitrite) varies between strains (40). Several Anaeromyxobacter isolates were isolated based on their ability to perform reductive dechlorination, but putative reductive dehalogenase genes are lacking in the strain FW109-5 genome (GenBank accession number CP000769), and in contrast to other strains, this isolate cannot grow aerobically (M. Fields, personal communication). The dominance of cluster C representatives and the apparent absence of cluster A and B representatives following ethanol biostimulation in area 3 suggest that environmental conditions in area 3 may exclude some Anaeromyxobacter strains (51-53). Alternatively, cluster C Anaeromyxobacter strains may be capable of using ethanol directly as an electron donor and thus may have a competitive advantage over strains that utilize only ethanol fermentation products (i.e., hydrogen and acetate) as electron donors (40, 50). Cluster C sequences recovered from area 3 share 99.4 to 100% sequence similarity (7; this study), suggesting that one group of closely related strains dominates the Anaeromyxobacter community in this treatment zone.
Successful dispersal of organisms in the environment requires not only appropriate metabolic adaptations but also transport of organisms from one location to another (37). In spite of cross-plane fracture connectivity between areas 1 and 3, the heterogeneous distribution of Anaeromyxobacter strains may be based on hydrologic isolation between both areas. Groundwater flow through fractures along dipping bedding planes connects the two areas. However, flow in the dip direction is small relative to strike parallel flow through the high-conductivity transition zone between overlying unconsolidated saprolite and underlying, less-weathered bedrock (35). Thus, while areas 1 and 3 may be connected for the purposes of contaminant transport, transport of bacterial cells may be subject to limitations that play a role in the observed heterogeneous distributions (20-22, 35, 47). Tracer tests have been conducted in the area 1 and area 3 treatment zones (19, 27), but no analyses have been conducted to elucidate transport between the two treatment zones. Detailed studies that will explore chemical and bacterial transport between areas 1 and 3 are needed to clarify this issue and to contribute to scaled-up remediation efforts.
While the reasons and mechanisms for the observed heterogeneous distribution of Anaeromyxobacter populations at the Oak Ridge IFC site require further investigation, this study demonstrates that strain-specific monitoring provides valuable information about the microbiology contributing to U(VI) reduction. Apparently, cluster C Anaeromyxobacter populations are involved in U(VI) reduction following ethanol biostimulation in area 3, and future efforts should focus on the isolation and characterization of those strains that are contributors to the process of interest under in situ conditions.
Supplementary Material
Acknowledgments
This research was supported by the Environmental Remediation Science Division (ERSD), Biological and Environmental Research (BER), U.S. Department of Energy, and by NSF IGERT (grant DGE 0114400) and NSF GK-12 (grant 0338261) fellowships to S.H.T. E.P.-C. also acknowledges partial support through an NSF IGERT fellowship and is the recipient of an NSF Graduate Research Fellowship.
We thank Joy Van Nostrand, Jizhong Zhou, Mary Beth Leigh, and Erick Cardenas for DNA samples extracted from the area 3 treatment zone; Joel Kostka and Denise Akob for soil, DNA, and enrichment samples from area 1; Qingzhong Wu for providing isolate FRC-W; and Youlboong Sung for providing DNAs from area 3 site samples and microcosms.
Footnotes
Published ahead of print on 3 April 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.ABI. 2005. Applied Biosystems 7900HT fast real-time PCR system and 7300/7500 real-time PCR systems: chemistry guide 4348358, revision E. ABI, Foster City, CA.
- 2.Afonina, I., M. Zivarts, I. Kutyavin, E. Lukhtanov, H. Gamper, and R. B. Meyer. 1997. Efficient priming of PCR with short oligonucleotides conjugated to a minor groove binder. Nucleic Acids Res. 25:2657-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akob, D. M., H. J. Mills, and J. E. Kostka. 2007. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95-107. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Amos, B. K., Y. Sung, K. E. Fletcher, T. J. Gentry, W. M. Wu, C. S. Criddle, J. Zhou, and F. E. Löffler. 2007. Detection and quantification of Geobacter lovleyi strain SZ: implications for bioremediation at tetrachloroethene- and uranium-impacted sites. Appl. Environ. Microbiol. 73:6898-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, S. C. 2001. Waste characteristics of the former S-3 ponds and outline of uranium chemistry relevant to NABIR Field Research Center studies. U.S. Department of Energy report ORNL/TM-2001/27. U.S. Department of Energy, Oak Ridge, TN.
- 7.Cardenas, E., W. M. Wu, M. B. Leigh, J. Carley, L. S. Carrol, T. Gentry, J. Luo, D. Watson, B. Gu, M. Ginder-Vogel, P. K. Kitanidis, P. M. Jardine, J. Zhou, C. S. Criddle, T. L. Marsh, and J. M. Tiedje. 2008. Microbial communities in contaminated sediments, associated with bioremediation of uranium to submicromolar levels. Appl. Environ. Microbiol. 74:3718-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carney, K. M., P. A. Matson, and B. J. M. Bohannan. 2004. Diversity and composition of tropical soil nitrifiers across a plant diversity gradient and among land-use types. Ecol. Lett. 7:684-694. [Google Scholar]
- 9.Chakravorty, S., D. Helb, M. Burday, N. Connell, and D. Alland. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, J. C., and J. M. Tiedje. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, J. R., A. L. Cascarelli, W. W. Mohn, and J. M. Tiedje. 1994. Isolation and characterization of a novel bacterium growing via reductive dehalogenation of 2-chlorophenol. Appl. Environ. Microbiol. 60:3536-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreier, R. B., D. K. Solomon, and C. M. Beaudoin. 1987. Fracture characterization in the unsaturated zone of a shallow land burial facility, p. 51-59. In D. D. Evans and T. J. Nicholson (ed.), Flow and transport through unsaturated rock, vol. 42. American Geophysical Union, Washington, DC. [Google Scholar]
- 14.Felis, G. E., and F. Dellaglio. 2007. On species descriptions based on a single strain: proposal to introduce the status species proponenda (sp. pr.). Int. J. Syst. Evol. Microbiol. 57:2185-2187. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 16.Green, J., and B. J. M. Bohannan. 2006. Spatial scaling of microbial biodiversity. Trends Ecol. Evol. 21:501-507. [DOI] [PubMed] [Google Scholar]
- 17.He, J. Z., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, Q., and R. A. Sanford. 2003. Characterization of Fe(III) reduction by chlororespiring Anaeromyxobacter dehalogenans. Appl. Environ. Microbiol. 69:2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Istok, J. D., J. M. Senko, L. R. Krumholz, D. Watson, M. A. Bogle, A. Peacock, Y. J. Chang, and D. C. White. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468-475. [DOI] [PubMed] [Google Scholar]
- 20.Jardine, P. M., T. L. Mehlhorn, I. L. Larsen, W. B. Bailey, S. C. Brooks, Y. Roh, and J. P. Gwo. 2002. Influence of hydrological and geochemical processes on the transport of chelated-metals and chromate in fractured shale bedrock. J. Contam. Hydrol. 55:137-159. [DOI] [PubMed] [Google Scholar]
- 21.Jardine, P. M., W. E. Sanford, J. P. Gwo, O. C. Reedy, D. S. Hicks, R. J. Riggs, and W. B. Bailey. 1999. Quantifying diffusive mass transfer in fractured shale bedrock. Water Resour. Res. 35:2015-2030. [Google Scholar]
- 22.Jiang, H. L., A. M. Maszenan, and J. H. Tay. 2007. Bioaugmentation and coexistence of two functionally similar bacterial strains in aerobic granules. Appl. Microbiol. Biotechnol. 75:1191-1200. [DOI] [PubMed] [Google Scholar]
- 23.Konstantinidis, K. T., and J. M. Tiedje. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konstantinidis, K. T., and J. M. Tiedje. 2005. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187:6258-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, D., M. L. Lawrence, F. W. Austin, and A. J. Ainsworth. 2007. A multiplex PCR for species- and virulence-specific determination of Listeria monocytogenes. J. Microbiol. Methods 71:133-140. [DOI] [PubMed] [Google Scholar]
- 26.Löffler, F. E., R. A. Sanford, and J. M. Tiedje. 1996. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl. Environ. Microbiol. 62:3809-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, J., O. A. Cirpka, M. N. Fienen, W. M. Wu, T. L. Mehlhorn, J. Carley, P. M. Jardine, C. S. Criddle, and P. K. Kitanidis. 2006. A parametric transfer function methodology for analyzing reactive transport in nonuniform flow. J. Contam. Hydrol. 83:27-41. [DOI] [PubMed] [Google Scholar]
- 28.Luo, J., F.-A. Weber, O. A. Cirpka, W.-M. Wu, J. L. Nyman, J. Carley, P. M. Jardine, C. S. Criddle, and P. K. Kitanidis. 2007. Modeling in situ uranium(VI) bioreduction by sulfate-reducing bacteria. J. Contam. Hydrol. 92:129-148. [DOI] [PubMed] [Google Scholar]
- 29.Mackay, I. M. 2004. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10:190-212. [DOI] [PubMed] [Google Scholar]
- 30.Marshall, M. J., A. C. Dohnalkova, D. W. Kennedy, A. E. Plymale, S. H. Thomas, F. E. Löffler, R. A. Sanford, J. M. Zachara, J. K. Fredrickson, and A. S. Beliaev. 2008. Electron donor-dependent radionuclide reduction and nanoparticle formation by Anaeromyxobacter dehalogenans strain 2CP-C. Environ. Microbiol. 11:534-543. [DOI] [PubMed] [Google Scholar]
- 31.Martiny, J. B. H., B. J. M. Bohannan, J. H. Brown, R. K. Colwell, J. A. Fuhrman, J. L. Green, M. C. Horner-Devine, M. Kane, J. A. Krumins, C. R. Kuske, P. J. Morin, S. Naeem, L. Ovreas, A.-L. Reysenbach, V. H. Smith, and J. T. Staley. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 32.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrie, L., N. N. North, S. L. Dollhopf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, D. H., D. B. Watson, Y. Roh, T. L. Mehlhorn, J. W. Moon, and P. M. Jardine. 2006. Distribution of uranium contamination in weathered fractured saprolite/shale and ground water. J. Environ. Qual. 35:1715-1730. [DOI] [PubMed] [Google Scholar]
- 36.Ramette, A., J. J. LiPuma, and J. M. Tiedje. 2005. Species abundance and diversity of Burkholderia cepacia complex in the environment. Appl. Environ. Microbiol. 71:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramette, A., and J. M. Tiedje. 2007. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol. 53:197-207. [DOI] [PubMed] [Google Scholar]
- 38.Ritalahti, K. M., B. K. Amos, Y. Sung, Q. Z. Wu, S. S. Koenigsberg, and F. E. Löffler. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 40.Sanford, R. A., J. R. Cole, and J. M. Tiedje. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanford, R. A., Q. Wu, Y. Sung, S. H. Thomas, B. K. Amos, E. K. Prince, and F. E. Löffler. 2007. Hexavalent uranium supports growth of Anaeromyxobacter dehalogenans and Geobacter spp. with lower than predicted biomass yields. Environ. Microbiol. 9:2885-2893. [DOI] [PubMed] [Google Scholar]
- 42.Scott, J. C., M. S. Koylass, M. R. Stubberfield, and A. M. Whatmore. 2007. Multiplex assay based on single-nucleotide polymorphisms for rapid identification of Brucella isolates at the species level. Appl. Environ. Microbiol. 73:7331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 44.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, S. H., R. D. Wagner, A. K. Arakaki, J. Skolnick, J. R. Kirby, L. J. Shimkets, R. A. Sanford, and F. E. Löffler. 2008. The mosaic genome of Anaeromyxobacter dehalogenans strain 2CP-C suggests an aerobic common ancestor to the delta-proteobacteria. PLoS ONE 3:e2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Treude, N., D. Rosencrantz, W. Liesack, and S. Schnell. 2003. Strain FAc12, a dissimilatory iron-reducing member of the Anaeromyxobacter subgroup of Myxococcales. FEMS Microbiol. Ecol. 44:261-269. [DOI] [PubMed] [Google Scholar]
- 47.Treves, D. S., B. Xia, J. Zhou, and J. M. Tiedje. 2003. A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microb. Ecol. 45:20-28. [DOI] [PubMed] [Google Scholar]
- 48.Varma-Basil, M., H. El-Hajj, S. A. E. Marras, M. H. Hazbon, J. M. Mann, N. D. Connell, F. R. Kramer, and D. Alland. 2004. Molecular beacons for multiplex detection of four bacterial bioterrorism agents. Clin. Chem. 50:1060-1063. [DOI] [PubMed] [Google Scholar]
- 49.Watson, D. B., J. E. Kostka, M. W. Fields, and P. M. Jardine. 2004. The Oak Ridge Field Research Center conceptual model. U.S. Department of Energy, Oak Ridge, TN. http://www.esd.ornl.gov/orifrc/.
- 50.Wu, Q., R. A. Sanford, and F. E. Löffler. 2006. Uranium(VI) reduction by Anaeromyxobacter dehalogenans strain 2CP-C. Appl. Environ. Microbiol. 72:3608-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, W.-M., J. Carley, M. Fienen, T. Mehlhorn, K. Lowe, J. Nyman, J. Luo, M. E. Gentile, R. Rajan, D. Wagner, R. F. Hickey, B. H. Gu, D. Watson, O. A. Cirpka, P. K. Kitanidis, P. M. Jardine, and C. S. Criddle. 2006. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 1. Conditioning of a treatment zone. Environ. Sci. Technol. 40:3978-3985. [DOI] [PubMed] [Google Scholar]
- 52.Wu, W.-M., J. Carley, J. Luo, M. A. Ginder-Vogel, E. Cardenas, M. B. Leigh, C. A. C. Hwang, S. D. Kelly, C. M. Ruan, L. Wu, J. V. Nostrand, T. Gentry, K. Lowe, T. Mehlhorn, S. Carroll, W. Luo, M. W. Fields, B. H. Gu, D. Watson, K. M. Kemner, T. Marsh, J. Tiedje, J. Zhou, S. Fendorf, P. K. Kitanidis, P. M. Jardine, and C. S. Criddle. 2007. In situ bioreduction of uranium (VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ. Sci. Technol. 41:5716-5723. [DOI] [PubMed] [Google Scholar]
- 53.Wu, W.-M., J. Carley, T. Gentry, M. A. Ginder-Vogel, M. Fienen, T. Mehlhorn, H. Yan, S. Caroll, M. N. Pace, J. Nyman, J. Luo, M. E. Gentile, M. W. Fields, R. F. Hickey, B. H. Gu, D. Watson, O. A. Cirpka, J. Z. Zhou, S. Fendorf, P. K. Kitanidis, P. M. Jardine, and C. S. Criddle. 2006. Pilot-scale in situ bioremedation of uranium in a highly contaminated aquifer. 2. Reduction of U(VI) and geochemical control of U(VI) bioavailability. Environ. Sci. Technol. 40:3986-3995. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, J., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil. Microbiology (United Kingdom) 143:3913-3919. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, J. Z., B. C. Xia, H. Huang, A. V. Palumbo, and J. M. Tiedje. 2004. Microbial diversity and heterogeneity in sandy subsurface soils. Appl. Environ. Microbiol. 70:1723-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.